Can Phytoremediation-Induced Changes in the Microbiome Improve Saline/Sodic Soil and Plant Health?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Soil Samples for Microbiome Analysis
2.3. Root Sample Preparation for Microbiome Analysis
2.4. DNA Extraction for Microbiome Analysis
2.5. Microbiome Data Analysis
2.6. Plant Materials and Preparation for Metabolite Analysis
2.7. Sample Culturing and Isolation for Root-Associated Microbial Inoculation of Seeds
2.8. Seed Inoculation and NaCl Challenge and Data Analysis
3. Results
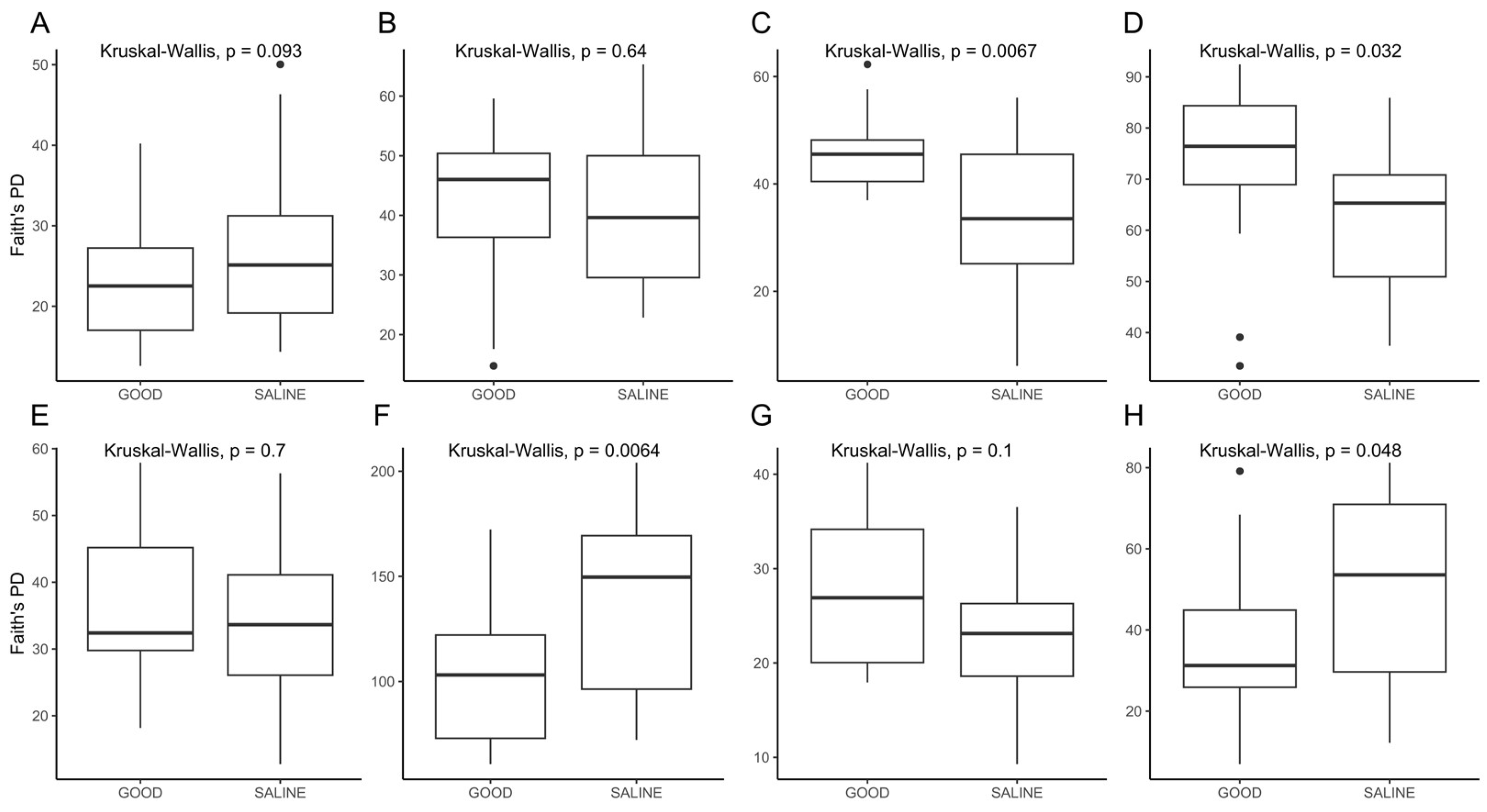
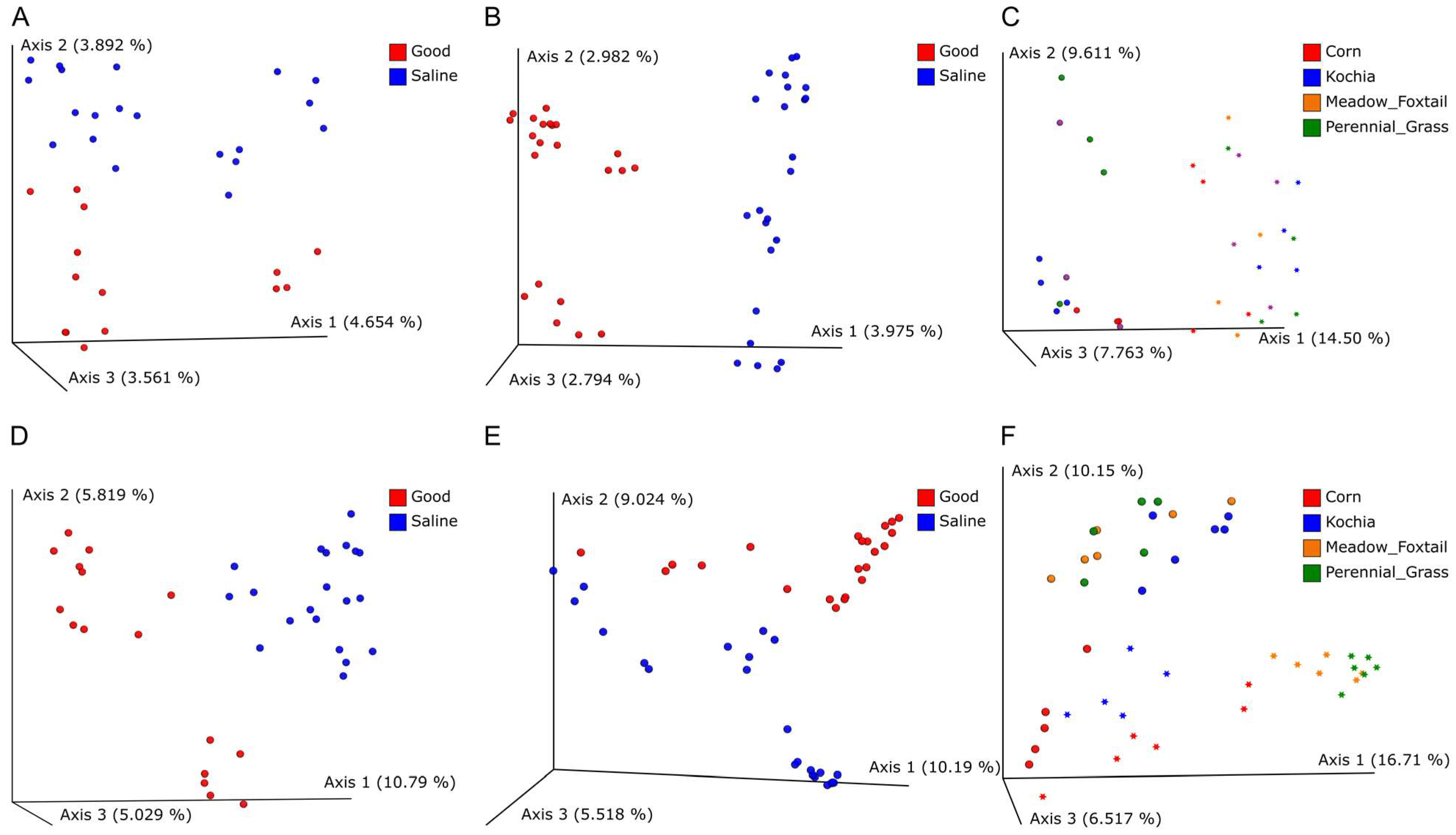
3.1. Microbiome Analysis of Bulk Soil and Root Samples
3.2. Root Metabolite Analysis
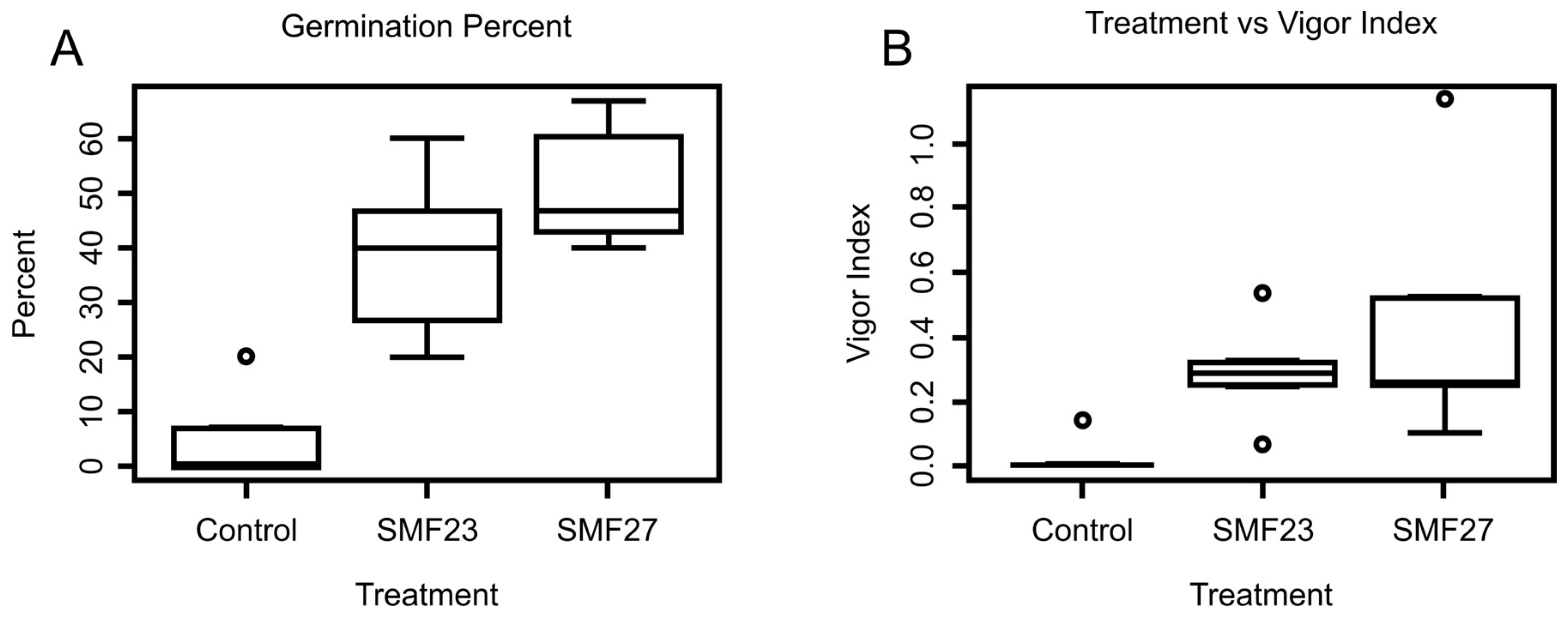
3.3. Endophyte Inoculation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fiedler, D.J.; Clay, D.E.; Joshi, D.R.; Engel, A.; Marzano, S.Y.; Jakubowski, D.; Bhattarai, D.; Reese, C.L.; Bruggeman, S.A.; Clay, S.A. CO2 and N2O emissions and microbial community structure from fields that include salt-affected soils. J. Environ. Qual. 2021, 50, 567–579. [Google Scholar] [CrossRef]
- Fiedler, D.; Clay, S.; Westhoff, S.; Reese, C.; Bruggeman, S.; Moriles-Miller, J.; Perkins, L.; Joshi, D.; Marzano, S.-Y.; Clay, D. Phytoremediation and high rainfall combine to improve soil and plant health in a North America Northern Great Plains saline sodic soil. J. Soil Water Conserv. 2022, 77, 381–388. [Google Scholar] [CrossRef]
- Clay, S.A.; Fiedler, D.; Reese, C.; Clay, D.E. Restoring ecological function to saline–sodic soils in South Dakota with perennial grass mixtures. Agron. J. 2023, 115, 135–146. [Google Scholar] [CrossRef]
- Blanchard, A.P. Revegetating Salt-Impacted Soils in the Northern Great Plains; South Dakota State University: Brookings, SD, USA, 2021; Available online: https://openprairie.sdstate.edu/etd/5705 (accessed on 18 December 2023).
- Budak, M.; Clay, D.; Clay, S.; Reese, C.; Westhoff, S.; Howe, L.; Owen, R.; Birru, G.; He, Y.; Wang, Z. Increased rainfall may place saline/sodic soils on the tipping point of sustainability. J. Soil Water Conserv. 2022, 77, 418–425. [Google Scholar] [CrossRef]
- Wang, W.X.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Maggio, A.; De Pascale, S.; Angelino, G.; Ruggiero, C.; Barbieri, G. Physiological response of tomato to saline irrigation in long-term salinized soils. Eur. J. Agron. 2004, 21, 149–159. [Google Scholar] [CrossRef]
- Birru, G.A.; Clay, D.E.; DeSutter, T.M.; Reese, C.L.; Kennedy, A.C.; Clay, S.A.; Bruggeman, S.A.; Owen, R.K.; Malo, D.D. Chemical Amendments of Dryland Saline–Sodic Soils Did Not Enhance Productivity and Soil Health in Fields without Effective Drainage. Agron. J. 2019, 111, 496–508. [Google Scholar] [CrossRef]
- Blanchard, A.; Clay, S.; Perkins, L. The good, the bad, the salty: Investigation of native plants for revegetation of salt-impacted soil in the northern Great Plains, United States. J. Soil Water Conserv. 2023, 78, 479–485. [Google Scholar] [CrossRef]
- Rosenblueth, M.; Martinez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant-Microbe Interact. 2006, 19, 827–837. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef]
- Sgroy, V.; Cassan, F.; Masciarelli, O.; Del Papa, M.F.; Lagares, A.; Luna, V. Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Appl. Microbiol. Biotechnol. 2009, 85, 371–381. [Google Scholar] [CrossRef]
- Rashid, S.; Charles, T.C.; Glick, B.R. Isolation and characterization of new plant growth-promoting bacterial endophytes. Appl. Soil Ecol. 2012, 61, 217–224. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Gans, J.; Wolinsky, M.; Dunbar, J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 2005, 309, 1387–1390. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. Fems Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef]
- Gonzalez-Garcia, S.; Alvarez-Perez, J.M.; de Miera LE, S.; Cobos, R.; Ibanez, A.; Diez-Galan, A.; Garzon-Jimeno, E.; Coque, J.J.R. Developing tools for evaluating inoculation methods of biocontrol Streptomyces sp. strains into grapevine plants. PLoS ONE 2019, 14, e0211225. [Google Scholar] [CrossRef]
- Garbaye, J. Helper Bacteria—A New Dimension to the Mycorrhizal Symbiosis. New Phytol. 1994, 128, 197–210. [Google Scholar] [CrossRef]
- Ashraf, M.; Hasnain, S.; Berge, O.; Mahmood, T. Inoculating wheat seedlings with exopolysaccharide-producing bacteria restricts sodium uptake and stimulates plant growth under salt stress. Biol. Fertil. Soils 2004, 40, 157–162. [Google Scholar] [CrossRef]
- Wang, C.H.; Wu, L.; Wang, Z.; Alabady, M.S.; Parson, D.; Molumo, Z.; Fankhauser, S.C. Characterizing changes in soil microbiome abundance and diversity due to different cover crop techniques. PLoS ONE 2020, 15, e0232453. [Google Scholar] [CrossRef]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 2015, 10, 1457. [Google Scholar] [CrossRef]
- Riesenfeld, C.S.; Goodman, R.M.; Handelsman, J. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ. Microbiol. 2004, 6, 981–989. [Google Scholar] [CrossRef]
- Estaki, M.; Jiang, L.; Bokulich, N.A.; McDonald, D.; Gonzalez, A.; Kosciolek, T.; Martino, C.; Zhu, Q.; Birmingham, A.; Vazquez-Baeza, Y.; et al. QIIME 2 Enables Comprehensive End-to-End Analysis of Diverse Microbiome Data and Comparative Studies with Publicly Available Data. Curr. Protoc. Bioinform. 2020, 70, e100. [Google Scholar] [CrossRef]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef]
- Tan, Y.; Li, K.; Hu, L.; Chen, S.; Gai, Y.; Jiang, X. Fast and simple droplet sampling of sap from plant tissues and capillary microextraction of soluble saccharides for picogram-scale quantitative determination with GC-MS. J. Agric. Food Chem. 2010, 58, 9931–9935. [Google Scholar] [CrossRef]
- He, F. E. coli Genomic DNA Extraction. Bio-101 2011, 1, e97. [Google Scholar] [CrossRef]
- Ab Rahman, S.F.S.; Singh, E.; Pieterse, C.M.; Schenk, P.M. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018, 267, 102–111. [Google Scholar] [CrossRef]
- Jakubowski, D. The Effects of Saline Soil on Microbiome and the Isolation of Root-Associated Microbes to Relieve Salinity Stress. Master’s Thesis, South Dakota State University, Brookings, SD, USA, 2021. [Google Scholar]
- Gamalero, E.; Berta, G.; Massa, N.; Glick, B.R.; Lingua, G. Synergistic interactions between the ACC deaminase-producing bacterium Pseudomonas putida UW4 and the AM fungus Gigaspora rosea positively affect cucumber plant growth. Fems Microbiol. Ecol. 2008, 64, 459–467. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Hsieh, T.C.; Chao, A. Rarefaction and Extrapolation: Making Fair Comparison of Abundance-Sensitive Phylogenetic Diversity among Multiple Assemblages. Syst. Biol. 2017, 66, 100–111. [Google Scholar] [CrossRef]
- Bouchard, M.; Jousselme, A.L.; Dore, P.E. A proof for the positive definiteness of the Jaccard index matrix. Int. J. Approx. Reason. 2013, 54, 615–626. [Google Scholar] [CrossRef]
- Chen, J.; Bittinger, K.; Charlson, E.S.; Hoffmann, C.; Lewis, J.; Wu, G.D.; Collman, R.G.; Bushman, F.D.; Li, H.Z. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 2012, 28, 2106–2113. [Google Scholar] [CrossRef]
- Raman, S.B.; Rathinasabapathi, B. Pantothenate synthesis in plants. Plant Sci. 2004, 167, 961–968. [Google Scholar] [CrossRef]
- Hanson, A.D.; Beaudoin, G.A.; McCarty, D.R.; Gregory, J.F. Does Abiotic Stress Cause Functional B Vitamin Deficiency in Plants? Plant Physiol. 2016, 172, 2082–2097. [Google Scholar] [CrossRef]
- Saleem, M.; Hu, J.; Jousset, A. More Than the Sum of Its Parts: Microbiome Biodiversity as a Driver of Plant Growth and Soil Health. Annu. Rev. Ecol. Evol. Syst. 2019, 50, 145. [Google Scholar] [CrossRef]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion. Front. Plant Sci. 2020, 10. [Google Scholar] [CrossRef]
- Hartmann, M.; Frey, B.; Mayer, J.; Mader, P.; Widmer, F. Distinct soil microbial diversity under long-term organic and conventional farming. Isme J. 2015, 9, 1177–1194. [Google Scholar] [CrossRef]
- Zhang, H.; Kim, M.S.; Sun, Y.; Dowd, S.E.; Shi, H.Z.; Pare, P.W. Soil bacteria confer plant salt tolerance by tissue-specific regulation of the sodium transporter HKT1. Mol. Plant-Microbe Interact. 2008, 21, 737–744. [Google Scholar] [CrossRef]
- Gururani, M.A.; Upadhyaya, C.P.; Baskar, V.; Venkatesh, J.; Nookaraju, A.; Park, S.W. Plant Growth-Promoting Rhizobacteria Enhance Abiotic Stress Tolerance in Solanum tuberosum Through Inducing Changes in the Expression of ROS-Scavenging Enzymes and Improved Photosynthetic Performance. J. Plant Growth Regul. 2013, 32, 245–258. [Google Scholar] [CrossRef]



| Sample Name | Suggested Species |
|---|---|
| SMF22 |
|
| SMF23 |
|
| SMF24 |
|
| SMF25 |
|
| SMF26 |
|
| SMF27 |
|
| SMF28 |
|
| SMF29 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neupane, A.; Jakubowski, D.; Fiedler, D.; Gu, L.; Clay, S.A.; Clay, D.E.; Lee Marzano, S.-Y. Can Phytoremediation-Induced Changes in the Microbiome Improve Saline/Sodic Soil and Plant Health? Agronomy 2024, 14, 29. https://doi.org/10.3390/agronomy14010029
Neupane A, Jakubowski D, Fiedler D, Gu L, Clay SA, Clay DE, Lee Marzano S-Y. Can Phytoremediation-Induced Changes in the Microbiome Improve Saline/Sodic Soil and Plant Health? Agronomy. 2024; 14(1):29. https://doi.org/10.3390/agronomy14010029
Chicago/Turabian StyleNeupane, Achal, Duncan Jakubowski, Douglas Fiedler, Liping Gu, Sharon A. Clay, David E. Clay, and Shin-Yi Lee Marzano. 2024. "Can Phytoremediation-Induced Changes in the Microbiome Improve Saline/Sodic Soil and Plant Health?" Agronomy 14, no. 1: 29. https://doi.org/10.3390/agronomy14010029
APA StyleNeupane, A., Jakubowski, D., Fiedler, D., Gu, L., Clay, S. A., Clay, D. E., & Lee Marzano, S.-Y. (2024). Can Phytoremediation-Induced Changes in the Microbiome Improve Saline/Sodic Soil and Plant Health? Agronomy, 14(1), 29. https://doi.org/10.3390/agronomy14010029





