A Hierarchical Feature-Aware Model for Accurate Tomato Blight Disease Spot Detection: Unet with Vision Mamba and ConvNeXt Perspective
Abstract
1. Introduction
2. Materials and Methods
2.1. Image Acquisition and Annotation Preparation
2.2. Data Augmentation
2.3. Unet
2.4. Proposed Model
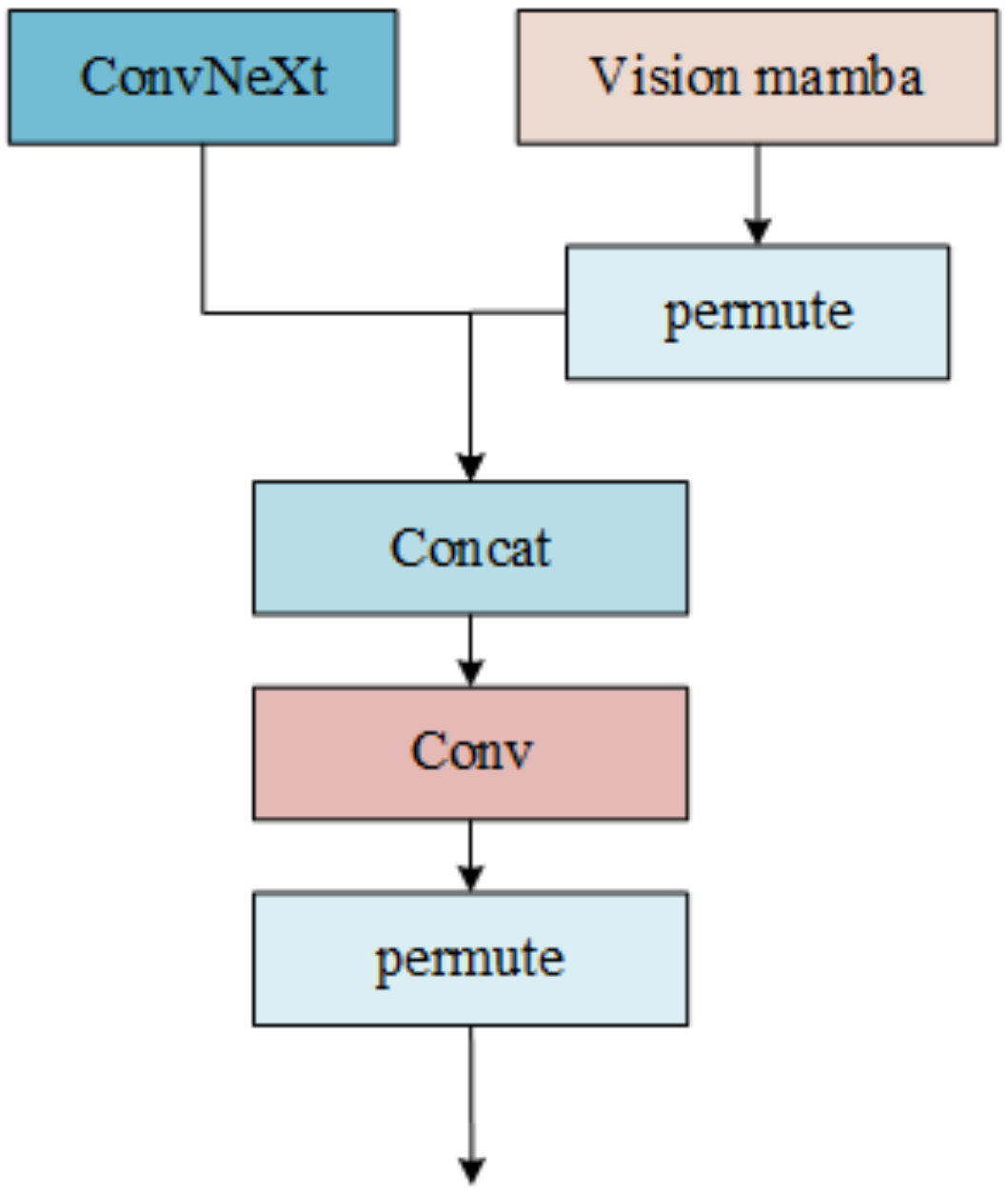
2.4.1. Parallel Feature-Aware Backbone
2.4.2. Mamba Decoder Module
2.4.3. Feature Fusion Unit
2.5. Loss Function
2.6. Experimental Environment and Evaluation Metrics
3. Results
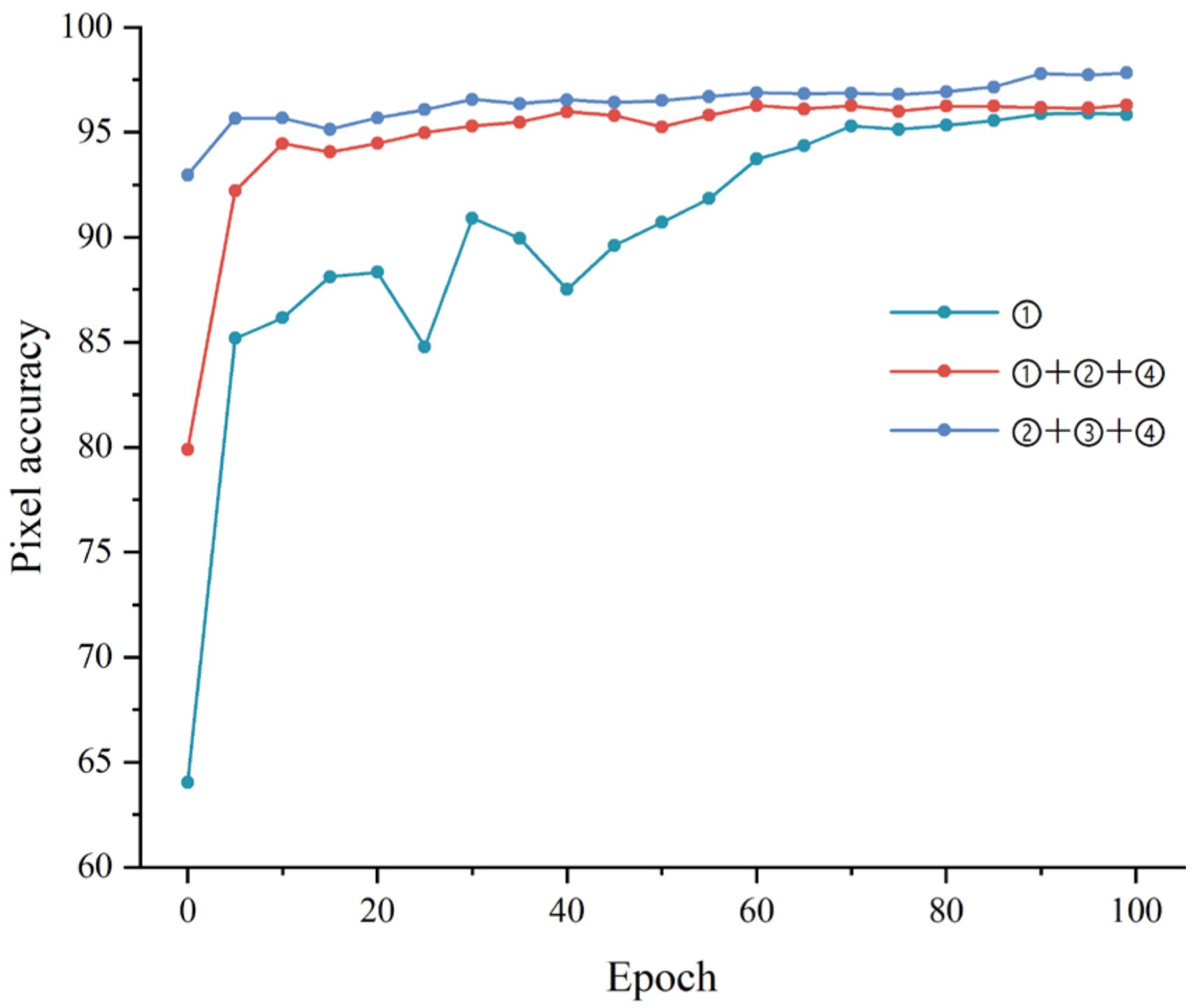
3.1. Impact of Loss Functions
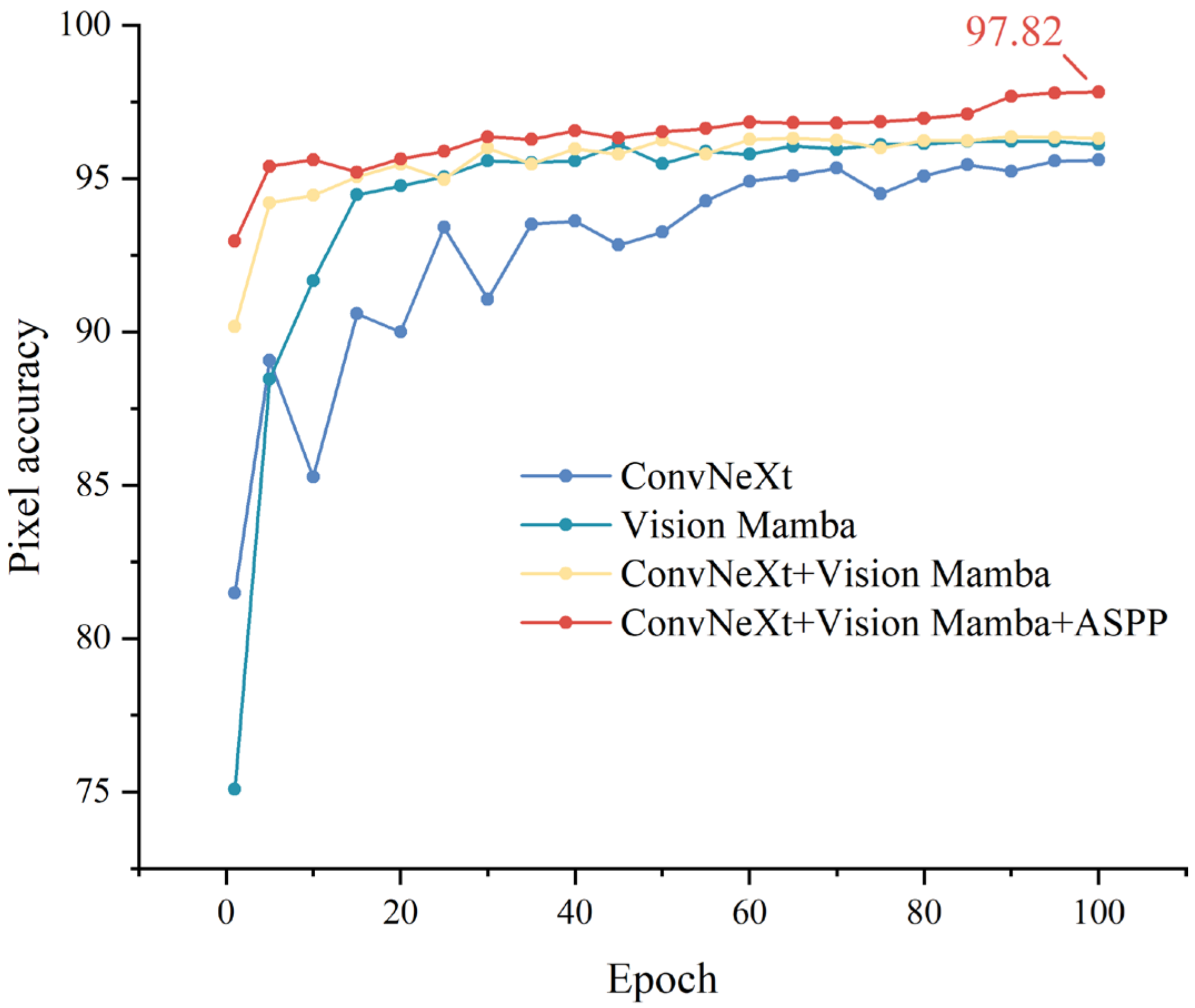
3.2. Ablation Study
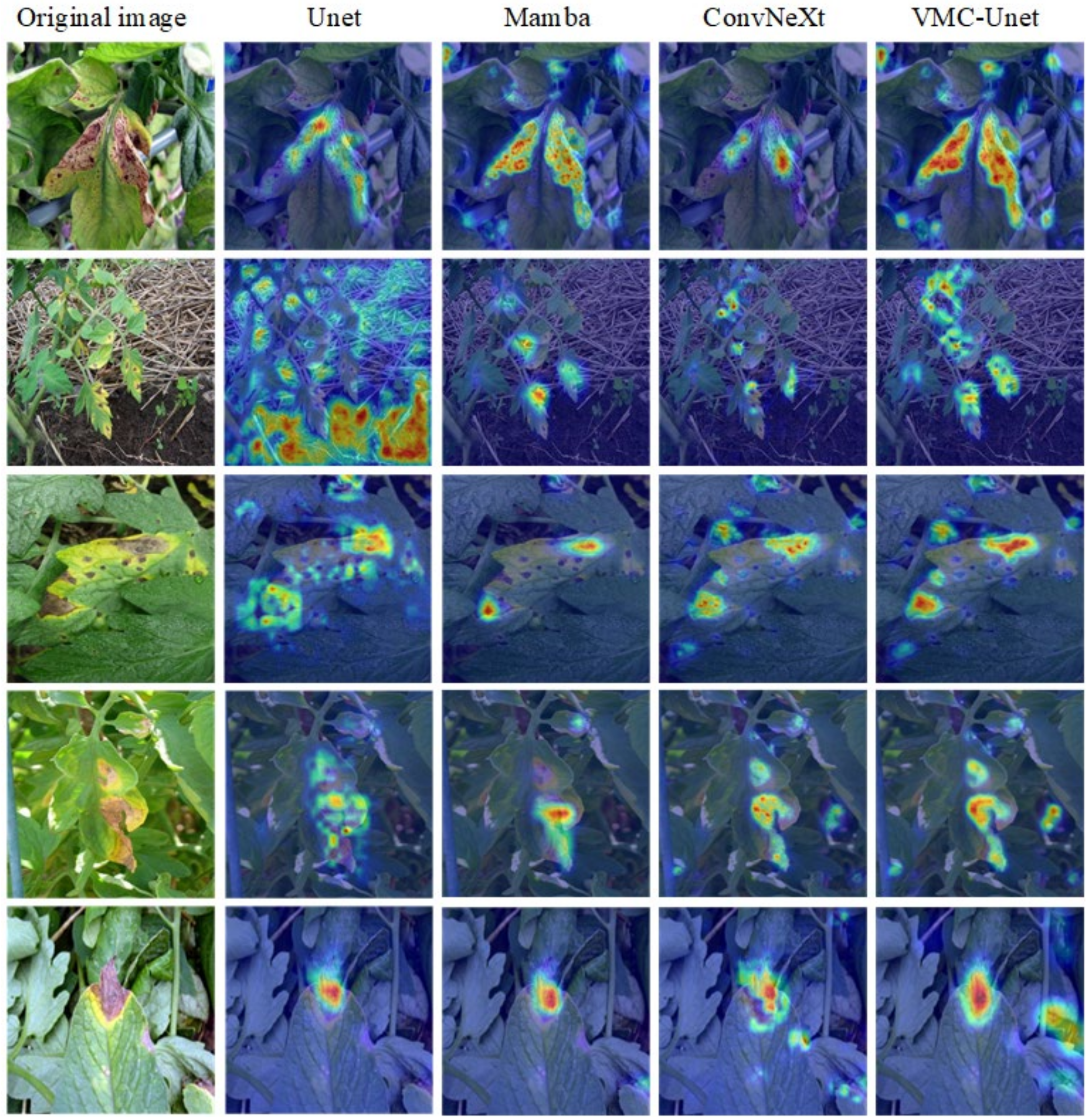
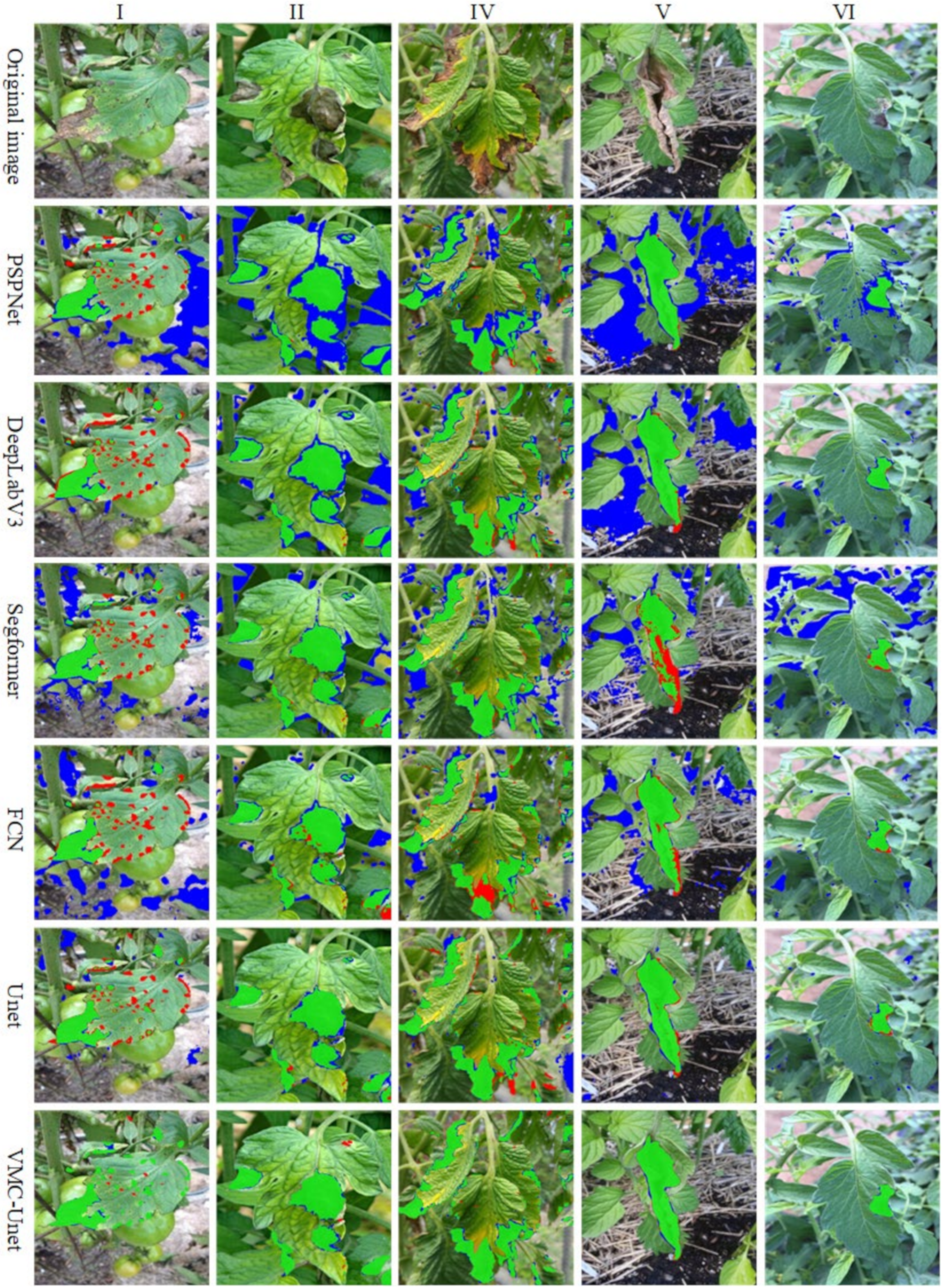
3.3. Model Interpretability Analysis
3.4. Comprehensive Model Performance Evaluation
4. Discussion
5. Conclusions
- This study designed the VMC-Unet model, an advanced asymmetric lesion segmentation model tailored for the complex task of tomato blight detection. By integrating ConvNeXt and Vision Mamba as dual backbone networks and employing the ASPP module, the model significantly improved segmentation accuracy in challenging agricultural environments.
- Extensive experiments demonstrated that VMC-Unet outperforms traditional segmentation models across multiple key metrics. Specifically, the model achieved a pixel accuracy of 97.82%, an F1 score of 87.94%, and an mIoU of 86.75% on the tomato blight dataset.
- We designed the joint loss function for training the lesion segmentation model and achieved satisfactory results.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fatima, A.; Ali, S.; Azmat, S.; Amrao, L.; Ghani, M.U.; Iftikhar, Y.; Zeshan, M.A.; Ahmad, A.; Kalsoom, H. Morphological assessment of resistance potential in tomato against early blight disease and its management. Pak. J. Agric. Res. 2024, 37, 88–189. [Google Scholar] [CrossRef]
- Sundararaman, B.; Jagdev, S.; Khatri, N. Transformative role of artificial intelligence in advancing sustainable tomato (Solanum lycopersicum) disease management for global food security: A comprehensive review. Sustainability 2023, 15, 11681. [Google Scholar] [CrossRef]
- Thangaraj, R.; Anandamurugan, S.; Pandiyan, P.; Kaliappan, V.K. Artificial intelligence in tomato leaf disease detection: A comprehensive review and discussion. J. Plant Dis. Prot. 2022, 129, 469–488. [Google Scholar] [CrossRef]
- Al Hiary, H.; Ahmad, S.B.; Reyalat, M.; Braik, M.; Alrahamneh, Z. Fast and accurate detection and classification of plant diseases. Int. J. Comput. Appl. 2011, 17, 31–38. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Zhang, Y.; Wa, S.; Liu, Y.; Zhou, X.; Sun, P.; Ma, Q. High-accuracy detection of maize leaf diseases CNN based on multi-pathway activation function module. Remote Sens. 2021, 13, 4218. [Google Scholar] [CrossRef]
- Alzahrani, M.S.; Alsaade, F.W. Transform and deep learning algorithms for the early detection and recognition of tomato leaf disease. Agronomy 2023, 13, 1184. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef]
- Li, L.; Zhang, S.; Wang, B. Plant disease detection and classification by deep learning—A review. IEEE Access 2021, 9, 56683–56698. [Google Scholar] [CrossRef]
- Hu, X.; Wang, R.; Du, J.; Hu, Y.; Jiao, L.; Xu, T. Class-attention-based lesion proposal convolutional neural network for strawberry diseases identification. Front. Plant Sci. 2023, 14, 1091600. [Google Scholar] [CrossRef]
- Bhujel, A.; Kim, N.-E.; Arulmozhi, E.; Basak, J.K.; Kim, H.-T. A lightweight attention-based convolutional neural networks for tomato leaf disease classification. Agriculture 2022, 12, 228. [Google Scholar] [CrossRef]
- Liu, Y.; Song, Y.; Ye, R.; Zhu, S.; Huang, Y.; Chen, T.; Zhou, J.; Li, J.; Li, M.; Lv, C. High-Precision Tomato Disease Detection Using NanoSegmenter Based on Transformer and Lightweighting. Plants 2023, 12, 2559. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Xi, H.; Zhou, G.; Chen, A.; Wang, Y.; Li, L.; Hu, Y. An effective image-based tomato leaf disease segmentation method using MC-UNet. Plant Phenomics 2023, 5, 0049. [Google Scholar] [CrossRef]
- Perveen, K.; Debnath, S.; Pandey, B.; Chand, S.P.; Bukhari, N.A.; Bhowmick, P.; Alshaikh, N.A.; Arzoo, S.; Batool, S. Deep learning-based multiscale CNN-based U network model for leaf disease diagnosis and segmentation of lesions in tomato. Physiol. Mol. Plant Pathol. 2023, 128, 102148. [Google Scholar] [CrossRef]
- Kaur, P.; Harnal, S.; Gautam, V.; Singh, M.P.; Singh, S.P. Performance analysis of segmentation models to detect leaf diseases in tomato plant. Multimed. Tools Appl. 2024, 83, 16019–16043. [Google Scholar] [CrossRef]
- Zhao, X.; Li, X.; Ye, S.; Feng, W.; You, X. Multi-Scale Tomato Disease Segmentation Algorithm Based on Improved U-Net Network. J. Comput. Eng. Appl. 2022, 58, 216. [Google Scholar]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef]
- Lu, Y.; Yi, S.; Zeng, N.; Liu, Y.; Zhang, Y. Identification of rice diseases using deep convolutional neural networks. Neurocomputing 2017, 267, 378–384. [Google Scholar] [CrossRef]
- Yadav, S.; Shukla, S. Analysis of k-fold cross-validation over hold-out validation on colossal datasets for quality classification. In Proceedings of the 2016 IEEE 6th International Conference on Advanced Computing (IACC), Bhimavaram, India, 27–28 February 2016; pp. 78–83. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 11976–11986. [Google Scholar]
- Zhu, L.; Liao, B.; Zhang, Q.; Wang, X.; Liu, W.; Wang, X. Vision mamba: Efficient visual representation learning with bidirectional state space model. arXiv 2024, arXiv:2401.09417. [Google Scholar]
- Wang, Y.; Liang, B.; Ding, M.; Li, J. Dense semantic labeling with atrous spatial pyramid pooling and decoder for high-resolution remote sensing imagery. Remote Sens. 2018, 11, 20. [Google Scholar] [CrossRef]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Jorge Cardoso, M. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: Third International Workshop, DLMIA 2017, Proceedings of the 7th International Workshop, ML-CDS 2017, Held in Conjunction with MICCAI 2017, Québec City, QC, Canada, 14 September 2017; Proceedings 3; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 240–248. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollar, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Müller, R.; Kornblith, S.; Hinton, G.E. When does label smoothing help? In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; p. 32. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and efficient design for semantic segmentation with transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 12077–12090. [Google Scholar]
- Chen, L.C. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Sun, L.; He, J.; Zhang, L. CASF-MNet: Multi-scale network with cross attention mechanism and spatial dimension feature fusion for maize leaf disease detection. Crop. Prot. 2024, 180, 106667. [Google Scholar] [CrossRef]
- Fu, L.; Li, S.; Sun, Y.; Mu, Y.; Hu, T.; Gong, H. Lightweight-convolutional neural network for apple leaf disease identification. Front. Plant Sci. 2022, 13, 831219. [Google Scholar] [CrossRef]
- Paul, S.G.; Biswas, A.A.; Saha, A.; Zulfiker, S.; Ritu, N.A.; Zahan, I.; Rahman, M.; Islam, M.A. A real-time application-based convolutional neural network approach for tomato leaf disease classification. Array 2023, 19, 100313. [Google Scholar] [CrossRef]
- Momeny, M.; Jahanbakhshi, A.; Neshat, A.A.; Hadipour-Rokni, R.; Zhang, Y.-D.; Ampatzidis, Y. Detection of citrus black spot disease and ripeness level in orange fruit using learning-to-augment incorporated deep networks. Ecol. Inform. 2022, 71, 101829. [Google Scholar] [CrossRef]






| Dataset Category | Original Image | Data Augmentation | Dataset Division | ||
|---|---|---|---|---|---|
| Training Set | Training Set | Training Set | |||
| Tomato blight dataset | 630 | 1328 | 930 | 930 | 930 |
| Kaggle dataset | 588 | 2940 | 2058 | 2058 | 2058 |
| Loss Function | Precision | Recall | F1 | mIoU | Accuracy |
|---|---|---|---|---|---|
| ➀ | 77.27 | 50.54 | 61.11 | 65.28 | 87.84 |
| ➀ + ➁ + ➃ | 85.39 | 81.77 | 83.51 | 83.84 | 96.33 |
| ➁ + ➂ + ➃ | 90.03 | 85.94 | 87.94 | 86.75 | 97.82 |
| Module | Precision | Recall | F1 | mIoU | PA | ||
|---|---|---|---|---|---|---|---|
| ASPP | ConvNeXt | Vision Mamba | |||||
| × | × | × | 74.44 | 63.30 | 68.42 | 71.93 | 92.52 |
| × | √ | × | 85.90 | 80.11 | 82.91 | 82.94 | 95.6 |
| × | × | √ | 85.31 | 79.29 | 82.23 | 82.72 | 96.23 |
| × | √ | √ | 85.37 | 81.77 | 83.53 | 83.94 | 96.35 |
| √ | √ | √ | 90.03 | 85.94 | 87.94 | 86.75 | 97.82 |
| × | × | × | 74.44 | 63.30 | 68.42 | 71.93 | 92.52 |
| Model | Precision | Recall | F1 | mIoU | PA |
|---|---|---|---|---|---|
| FCN | 76.93 | 66.66 | 71.43 | 73.55 | 92.35 |
| PSPNet | 75.87 | 62.99 | 68.83 | 71.52 | 91.46 |
| SegFormer | 75.14 | 54.60 | 63.24 | 67.90 | 90.42 |
| DeepLabV3 | 85.53 | 56.30 | 67.90 | 70.81 | 91.13 |
| Unet | 74.44 | 63.30 | 68.42 | 71.93 | 92.52 |
| VMC-Unet | 90.03 | 85.94 | 87.94 | 86.75 | 97.82 |
| Model | Presion | Recall | F1 | mIoU | PA |
|---|---|---|---|---|---|
| FCN | 87.32 | 71.51 | 78.63 | 77.62 | 91.88 |
| PSPNet | 88.32 | 65.53 | 75.24 | 74.30 | 90.06 |
| SegFormer | 72.88 | 60.24 | 65.96 | 67.26 | 87.14 |
| DeepLabV3 | 93.42 | 67.92 | 78.66 | 77.25 | 91.33 |
| Unet | 91.40 | 82.12 | 86.57 | 85.28 | 95.15 |
| VMC-Unet | 94.15 | 90.18 | 92.12 | 91.90 | 97.33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, D.; Li, C.; Shi, H.; Liang, L.; Liu, H.; Diao, M. A Hierarchical Feature-Aware Model for Accurate Tomato Blight Disease Spot Detection: Unet with Vision Mamba and ConvNeXt Perspective. Agronomy 2024, 14, 2227. https://doi.org/10.3390/agronomy14102227
Shi D, Li C, Shi H, Liang L, Liu H, Diao M. A Hierarchical Feature-Aware Model for Accurate Tomato Blight Disease Spot Detection: Unet with Vision Mamba and ConvNeXt Perspective. Agronomy. 2024; 14(10):2227. https://doi.org/10.3390/agronomy14102227
Chicago/Turabian StyleShi, Dongyuan, Changhong Li, Hui Shi, Longwei Liang, Huiying Liu, and Ming Diao. 2024. "A Hierarchical Feature-Aware Model for Accurate Tomato Blight Disease Spot Detection: Unet with Vision Mamba and ConvNeXt Perspective" Agronomy 14, no. 10: 2227. https://doi.org/10.3390/agronomy14102227
APA StyleShi, D., Li, C., Shi, H., Liang, L., Liu, H., & Diao, M. (2024). A Hierarchical Feature-Aware Model for Accurate Tomato Blight Disease Spot Detection: Unet with Vision Mamba and ConvNeXt Perspective. Agronomy, 14(10), 2227. https://doi.org/10.3390/agronomy14102227







