A Soybean Pyrroline-5-Carboxylate Dehydrogenase GmP5CDH1 Modulates Plant Growth and Proline Sensitivity
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Extraction and Reverse Transcription of Total RNA from Soybean
2.3. Cloning and Bioinformatics Analysis of GmP5CDH1
2.4. Gene Expression Analysis
2.5. Subcellular Localization of GmP5CDH1
2.6. Arabidopsis Transformation
2.7. Phenotypic Analysis of Transgenic Arabidopsis
- (1)
- Germination rate statistics
- (2)
- Determination of root length and fresh weight of Arabidopsis thaliana
- (3)
- Determination of amino acid content in Arabidopsis thaliana
- (4)
- Detection of Pro content in Arabidopsis thaliana
- where C = proline content obtained from the standard curve (ug);
- VT = total volume of proline extract (ml);
- W = fresh weight of sample (g);
- VS = volume of extraction solution added during measurement (ml).
- (5)
- Analysis of GmP5CDH1 expression under abiotic stress
2.8. Yeast Two-Hybrid Assay
2.9. Luciferase (LUC) Fluorescence Complementary Assay
2.10. Bimolecular Fluorescence Complementation (BiFC) Assay
3. Results
3.1. Phylogenetic Analysis of the Pyrroline-5-Carboxylate Dehydrogenase Gene Families
3.2. Gene Structure and Conserved Motif Analysis of GmP5CDH1
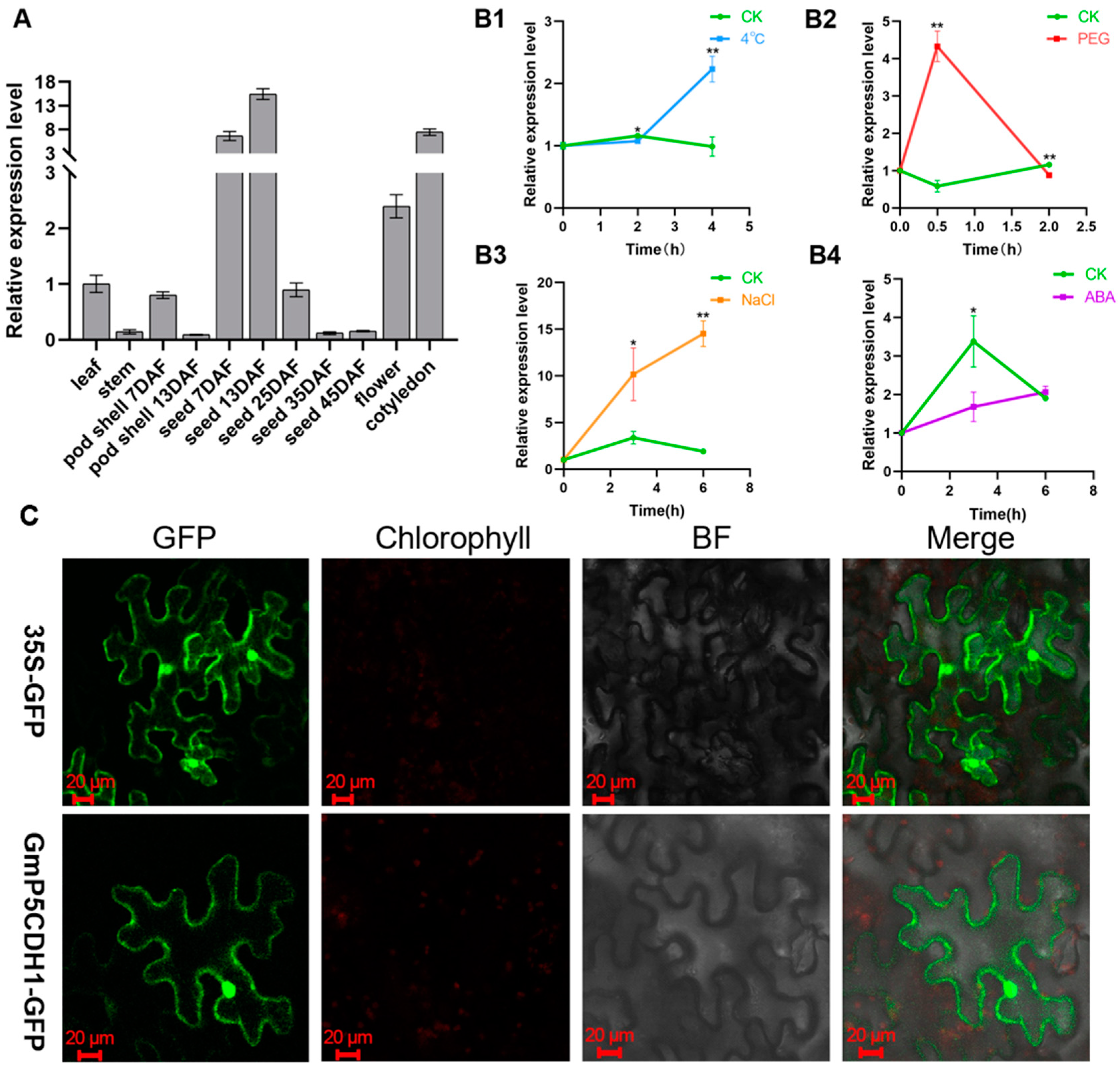
3.3. Expression Pattern Analysis of GmP5CDH1 in Soybean
3.4. GmP5CDH1 Is a Co-Localization Protein of the Nucleus and Cell Membrane
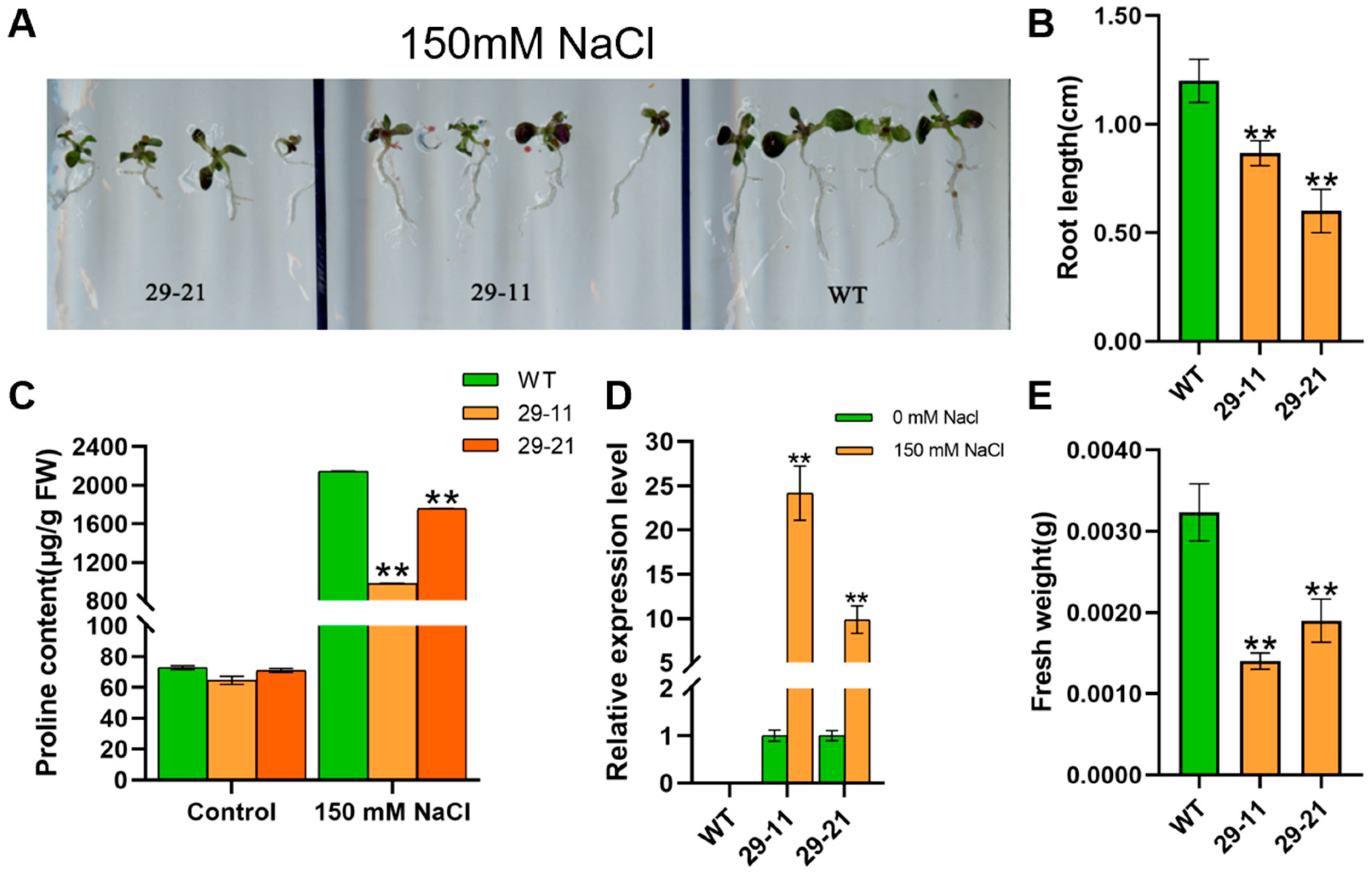
3.5. Heterologous Overexpression GmP5CDH1 in Arabidopsis Has a Pleiotropic Effect on Seed Germination Rate and Amino Acid Content
3.6. Induction of GmP5CDH1 Expression in Response to Salt Stress in Arabidopsis
3.7. GmP5CDH1 Responded to Exogenous Proline Induction and Participated in Proline Synthesis
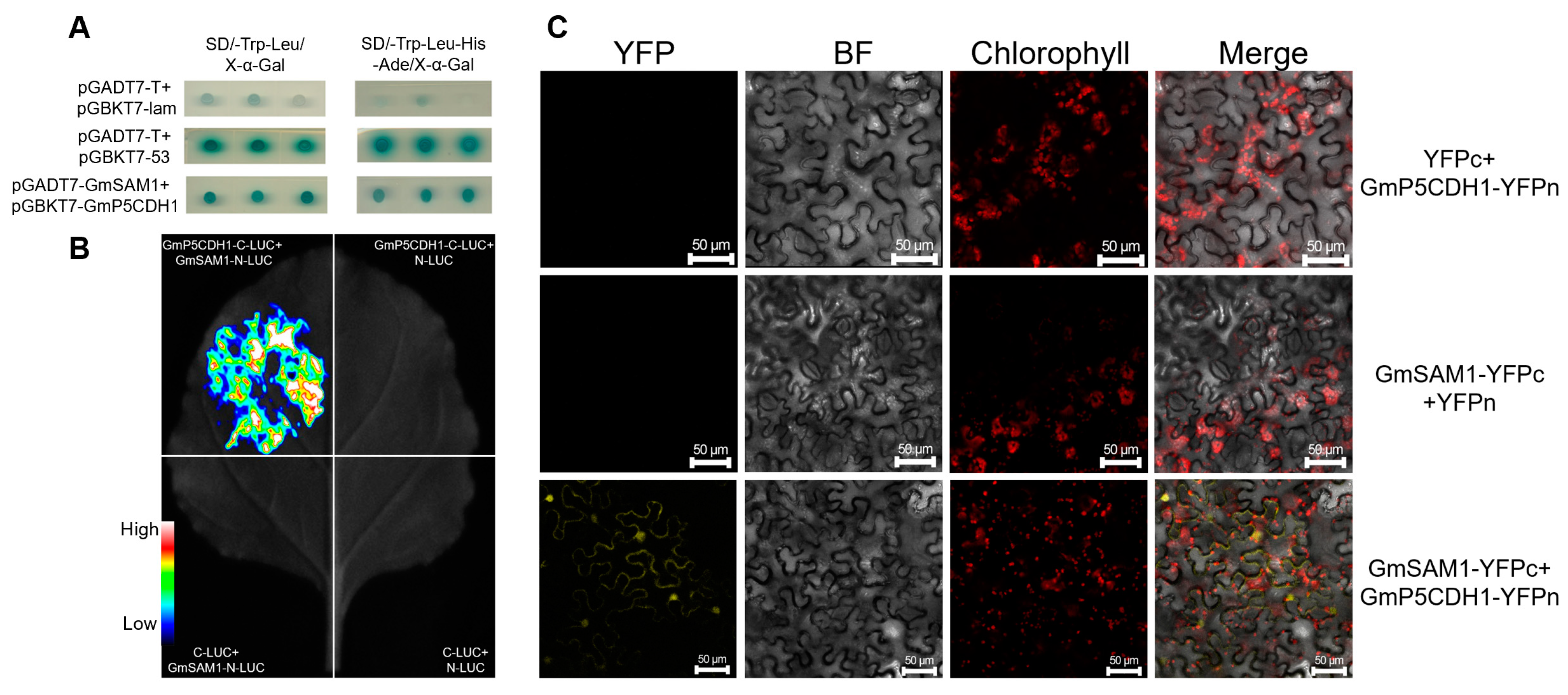
3.8. Interaction between GmP5CDH1 and GmSAM1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, S.; Liu, S.; Wang, J.; Yokosho, K.; Zhou, B.; Yu, Y.C.; Liu, Z.; Frommer, W.B.; Ma, J.F.; Chen, L.Q.; et al. Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl. Sci. Rev. 2020, 7, 1776–1786. [Google Scholar] [CrossRef] [PubMed]
- Hanly, A.; Karagiannis, J.; Lu, Q.S.M.; Tian, L.; Hannoufa, A. Characterization of the Role of SPL9 in Drought Stress Tolerance in Medicago sativa. Int. J. Mol. Sci. 2020, 21, 6003. [Google Scholar] [CrossRef] [PubMed]
- Chebrolu, K.K.; Fritschi, F.B.; Krishnan, H.B.; Smith, J.R.; Gillman, J.D. Impact of heat stress during seed development on soybean seed metabolome. Metabolomics 2016, 12, 1–14. [Google Scholar] [CrossRef]
- Clarke, J.D.; Alexander, D.C.; Ward, D.P.; Ryals, J.A.; Mitchell, M.W.; Wulff, J.E.; Guo, L. Assessment of genetically modified soybean in relation to natural variation in the soybean seed metabolome. Sci. Rep. 2013, 3, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Ding, C.; Li, W.; Wang, D.; Cui, D. Applications of metabolomics in the research of soybean plant under abiotic stress. Food Chem. 2020, 310, 125914. [Google Scholar] [CrossRef]
- Manavalan, L.P.; Guttikonda, S.K.; Phan Tran, L.S.; Nguyen, H.T. Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol. 2009, 50, 1260–1276. [Google Scholar] [CrossRef]
- Kemble, A.R.; Macpherson, H.T. Liberation of amino acids in perennial rye grass during wilting. Biochem. J. 1954, 58, 46–49. [Google Scholar] [CrossRef]
- Hu, C.A.; Delauney, A.J.; Verma, D.P. A bifunctional enzyme (delta 1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proc. Natl. Acad. Sci. USA 1992, 89, 9354–9358. [Google Scholar] [CrossRef]
- Verbruggen, N.; Hermans, C. Proline accumulation in plants: A review. Amino Acids 2008, 35, 753–759. [Google Scholar] [CrossRef]
- Mattioli, R.; Costantino, P.; Trovato, M. Proline accumulation in plants: Not only stress. Plant Signal. Behav. 2009, 4, 1016–1018. [Google Scholar] [CrossRef]
- Trinei, M.; Migliaccio, E.; Bernardi, P.; Paolucci, F.; Pelicci, P.; Giorgio, M. p66Shc, mitochondria, and the generation of reactive oxygen species. Methods Enzymol. 2013, 528, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhao, L.; Jing, R.; Wang, S.; Lu, Y. Advance in genetic engineering of proline synthetase in plant. Biotechnol. Bull. 2010, 20, 8–10. [Google Scholar] [CrossRef]
- Mishra, S.; Dubey, R.S. Inhibition of ribonuclease and protease activities in arsenic exposed rice seedlings: Role of proline as enzyme protectant. J. Plant Physiol. 2006, 163, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, P.; Sharmila, P.; Pardha Saradhi, P. Proline alleviates salt-stress-induced enhancement in ribulose-1, 5-bisphosphate oxygenase activity. Biochem. Biophys. Res. Commun. 2000, 279, 512–515. [Google Scholar] [CrossRef]
- Lehmann, S.; Funck, D.; Szabados, L.; Rentsch, D. Proline metabolism and transport in plant development. Amino Acids 2010, 39, 949–962. [Google Scholar] [CrossRef]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef]
- Zhang, M.; Huang, H.; Dai, S. Isolation and expression analysis of proline metabolism-related genes in Chrysanthemum lavandulifolium. Gene 2014, 537, 203–213. [Google Scholar] [CrossRef]
- Elthon, T.E.; Stewart, C.R. Submitochondrial location and electron transport characteristics of enzymes involved in proline oxidation. Plant Physiol. 1981, 67, 780–784. [Google Scholar] [CrossRef]
- Deuschle, K.; Funck, D.; Hellmann, H.; Däschner, K.; Binder, S.; Frommer, W.B. A nuclear gene encoding mitochondrial Delta-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. Plant J. Cell Mol. Biol. 2001, 27, 345–356. [Google Scholar] [CrossRef]
- Forlani, G.; Scainelli, D.; Nielsen, E. [delta]1-Pyrroline-5-Carboxylate Dehydrogenase from Cultured Cells of Potato (Purification and Properties). Plant Physiol. 1997, 113, 1413–1418. [Google Scholar] [CrossRef][Green Version]
- Deuschle, K.; Funck, D.; Forlani, G.; Stransky, H.; Biehl, A.; Leister, D.; van der Graaff, E.; Kunze, R.; Frommer, W.B. The role of [Delta]1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell 2004, 16, 3413–3425. [Google Scholar] [CrossRef] [PubMed]
- Rizzi, Y.S.; Monteoliva, M.I.; Fabro, G.; Grosso, C.L.; Laróvere, L.E.; Alvarez, M.E. P5CDH affects the pathways contributing to Pro synthesis after ProDH activation by biotic and abiotic stress conditions. Front. Plant Sci. 2015, 6, 572. [Google Scholar] [CrossRef] [PubMed]
- Dourmap, C.; Marmagne, A.; Lebreton, S.; Clément, G.; Guivarc’h, A.; Savouré, A.; Masclaux-Daubresse, C. Carbon and nitrogen remobilization during seed filling in Arabidopsis is strongly impaired in the pyrroline-5-carboxylate dehydrogenase mutant. J. Exp. Bot. 2023, 74, 1489–1500. [Google Scholar] [CrossRef] [PubMed]
- Min, Y.; Yu, D.; Yang, J.; Zhao, W.; Zhang, L.; Bai, Y.; Guo, C. Bioinformatics and expression analysis of proline metabolism-related gene families in alfalfa under saline-alkali stress. Plant Physiol. Biochem. PPB 2023, 205, 108182. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, P.; Duan, Z.; Lu, L.; Nan, Z.; Zhang, J. Overexpression of P5CDH from Cleistogenes songorica improves alfalfa growth performance under field drought conditions. Plant Physiol. Biochem. PPB 2024, 209, 108551. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Shi, G.; Du, H.; Wang, H.; Zhang, Z.; Hu, D.; Wang, J.; Huang, F.; Yu, D. Genome-Wide Analysis of Soybean LATERAL ORGAN BOUNDARIES Domain-Containing Genes: A Functional Investigation of GmLBD12. Plant Genome 2017, 10. [Google Scholar] [CrossRef]
- Yang, H.; Xue, Q.; Zhang, Z.; Du, J.; Yu, D.; Huang, F. GmMYB181, a soybean R2R3-MYB protein, increases branch number in transgenic Arabidopsis. Front. Plant Sci. 2018, 9, 1027. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Mara, C.; Grigorova, B.; Liu, Z. Floral-dip transformation of Arabidopsis thaliana to examine pTSO2::beta-glucuronidase reporter gene expression. J. Vis. Exp. JoVE 2010, 2010, 1952. [Google Scholar] [CrossRef]
- Cecchini, N.M.; Monteoliva, M.I.; Alvarez, M.E. Proline dehydrogenase contributes to pathogen defense in Arabidopsis. Plant Physiol. 2011, 155, 1947–1959. [Google Scholar] [CrossRef]
- Funck, D.; Eckard, S.; Müller, G. Non-redundant functions of two proline dehydrogenase isoforms in Arabidopsis. BMC Plant Biol. 2010, 10, 70. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.; Van De Cotte, B.; Van Montagu, M.; Verbruggen, N. Altered levels of proline dehydrogenase cause hypersensitivity to proline and its analogs in Arabidopsis. Plant Physiol. 2002, 128, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Huang, J.; Li, H.; Ma, Y.; Gui, C.; Huang, F.; Feng, X.; Yu, D.; Wang, H.; Kan, G. Genetic dissection reveals the complex architecture of amino acid composition in soybean seeds. TAG. Theor. Appl. Genetics. Theor. Angew. Genet. 2023, 136, 17. [Google Scholar] [CrossRef] [PubMed]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Prerostova, S.; Dobrev, P.I.; Gaudinova, A.; Hosek, P.; Soudek, P.; Knirsch, V.; Vankova, R. Hormonal dynamics during salt stress responses of salt-sensitive Arabidopsis thaliana and salt-tolerant Thellungiella salsuginea. Plant Sci. Int. J. Exp. Plant Biol. 2017, 264, 188–198. [Google Scholar] [CrossRef]
- Forlani, G.; Bertazzini, M.; Zarattini, M.; Funck, D. Functional characterization and expression analysis of rice δ(1)-pyrroline-5-carboxylate dehydrogenase provide new insight into the regulation of proline and arginine catabolism. Front. Plant Sci. 2015, 6, 591. [Google Scholar] [CrossRef][Green Version]
- Cacefo, V.; Ribas, A.F.; Vieira, L.G.E. Proline metabolism as a mechanism for the energy dissipation in VaP5CSF129A transgenic tobacco plants under water deficit. J. Plant Physiol. 2023, 283, 153964. [Google Scholar] [CrossRef]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef]
- Santarius, K.A. Freezing of isolated thylakoid membranes in complex media. viii. differential cryoprotection by sucrose, proline and glycerol. Physiol. Plant. 1992, 84, 87–93. [Google Scholar] [CrossRef]
- Ben Ahmed, C.; Magdich, S.; Ben Rouina, B.; Sensoy, S.; Boukhris, M.; Ben Abdullah, F. Exogenous proline effects on water relations and ions contents in leaves and roots of young olive. Amino Acids 2011, 40, 565–573. [Google Scholar] [CrossRef]
- Hoque, M.A.; Banu, M.N.; Nakamura, Y.; Shimoishi, Y.; Murata, Y. Proline and glycinebetaine enhance antioxidant defense and methylglyoxal detoxification systems and reduce NaCl-induced damage in cultured tobacco cells. J. Plant Physiol. 2008, 165, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.M.; Sun, J.; Guo, S.R. Effects of exogenous proline on nitrate reduction in melon seedlings under salt stress. Plant Sci. J. 2011, 29, 118–123. [Google Scholar] [CrossRef]
- Bai, H.; Guo, W.Y. Effect of exogenous proline on sod and pod activity for soybean callus under salt stress. Acta Agric. Boreall-Sin. 2002, 17, 37–40. [Google Scholar] [CrossRef]
- Yoon, K.A.; Nakamura, Y.; Arakawa, H. Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J. Hum. Genet. 2004, 49, 134–140. [Google Scholar] [CrossRef]
- Hellmann, H.; Funck, D.; Rentsch, D.; Frommer, W.B. Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiol. 2000, 122, 357–368. [Google Scholar] [CrossRef]
- Hare, P.D.; Cress, W.A.; Van, S.J. Disruptive effects of exogenous proline on chloroplast and mitochondrial ultrastructure in arabidopsis leaves. S. Afr. J. Bot. 2002, 68, 68. [Google Scholar] [CrossRef]
- Yuan, Z.L.; Liu, X.M.; Li, H.X. Review on the relationship between polyamines and environmental stress. Acta Bot. Boreali-Occident. Sin. 2008, 28, 1912–1919. [Google Scholar]
- Lyon, B.R.; Lee, P.A.; Bennett, J.M.; DiTullio, G.R.; Janech, M.G. Proteomic analysis of a sea-ice diatom: Salinity acclimation provides new insight into the dimethylsulfoniopropionate production pathway. Plant Physiol. 2011, 157, 1926–1941. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zou, T.; McCormick, S. S-Adenosylmethionine Synthetase 3 Is Important for Pollen Tube Growth. Plant Physiol. 2016, 172, 244–253. [Google Scholar] [CrossRef]
- Meng, J.; Wang, L.; Wang, J.; Zhao, X.; Cheng, J.; Yu, W.; Jin, D.; Li, Q.; Gong, Z. METHIONINE ADENOSYLTRANSFERASE4 Mediates DNA and Histone Methylation. Plant Physiol. 2018, 177, 652–670. [Google Scholar] [CrossRef]
- Ji, D.; Cui, X.; Qin, G.; Chen, T.; Tian, S. SlFERL Interacts with S-Adenosylmethionine Synthetase to Regulate Fruit Ripening. Plant Physiol. 2020, 184, 2168–2181. [Google Scholar] [CrossRef] [PubMed]

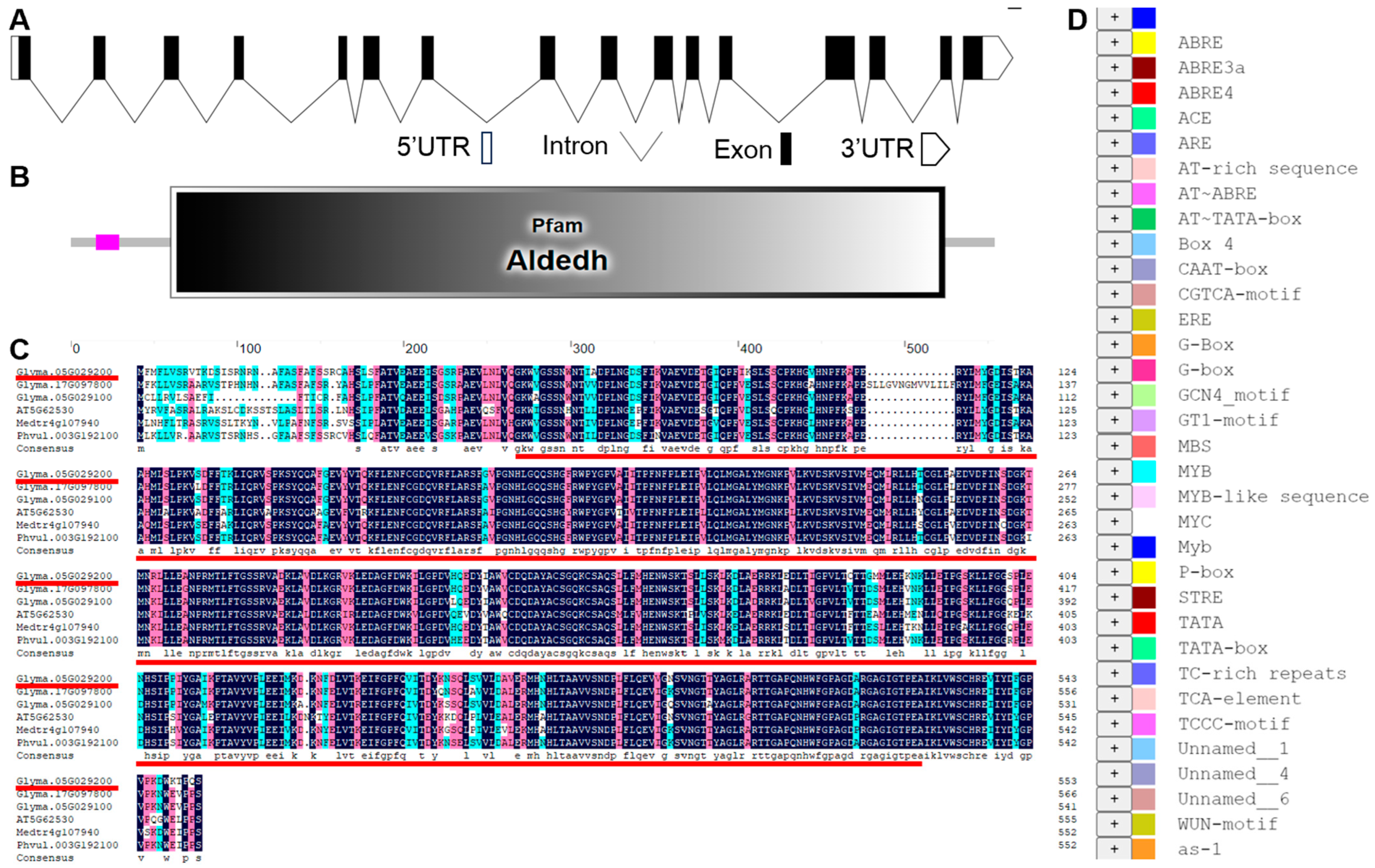


| Cis-Acting Regulatory Elements | Sequence | Function |
|---|---|---|
| ABRE | ACGTG/CACGTG | cis-acting element involved in the abscisic acid responsiveness |
| ACE | CTAACGTATT/GACACGTATG | cis-acting element involved in light responsiveness |
| ARE | AAACCA | cis-acting regulatory element essential for the anaerobic induction |
| AT-rich sequence | TAAAATACT | element for maximal elicitor-mediated activation (2 copies) |
| Box 4 | ATTAAT | part of a conserved DNA module involved in light responsiveness |
| CAAT-box | CCAAT | common cis-acting element in promoter and enhancer regions |
| CGTCA-motif | CGTCA | cis-acting regulatory element involved in the MeJA-responsiveness |
| G-Box | CACGTT | cis-acting regulatory element involved in light responsiveness |
| G-box | CACGTG | cis-acting regulatory element involved in light responsiveness |
| GCN4_motif | TGAGTCA | cis-regulatory element involved in endosperm expression |
| GT1-motif | GGTTAAT/GTGTGTGAA | light responsive element |
| MBS | CAACTG | MYB binding site involved in drought-inducibility |
| P-box | CCTTTTG | gibberellin-responsive element |
| TATA-box | TATA/TATAA/TATTTAAA | core promoter element around −30 of transcription start |
| TCA-element | CCATCTTTTT | cis-acting element involved in salicylic acid responsiveness |
| TCCC-motif | TCTCCCT | part of a light-responsive element |
| TC-rich repeats | ATTCTCTAAC | cis-acting element involved in defense and stress responsiveness |
| TGACG-motif | TGACG | cis-acting regulatory element involved in the MeJA-responsiveness |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, S.; Mao, Z.; Yang, Z.; Li, X.; Hu, D.; Wu, F.; Yu, D.; Huang, F. A Soybean Pyrroline-5-Carboxylate Dehydrogenase GmP5CDH1 Modulates Plant Growth and Proline Sensitivity. Agronomy 2024, 14, 2411. https://doi.org/10.3390/agronomy14102411
Dong S, Mao Z, Yang Z, Li X, Hu D, Wu F, Yu D, Huang F. A Soybean Pyrroline-5-Carboxylate Dehydrogenase GmP5CDH1 Modulates Plant Growth and Proline Sensitivity. Agronomy. 2024; 14(10):2411. https://doi.org/10.3390/agronomy14102411
Chicago/Turabian StyleDong, Shupeng, Zhuozhuo Mao, Zhongyi Yang, Xiao Li, Dezhou Hu, Fei Wu, Deyue Yu, and Fang Huang. 2024. "A Soybean Pyrroline-5-Carboxylate Dehydrogenase GmP5CDH1 Modulates Plant Growth and Proline Sensitivity" Agronomy 14, no. 10: 2411. https://doi.org/10.3390/agronomy14102411
APA StyleDong, S., Mao, Z., Yang, Z., Li, X., Hu, D., Wu, F., Yu, D., & Huang, F. (2024). A Soybean Pyrroline-5-Carboxylate Dehydrogenase GmP5CDH1 Modulates Plant Growth and Proline Sensitivity. Agronomy, 14(10), 2411. https://doi.org/10.3390/agronomy14102411




