Agronomic Performance and Resistance to Maize Lethal Necrosis in Maize Hybrids Derived from Doubled Haploid Lines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Parental Selection and Hybrid Formation
2.2. Environments and Trial Management
2.3. Inoculum Production and Inoculation
2.4. Trait Assessment
2.5. Statistical Analysis
3. Results
3.1. Analysis of Variance and Hybrid Performance
3.2. Combining Ability Analysis
3.3. Estimates of GCA Effects
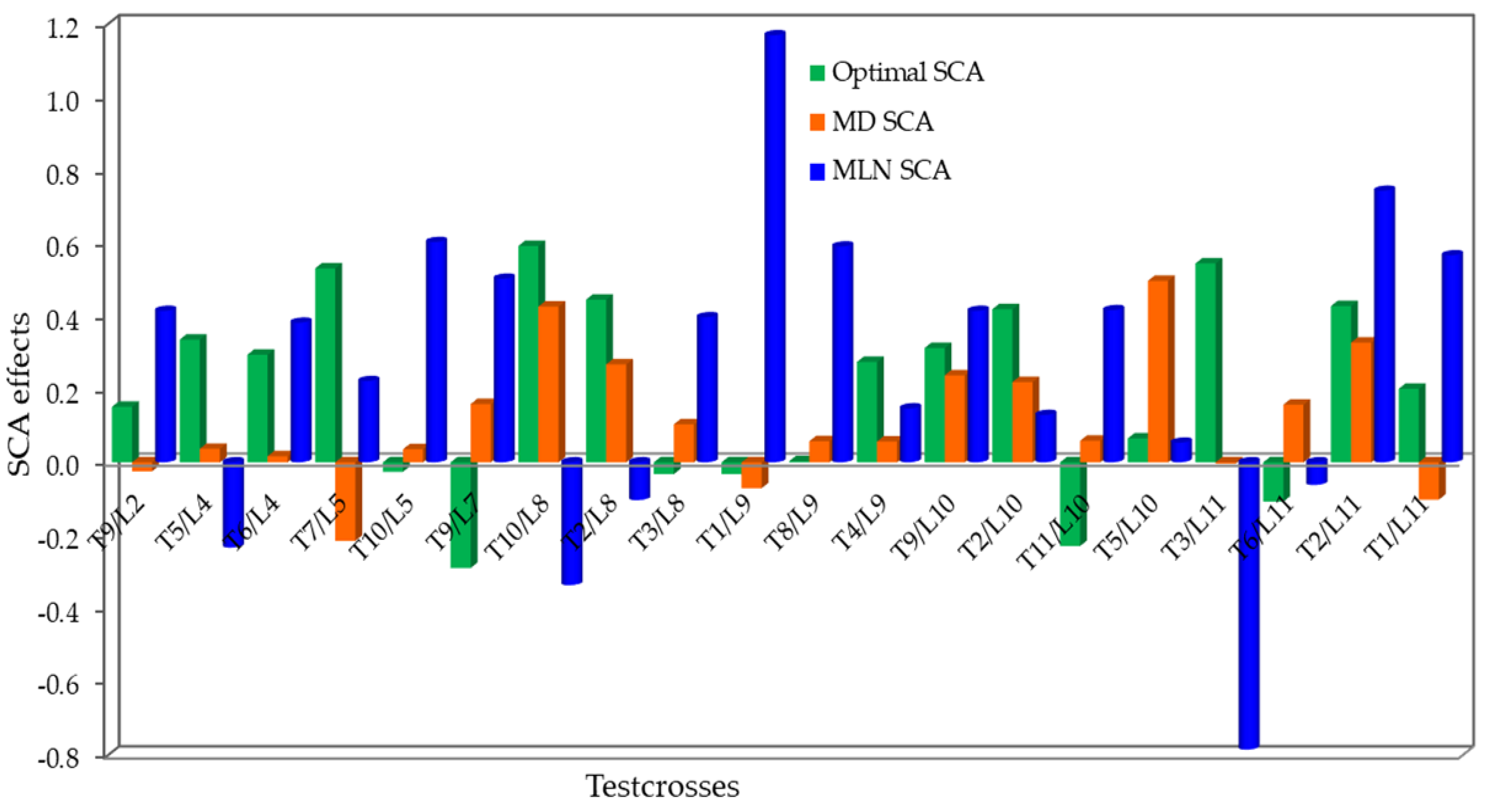
3.4. Estimates of SCA Effects
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AARC | Ambo Agricultural Research Center |
| ASI | anthesis-silking interval |
| CIMMYT | Centro Internacional de Mejoramiento de Maíz y Trigo (International Maize and Wheat Improvement Center) |
| DA | days to anthesis |
| DS | days to silking |
| EH | ear height |
| EIAR | Ethiopia Institute of Agricultural Research |
| EPP | ears per plant |
| GCA | general combining ability |
| GY | grain yield |
| H2 | broad-sense heritability |
| HC | bad husk cover |
| KALRO | Kenya Agricultural and Livestock Research Organization |
| L | line |
| L × T | line-by-tester |
| MCMV | maize chlorotic mottle virus |
| MLN | maize lethal necrosis |
| MLN-AI | maize lethal necrosis artificial inoculation |
| MLN-DS | maize lethal necrosis disease severity |
| PH | plant height |
| SCA | specific combining ability |
| SCMV | sugarcane mosaic virus |
| SEN | leaf senescence |
| SSA | sub-Sahara Africa |
| T | tester |
| UG | University of Ghana |
| WACCI | West Africa Center for Crop Improvement |
References
- Hellin, J.J.; Shiferaw, B.; Cairns, J.E.; Reynolds, M.P.; Ortíz-Monasterio, I.; Bänziger, M.; Sonder, K.; La Rovere, R. Climate change and food security in the developing world: The potential of maize and wheat research to expand options for adaptation and mitigation. J. Dev. Agric. Econ. 2012, 4, 311–321. [Google Scholar]
- Bänziger, M.; Setimela, P.S.; Hodson, D.; Vivek, B. Breeding for improved abiotic stress tolerance in maize adapted to southern Africa. Agric. Water Manag. 2006, 80, 212–224. [Google Scholar] [CrossRef]
- Salami, A.; Kamara, A.B.; Brixiova, Z. Smallholder Agriculture in East Africa: Trends, Constraints and Opportunities; Working Papers Series N° 105; African Development Bank: Tunis, Tunisia, 2010. [Google Scholar]
- Matongera, N.; Ndhlela, T.; van Biljon, A.; Kamutando, C.N.; Labuschagne, M. Combining ability and testcross performance of multi-nutrient maize under stress and non-stress environments. Front. Plant Sci. 2023, 14, 1070302. [Google Scholar] [CrossRef]
- Prasanna, B.; Suresh, L.M.; Francis, M.; Yoseph, B.; Dan, M.; Manje, G.; Mike, O.; David, H.; Mosisa, W.; Monica, M.; et al. Maize lethal necrosis (MLN): Efforts toward containing the spread and impact of a devastating transboundary disease in sub-Saharan Africa. Virus Res. 2020, 282, 197943. [Google Scholar]
- Wangai, A.W.; Redinbaugh, M.G.; Kinyua, Z.M.; Miano, D.W.; Leley, P.K.; Kasina, M.; Mahuku, G.; Scheets, K.; Jeffers, D. First Report of Maize chlorotic mottle virus and Maize Lethal Necrosis in Kenya. Plant Dis. 2012, 96, 1582. [Google Scholar] [CrossRef]
- Mahuku, G.; Lockhart, B.E.; Wanjala, B.J.; Mark, W.K.; Janet, N.S.; Lucy, R.C.; Bryan, J.S.; Subramanian, N.; Johnson, O.K.; Elizabeth Kumar, P.; et al. Maize lethal necrosis (MLN), an emerging threat to maize-based food security in sub-Saharan Africa. Phytopathology 2015, 105, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Kabululu, M.S.; Ndunguru, J.; Ndakidemi, P.; Feyissa, T. Genetic diversity of maize accessions for maize lethal necrosis disease resistance. Indian J. Agric. Res. 2017, 51, 17–24. [Google Scholar]
- Awata, L.; Ifie, B.E.; Danquah, E.; Jumbo, M.; Suresh, L.; Gowda, M.; Marchelo-Dragga, P.; Olsen, M.; Shorinola, O.; Nasser, K.; et al. Introgression of Maize Lethal Necrosis Resistance Quantitative Trait Loci Into Susceptible Maize Populations and Validation of the Resistance Under Field Conditions in Naivasha, Kenya. Front. Plant Sci. 2021, 12, 649308. [Google Scholar] [CrossRef] [PubMed]
- Ohlson, E.W.; Margaret, G.R.; Mark, W.J. Mapping maize chlorotic mottle virus tolerance loci in the maize 282 association panel. Crop Sci. 2022, 62, 1234–1245. [Google Scholar] [CrossRef]
- Redinbaugh, M.G.; Stewart, L.R. Maize lethal necrosis: An emerging, synergistic viral disease. Annu. Rev. Virol. 2018, 5, 301–322. [Google Scholar] [CrossRef]
- Nyaga, C.; Gowda, M.; Beyene, Y.; Murithi, W.; Burgueno, J.; Toledo, F.; Makumbi, D.; Olsen, M.; Das, B.; Suresh, L.; et al. Hybrid breeding for MLN resistance: Heterosis, combining ability, and hybrid prediction. Plants 2020, 9, 468. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Tinker, N.A. Biplot analysis of multi-environment trial data: Principles and applications. Can. J. Plant Sci. 2006, 86, 623–645. [Google Scholar] [CrossRef]
- Yan, W. Analysis and handling of G × E in a practical breeding program. Crop Sci. 2016, 56, 2106–2118. [Google Scholar] [CrossRef]
- Bänziger, M.; Betrán, F.J.; Lafitte, H.R. Efficiency of high-nitrogen selection environments for improving maize for low-nitrogen target environments. Crop Sci. 1997, 37, 1103–1109. [Google Scholar] [CrossRef]
- Bänziger, M.; Setimela, P.S.; Hodson, D.; Vivek, B.; Bänziger, M.; Setimela, P.S.; Hodson, D.; Vivek, B. Breeding for improved drought tolerance in maize adapted to southern Africa. Crop Sci. 2004, 80, 212–224. [Google Scholar]
- Annor, B.; Badu-Apraku, B.; Nyadanu, D.; Akromah, R.; Fakorede, M.A.B. Testcross performance and combining ability of early maturing maize inbreds under multiple-stress environments. Sci. Rep. 2019, 9, 13809. [Google Scholar] [CrossRef] [PubMed]
- Kempthorne, O. An Introduction to Genetics Statistics, In Search. 1957, 143. Available online: https://int.search.myway.com/web (accessed on 12 August 2024).
- Vasal, S.; Srinivasan, G.; González, F.; Han, G.; Pandey, S.; Beck, D.; Crossa, J. Heterosis and Combining Ability of CIMMYT’s Tropical × Subtropical Maize Germplasm. Crop Sci. 1992, 32, 1483–1489. [Google Scholar] [CrossRef]
- Vacaro, E.; Barbosa, N.J.; Pegoraro, D.G.; Nuss, C.N.; Haa Conceição, L.D. Combining ability of twelve maize populations. Pesqui. Agropecu. Bras. 2002, 37, 67–72. [Google Scholar] [CrossRef]
- Kassahun, S.; Yoseph, B.; Beatrice, E.I.; Suresh, L.M.; Michael, S.O.; Veronica, O.; Dagne, W.; Pangirayi, T.; Eric, D.; Samuel, K.O.; et al. Identification of Genomic Regions Associated with Agronomic and Disease Resistance Traits in a Large Set of Multiple DH Populations. Genes 2022, 13, 351. [Google Scholar] [CrossRef]
- Banziger, M.; Edmeades, G.O.; Beck, D.; Bellon, M. Breeding for Drought and Nitrogen Stress Tolerance in Maize: From Theory to Practice; CIMMYT: Texcoco, Mexico, 2000. [Google Scholar]
- Prasanna, B.M. (Ed.) Maize Lethal Necrosis (MLN): A Technical Manual for Disease Management; CIMMYT: Texcoco, Mexico, 2021. [Google Scholar]
- Roger, P.; Darren, M.; Simon, H.; David, B.; Duncan, S. GenStat for Windows 12th Edition Introduction®TM; VSN International Ltd.: Hempstead, UK, 2012. [Google Scholar]
- Alvarado, G.; Vargas, M.; Combs, E.; Atlin, G.; Mathews, K.; Crossa, J. META: A Suite of SAS Programs to Analyze Multienvironment Breeding Trials. Agron. J. 2015, 1, 11–19. [Google Scholar]
- Souza, L.V.; Miranda, G.V.; Cardoso Galvão, J.C.; Moreira Guimarães, L.J.; Santos, I.C. Combining ability of maize grain yield under different levels of environmental stress. Pesqui. Agropecu. Bras. 2009, 44, 1297–1303. [Google Scholar] [CrossRef]
- Dan, M.; Silvano, A.; Alpha, D.; Cosmos, M.; Godfrey, A.; Mosisa, W.; Marianne, B. Genetic analysis of tropical midaltitude-adapted maize populations under stress and nonstress conditions. Crop Sci. 2018, 58, 1492–1507. [Google Scholar]
- Siphesihle, G.M.; John, D.; Fikile, Q.; Pedro, F.; Edmore, G.; Paramu, M. Heritability and genetic gain for grain yield and path coefficient analysis of some agronomic traits in early-maturing maize hybrids. Euphytica 2015, 206, 225–244. [Google Scholar]
- Sesay, S.; Ojo, D.K.; Ariyo, O.J.; Meseka, S. Genetic variability, heritability and genetic advance studies in top-cross and three-way cross maize (Zea mays L.) hybrids. Maydica 2016, 204, 225–244. [Google Scholar]
- Ayodeji, A.; Comfort, A.A. Genetic variability, heritability and genetic advance in shrunken-2 super-sweet corn (Zea mays L. saccharata) populations. J. Plant Breed. Crop Sci. 2019, 11, 100–105. [Google Scholar] [CrossRef]
- Yoseph, B.; Mugo, S.; Kassa, S.; Godfrey, A.; Walter, T.; Amsal, T.; Tadele, T.; James, G.; Barnabas, K.; John, G.; et al. Genetic distance among doubled haploid maize lines and their testcross performance under drought stress and non-stress conditions. Euphytica 2013, 192, 379–392. [Google Scholar]
- John, D.; Pangirayi, T.; Bindiganavile, S.V.; Mark, D.L. Gene action controlling grain yield and secondary traits in southern African maize hybrids under drought and non-drought environments. Euphytica 2008, 162, 411–422. [Google Scholar]
- John, D.; Pangirayi, T.; Kevin, V.P.; Bindiganavile, V.; Mark, D.L.; Neil, C. Gene action controlling gray leaf spot resistance in southern African maize germplasm. Crop Sci. 2008, 48, 93–98. [Google Scholar]
- Legesse, B.W.; Pixley, K.V.; Botha, A.M. Combining ability and heterotic grouping of highland transition maize inbred lines. Maydica 2009, 54, 1–9. [Google Scholar]
- Gurung, D.B.; George, M.L.C.; Delacruz, Q.D. Determination of Heterotic Groups in Nepalese Yellow Maize Populations. Nepal J. Sci. Technol. 2009, 10, 1–8. [Google Scholar] [CrossRef]
- Estakhr, A.; Heidari, B. Combining ability and gene action for maturity and agronomic traits in different heterotic groups of maize inbred lines and their diallel crosses. J. Crop Sci. Biotechnol. 2012, 15, 219–229. [Google Scholar] [CrossRef]
- Abebe, M.; Ivan, I.; Charles, T. Testcross performance and diversity analysis of white maize lines derived from backcrosses containing exotic germplasm. Euphytica 2007, 155, 417–428. [Google Scholar]
- Istipliler, D.; Ilker, E.; Aykut Tonk, F.; Civi, G.; Tosun, M. Line × tester analysis and estimating combining abilities for yield and some yield components in bread wheat. Turk. J. Field Crops 2015, 20, 72–77. [Google Scholar] [CrossRef]
- Bänziger, M.; Edmeades, G.O.; Lafitte, H.R. Selection for drought tolerance increases maize yields across a range of nitrogen levels. Crop Sci. 1999, 39, 1035–1040. [Google Scholar] [CrossRef]
- Simon, Z.; Kasozi, L.C.; Patrick, R.; Abubaker, M. Gene Action for Grain Yield and Agronomic Traits in Selected Maize Inbred Lines with Resistance to Striga Hermonthica in Uganda. J. Food Secur. 2018, 6, 155–162. [Google Scholar]
- Pearl, A.; Baffour, B.; Beatrice, E.I.; Pangirayi, T.; Leander, D.M.; Samuel, K.O. Genetic diversity and inter-trait relationship of tropical extra-early maturing quality protein maize inbred lines under low soil nitrogen stress. PLoS ONE 2021, 16, e0252506. [Google Scholar] [CrossRef]
- Makumbi, D.; Betrán, J.F.; Bänziger, M.; Ribaut, J.M. Combining ability, heterosis and genetic diversity in tropical maize (Zea mays L.) under stress and non-stress conditions. Euphytica 2011, 180, 143–162. [Google Scholar] [CrossRef]
- Ahmad, E.; Ansari, A.M. Study of combining ability and heterosis analysis for yield traits in maize (Zea mays L.). J. Pharmacogn. Phytochem. 2017, 924–927. Available online: https://www.phytojournal.com/archives/2017/vol6issue6S/PartV/SP-6-6-219.pdf (accessed on 17 October 2024).
- Worku, M.; Bänziger, M.; Friesen, D.; Schulte, G.; Horst, W.J.; Vivek, B.S. Relative importance of general combining ability and specific combining ability among tropical maize (Zea mays L.) inbreds under contrasting nitrogen environments. Maydica 2008, 53, 279–288. [Google Scholar]
- Tolera, K.; Mosisa, W.; Habtamu, Z. Combining ability and heterotic orientation of mid-altitude sub-humid tropical maize inbred lines for grain yield and related traits. Afr. J. Plant Sci. 2017, 11, 229–239. [Google Scholar] [CrossRef]
| Code | Name | Description |
|---|---|---|
| L1 | CKDHL64076 | Line |
| L2 | CKDHL10310 | Line |
| L3 | CKDHL64665 | Line |
| L4 | CKDHL64672 | Line |
| L5 | CKDHL63598 | Line |
| L6 | CKDHL63908 | Line |
| L7 | CKDHL63943 | Line |
| L8 | CKDHL64302 | Line |
| L9 | CKDHL42833 | Line |
| L10 | CKSBL10060 | Line |
| L11 | CKDHL63627 | Line |
| T1 | CKLTI0138/CKLMARSI0022 | Tester |
| T2 | CKLTI0227/CKLMARSI0022 | Tester |
| T3 | CKDHL10918/CKLMARSI0022 | Tester |
| T4 | CKLTI0138/CML550 | Tester |
| T5 | CKDHL10918/CML494 | Tester |
| T6 | CKLTI0139/CKDHL10918 | Tester |
| T7 | CKLTI0227/CKDHL10918 | Tester |
| T8 | CKLMARSI0037/CML543 | Tester |
| T9 | CML543/CML494 | Tester |
| T10 | CML322/CML543 | Tester |
| T11 | CKDHL0500/CML543 | Tester |
| CK1 | PH30G19 | Commercial MLN-susceptible check |
| CK2 | WH505 | Commercial MLN-susceptible check |
| CK3 | H516 | Commercial MLN-susceptible check |
| CK4 | DK8031 | Commercial MLN-susceptible check |
| CK5 | DK777 | MLN-tolerant check |
| Location | Management | Year | Altitude m.a.s.l. | Latitude | Longitude | Fertilization (kg ha−1) | Grain Yield (t/ha) | |
|---|---|---|---|---|---|---|---|---|
| Mean + SE | H2 | |||||||
| Kakamega | Optimum | 2019 | 1580 | 0°16′ N | 34°46′ E | 38 P, 93 N | 9.02 + 1.26 | 0.82 |
| Kiboko | Optimum | 2019 | 1020 | 2°15′ S | 37°75′ E | 60 P, 87 N | 7.67 + 0.86 | 0.77 |
| Kiboko | Managed drought | 2019 | 1020 | 2°15′ S | 37°75′ E | 60 P, 87 N | 4.86 + 0.74 | 0.69 |
| Kirinyaga | Optimum | 2019 | 1159 | 0°34′ S | 37°19′ E | 50 P, 138 N | 7.17 + 1.47 | 0.62 |
| Kaguru | Optimum | 2019 | 1460 | 0°02′ N | 37°39′ E | 50 P, 138 N | 6.54 + 0.98 | 0.61 |
| Naivasha | MLN-AI | 2019 | 1896 | 0°43′ N | 36°26′ E | 60 P, 87 N | 3.19 + 0.85 | 0.77 |
| Naivasha | MLN-AI | 2020 | 1896 | 0°43′ N | 36°26′ E | 60 P, 87 N | 2.63 + 0.65 | 0.76 |
| Source of Variation | Optimum Management | MLN-AI | Managed Drought | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DF | GY | AD | ASI | PH | EH | HC | EPP | DF | GY | AD | PH | MLN- DS | DF | GY | AD | ASI | PH | SEN | |
| Environment (E) | 3 | 128 ** | 8292.1 ** | 719.43 ** | 19,288.2 ** | 22,933.8 ** | 14,626.8 ** | 0.99 ** | 1 | 9.7 ** | 10,974.6 ** | 39,130.9 ** | 73.4 ** | - | - | - | - | - | - |
| Rep (Environment) | 4 | 1.6 | 10.82 | 2.39 | 327.35 | 326.5 | 35.03 | 0.01 | 2 | 4.0 ** | 1350.8 ** | 9436.5 ** | 1.1 | 1 | 2.3 ** | 2279 | 125.6 | 19,412.3 ** | 6.78 ** |
| Genotype (G) | 114 | 13.2 ** | 17.2 ** | 14.11 ** | 656.7 ** | 562.5 ** | 2094.45 ** | 0.07 ** | 114 | 3.0 ** | 39.7 ** | 458.7 * | 1.8 ** | 114 | 1.5 ** | 25.3 | 3.7 | 523.2 | 1.07 ** |
| GCALine | 10 | 86.5 ** | 35.4 ** | 16.61 ** | 1264.4 ** | 1550.5 ** | 1724.68 ** | 0.09 ** | 10 | 17.6 ** | 40.9 | 390.7 | 1.1 | 10 | 6.3 ** | 7.5 | 2.4 | 229.7 | 3.52 ** |
| GCATester | 10 | 49.6 ** | 116.7 ** | 96.97 ** | 4542.6 ** | 3614.7 ** | 19,718.12 ** | 0.58 ** | 10 | 5.4 ** | 267.9 ** | 2541.6 | 8.4 ** | 10 | 4.9 ** | 235.3 ** | 21.1 ** | 4174.2 ** | 2.31 ** |
| SCA | 94 | 1.5 * | 4.6 | 5.03 ** | 178.7 | 132.7 | 258.94 ** | 0.02 | 94 | 1.1 ** | 15.6 | 243.1 | 1.1 ** | 94 | 0.61 | 4.9 | 2 | 166 | 0.68 |
| G x E | 342 | 1.9 ** | 4.7 | 4.43 ** | 230.3 * | 169.6 | 293.54 ** | 0.02 * | 114 | 1.0 ** | 20.9 ** | 349.2 | 0.8 | - | - | - | - | - | - |
| GCALine × E | 30 | 4.5 ** | 6 | 3.77 ** | 282.1 | 204.3 | 256.43 | 0.02 | 10 | 3.1 ** | 21.3 | 214.3 | 1.9 | - | - | - | - | - | - |
| GCATester × E | 30 | 3.7 ** | 12.8 ** | 6.68 ** | 840.7 | 509.1 | 1196.62 ** | 0.06 ** | 10 | 1.8 ** | 96.3 ** | 1817.2 ** | 0.7 | - | - | - | - | - | - |
| SCA × E | 282 | 1.4 | 3.7 | 4.26 * | 159.8 | 129.8 | 201.41 | 0.02 | 94 | 0.7 | 12.9 | 207.8 | 0.7 | - | - | - | - | - | - |
| Residuals | 304 | 1.3 | 5.2 | 3.71 | 192 | 167 | 142.89 | 0.02 | 148 | 0.6 | 33 | 387.1 | 1.4 | 76 | 0.51 | 28.6 | 3.4 | 486.9 | 0.45 |
| %SS GCA | 0.91 | 0.78 | 0.71 | 0.78 | 0.81 | 0.83 | 0.78 | 0.69 | 0.68 | 0.56 | 0.48 | 0.66 | 0.84 | 0.56 | 0.74 | 0.48 | |||
| %SS SCA | 0.09 | 0.22 | 0.29 | 0.22 | 0.19 | 0.18 | 0.22 | 0.31 | 0.32 | 0.44 | 0.52 | 0.34 | 0.16 | 0.44 | 0.26 | 0.52 | |||
| Mean | 7.6 | 68.52 | −0.09 | 259.00 | 131.96 | 22.09 | 1.05 | 2.91 | 87.18 | 166.15 | 3.38 | 4.86 | 70.61 | −0.18 | 224.49 | 3.40 | |||
| Minimum | 4.69 | 63.67 | −1.54 | 227.51 | 109.06 | 2.59 | 0.95 | 0.96 | 81.03 | 143.92 | 2.30 | 3.27 | 66.99 | −1.95 | 194.82 | 2.46 | |||
| Maximum | 11.12 | 74.33 | 2.00 | 287.76 | 160.21 | 71.85 | 1.25 | 4.95 | 95.66 | 182.72 | 6.29 | 6.32 | 75.68 | 3.08 | 243.37 | 4.56 | |||
| LSD0.05 | 1.38 | 1.67 | 1.16 | 9.93 | 7.89 | 19.03 | 0.13 | 1.26 | 4.09 | 17.48 | 0.68 | 1.27 | 1.89 | 1.45 | 14.00 | 1.13 | |||
| CV (%) | 15.1 | 1.89 | 21.40 | 2.95 | 4.98 | 54.68 | 12.13 | 26.1 | 3.42 | 6.45 | 11.68 | 15.3 | 1.33 | 10.24 | 3.15 | 20.59 | |||
| Heritability (H2) | 0.85 | 0.93 | 0.76 | 0.94 | 0.96 | 0.86 | 0.70 | 0.74 | 0.83 | 0.66 | 0.90 | 0.69 | 0.89 | 0.79 | 0.81 | 0.60 | |||
| Hybrid Code | Tester/Line | Grain Yield Under | Agronomic Traits and MLN Disease Score | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Optimum | Managed Drought | MLN-AI | AD | ASI | EPP | HC | PH | EH | SEN | MLN-DS | ||
| H23 | T1/L3 | 10.1 | 5.4 | 4.2 | 70.1 | −0.8 | 1.22 | 8.6 | 287.8 | 152.9 | 3.1 | 3.7 |
| H24 | T2/L3 | 8.7 | 6.2 | 3.6 | 69.4 | −0.5 | 1.10 | 12.1 | 274.5 | 146.4 | 3.3 | 3.9 |
| H26 | T4/L3 | 9.2 | 5.4 | 3.5 | 70.0 | −0.8 | 1.20 | 11.0 | 281 | 148.7 | 3.7 | 3.8 |
| H27 | T5/L3 | 8.1 | 4.9 | 3.7 | 71.2 | −1.3 | 1.11 | 20.4 | 274.2 | 146.5 | 3.8 | 3.3 |
| H28 | T6/L3 | 8.1 | 5.4 | 3.5 | 71.6 | 0.4 | 1.08 | 15.3 | 278.9 | 145.9 | 3.5 | 3.5 |
| H29 | T7/L3 | 8.2 | 4.7 | 4.0 | 70.8 | 0.0 | 1.04 | 6.6 | 275.7 | 146 | 3.4 | 3.7 |
| H30 | T8/L3 | 9.1 | 6.3 | 3.4 | 70.5 | 0.8 | 1.07 | 11.9 | 272.8 | 144.6 | 3.1 | 4.2 |
| H31 | T9/L3 | 9.6 | 5.2 | 3.8 | 73.2 | 0.4 | 1.08 | 6.6 | 279.7 | 151.9 | 3.7 | 4.2 |
| H32 | T10/L3 | 10.0 | 6.1 | 2.8 | 71.8 | −0.4 | 1.05 | 7.7 | 276.1 | 150.4 | 3.6 | 4.7 |
| H33 | T11/L3 | 11.1 | 6.0 | 2.3 | 72.3 | −0.5 | 1.09 | 4.1 | 275.2 | 156.7 | 3.4 | 4.9 |
| H34 | T1/L4 | 8.5 | 5.5 | 3.0 | 68.7 | −0.8 | 1.22 | 5.0 | 270.1 | 137.5 | 3.3 | 4.4 |
| H35 | T2/L4 | 8.4 | 5.4 | 3.7 | 67.7 | −0.4 | 1.23 | 6.7 | 271.4 | 136.5 | 3.4 | 3.8 |
| H37 | T4/L4 | 8.3 | 5.3 | 3.9 | 68.3 | −0.7 | 1.25 | 7.9 | 266 | 137.1 | 4.0 | 3.9 |
| H42 | T9/L4 | 9.3 | 5.9 | 3.8 | 70.1 | −0.8 | 1.13 | 7.0 | 264.9 | 135.3 | 3.0 | 4.0 |
| H44 | T11/L4 | 9.6 | 5.6 | 3.6 | 69.7 | −1.2 | 1.09 | 4.9 | 270.0 | 146.0 | 3.0 | 3.7 |
| H59 | T9/L6 | 8.0 | 5.3 | 3.8 | 68.9 | −0.8 | 1.05 | 35.9 | 262.4 | 137.3 | 3.2 | 3.6 |
| H70 | T9/L7 | 7.8 | 5.5 | 4.0 | 67.5 | 0.0 | 1.02 | 25.3 | 263.0 | 135.9 | 3.5 | 3.6 |
| H72 | T11/L7 | 8.2 | 5.3 | 3.5 | 67.4 | −0.8 | 1.01 | 17.2 | 254.7 | 137.4 | 2.8 | 3.7 |
| H74 | T2/L8 | 8.3 | 4.9 | 3.4 | 64.5 | 0.4 | 1.0 | 43.2 | 259.6 | 130.9 | 2.8 | 3.9 |
| H80 | T9/L8 | 8.4 | 5.2 | 3.7 | 67.7 | 0.3 | 1.02 | 42.5 | 267.9 | 143.7 | 3.6 | 3.6 |
| H84 | T2/L9 | 8.7 | 4.9 | 3.1 | 70.5 | −1.3 | 1.24 | 3.5 | 282.2 | 152.3 | 3.1 | 4.8 |
| H105 | T1/L11 | 7.8 | 5.2 | 3.8 | 68.2 | −0.9 | 1.10 | 64.6 | 264.2 | 127.8 | 3.0 | 3.8 |
| H112 | T8/L11 | 8.1 | 5.4 | 3.5 | 67.2 | −0.5 | 1.00 | 46.5 | 260.1 | 128.1 | 2.9 | 4.0 |
| H114 | T10/L11 | 8.1 | 5.5 | 3.3 | 68.3 | −1.6 | 1.02 | 61.1 | 257.5 | 130.1 | 3.4 | 3.7 |
| H115 | T11/L11 | 8.3 | 5.7 | 3.3 | 69.5 | −1.9 | 1.06 | 22.5 | 261.9 | 137.5 | 2.8 | 4.5 |
| CK1 | PH30G19 | 8.8 | 3.8 | 1.2 | 68.2 | 3.1 | 1.0 | 6.5 | 265 | 117.7 | 3.1 | 6.1 |
| CK2 | WH505 | 8.7 | 4.6 | 1.3 | 73.1 | −0.1 | 1.0 | 5.6 | 282.7 | 148.7 | 2.9 | 5.3 |
| CK3 | H516 | 6.1 | 3.3 | 1.3 | 71.6 | 3.0 | 1.0 | 10.2 | 283.8 | 154.2 | 4.2 | 5.9 |
| CK4 | DK8031 | 6.5 | 3.3 | 1.0 | 67.1 | 1.5 | 1.0 | 10.0 | 252.2 | 123.0 | 4.5 | 7.1 |
| CK5 | DK777 | 7.5 | 4.5 | 2.4 | 70.8 | −1.2 | 1.0 | 11.1 | 255.8 | 123.9 | 3.2 | 5.2 |
| Mean | 8.7 | 5.4 | 3.5 | 69.4 | −0.5 | 1.1 | 19.9 | 270.1 | 141.7 | 3.3 | 4.0 | |
| Minimum | 7.8 | 4.7 | 2.3 | 64.5 | −1.9 | 1.0 | 3.5 | 254.7 | 127.8 | 2.8 | 3.3 | |
| Maximum | 11.1 | 6.3 | 4.2 | 73.2 | 0.8 | 1.3 | 64.6 | 287.8 | 156.7 | 4.0 | 4.9 | |
| LSD0.05 | 1.23 | 1.27 | 1.26 | 1.67 | 1.16 | 0.13 | 19.03 | 9.93 | 7.89 | 1.13 | 0.68 | |
| Line | Optimum Management | MLN-AI | Managed Drought Stress | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GY | AD | ASI | PH | EH | EPP | HC | GY | AD | PH | MLN-DS | GY | AD | ASI | PH | SEN | |
| L1 | −0.3 | −0.90 * | −0.05 | −4.87 * | −3.04 | −0.05 | −1.58 | 0.55 * | 0.23 | −1.63 | 0.18 | 0.53 * | −2.67 ** | −0.76 ** | −0.03 | 0.09 |
| L2 | −0.56 | 0.02 | 0.75 ** | 0.31 | −4.54 | −0.04 | −0.62 | −0.27 | −0.72 | 1.6 | −0.41 * | 0.49 * | −3.26 ** | 0.16 | 8.52 * | −0.18 |
| L3 | 1.19 ** | 1.75 ** | 0.00 | 12.63 ** | 10.47 ** | 0.04 | 5.73 * | 0.65 * | 2.87 ** | 8.14 * | −0.09 | 1.01 ** | −1.78 * | −0.80 * | 22.75 ** | −0.03 |
| L4 | 0.47 | 0.69 | −0.13 | 4.05 * | 1.64 | 0.06 ** | −2.63 | 0.35 | 1.42 * | 2.96 | 0.03 | 1.13 ** | −1.78 * | −1.17 ** | 17.9 ** | 0.42 * |
| L5 | −0.94 ** | −1.41 ** | −0.06 | −15.1 ** | −10.31 ** | −0.03 | 3.48 | −0.92 ** | −2.38 ** | −15.81 ** | 0.7 ** | −0.04 | −3.85 ** | −0.81 * | −1.07 | 0.12 |
| L6 | −0.22 | −0.44 | −0.45 ** | −2.52 | −4.48 | −0.04 | 6.36 ** | 0.37 | −1.98 * | 4.23 | −1.19 ** | 0.26 | −2.33 ** | −0.6 | 8.36 * | 0.52 ** |
| L7 | −0.18 | −1.21 ** | −0.38 * | −0.74 | 0.73 | −0.01 | 3.04 | 0.55 * | −4.00 ** | 3.91 | −0.59 * | −0.23 | 2.06 * | 0.36 | −12.87 ** | 0.18 |
| L8 | −0.01 | −0.65 | 0.23 | 1.17 | 1.41 | −0.02 | −2.55 | 0.01 | −2.18 * | 7.29 * | −0.36 | −0.88 ** | 3.54 ** | 1.26 ** | −7.38 | −0.64 ** |
| L9 | 0.57 * | 1.91 ** | −0.05 | 7.58 * | 8.05 ** | 0.08 ** | −1.75 | −0.96 ** | 5.92 ** | −2.13 | 0.92 ** | −0.85 ** | 4.47 ** | 1.16 ** | −11.31 ** | −0.11 |
| L10 | 0.22 | 0.12 | 0.29 * | −1.85 | 3.26 | 0 | 0.35 | −0.82 ** | 0.77 | −11.85 ** | 1.04 ** | −0.78 ** | 1.94 * | 1.16 ** | −7.66 * | 0.03 |
| L11 | −0.23 | 0.12 | −0.15 | −0.67 | −3.18 | −0.01 | −9.83 ** | 0.48 * | 0.04 | 3.29 | −0.23 | −0.64 ** | 3.67 ** | 0.05 | −17.20 ** | −0.39 * |
| SE± | 0.3 | 0.41 | 0.15 | 3.12 | 2.49 | 0.02 | 2.46 | 0.24 | 0.81 | 3.5 | 0.2 | 0.24 | 0.86 | 0.34 | 3.96 | 0.18 |
| Tester | Optimum Management | MLN-AI | Managed Drought Stress | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GY | AD | ASI | PH | EH | EPP | HC | GY | AD | PH | MLN-DS | GY | AD | ASI | PH | SEN | |
| T1 | 0.39 | −0.26 | 0.02 | 1.70 | 1.10 | 0.03 | −8.83 | 0.37 | −0.30 | 0.03 | 0.07 | −0.12 | −0.50 | 0.00 | 0.10 | −0.25 |
| T2 | −0.03 | −0.93 ** | 0.08 | 0.23 | −1.75 | 0.01 | −7.83 | 0.61 | 2.37 ** | −0.06 | 0.27 * | 0.16 | 0.51 | −0.09 | 0.44 | −0.32 |
| T3 | −0.74 * | −0.47 | 0.10 | −6.57 ** | −7.11 ** | −0.02 | −12.45 ** | 1.09 ** | 2.84 ** | −0.05 | 0.17 | −0.06 | 1.76 ** | −0.34 | −1.74 | 0.06 |
| T4 | −0.08 | −0.49 | −0.11 | 1.17 | 1.20 | 0.04 | −15.1 ** | 0.05 | −1.92 * | 0.04 | 0.16 | −0.14 | −1.46 ** | 0.07 | −1.65 | −0.11 |
| T5 | −0.55 | 0.06 | −0.16 | −2.01 | −2.05 | 0.00 | 3.29 | −0.93 ** | −1.66 * | 0.00 | −0.23 * | 0.01 | −0.23 | −0.10 | −2.06 | 0.46 * |
| T6 | −0.44 | 0.08 | −0.11 | −1.30 | −1.71 | −0.01 | 11.91 * | −0.03 | −2.13 * | 0.04 | −0.14 | −0.5 ** | −0.95 * | −0.05 | −5.02 | 0.24 |
| T7 | −0.50 | −0.20 | 0.06 | −1.60 | −1.61 | −0.03 | 6.17 | −0.81 * | −0.46 | 0.03 | −0.23 * | 0.00 | 0.44 | −0.28 | −0.12 | −0.41 * |
| T8 | 0.43 | −0.01 | 0.10 | 1.15 | −0.18 | 0.00 | 17.17 ** | −0.70 * | −0.34 | 0.02 | −0.11 | 0.30 | −0.03 | 0.04 | 2.23 | 0.05 |
| T9 | 0.36 | 0.87 * | 0.02 | 4.06 ** | 4.78 ** | 0.00 | −19.19 ** | −0.80 * | −1.36 | 0.02 | −0.24 * | 0.06 | −0.31 | −0.02 | 2.89 | 0.01 |
| T10 | 0.43 | 0.24 | 0.01 | −0.22 | 0.58 | −0.02 | −11.6 * | 0.56 | 0.47 | −0.07 | 0.15 | 0.05 | −0.16 | 0.61 ** | 0.68 | 0.12 |
| T11 | 0.73 * | 1.11 ** | −0.02 | 3.40 | 6.74 ** | 0.00 | 36.47 ** | 0.60 | 2.48 ** | 0.00 | 0.12 | 0.24 | 0.94 * | 0.17 | 4.26 | 0.15 |
| SE± | 0.27 | 0.3 | 0.1 | 1.83 | 1.73 | 0.16 | 4.65 | 0.31 | 0.77 | 0.04 | 0.14 | 0.19 | 0.34 | 0.18 | 2.23 | 0.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadessa, K.; Beyene, Y.; Ifie, B.E.; Gowda, M.; Suresh, L.M.; Olsen, M.S.; Tongoona, P.; Offei, S.K.; Danquah, E.; Prasanna, B.M.; et al. Agronomic Performance and Resistance to Maize Lethal Necrosis in Maize Hybrids Derived from Doubled Haploid Lines. Agronomy 2024, 14, 2443. https://doi.org/10.3390/agronomy14102443
Sadessa K, Beyene Y, Ifie BE, Gowda M, Suresh LM, Olsen MS, Tongoona P, Offei SK, Danquah E, Prasanna BM, et al. Agronomic Performance and Resistance to Maize Lethal Necrosis in Maize Hybrids Derived from Doubled Haploid Lines. Agronomy. 2024; 14(10):2443. https://doi.org/10.3390/agronomy14102443
Chicago/Turabian StyleSadessa, Kassahun, Yoseph Beyene, Beatrice E. Ifie, Manje Gowda, Lingadahalli M. Suresh, Michael S. Olsen, Pangirayi Tongoona, Samuel K. Offei, Eric Danquah, Boddupalli M. Prasanna, and et al. 2024. "Agronomic Performance and Resistance to Maize Lethal Necrosis in Maize Hybrids Derived from Doubled Haploid Lines" Agronomy 14, no. 10: 2443. https://doi.org/10.3390/agronomy14102443
APA StyleSadessa, K., Beyene, Y., Ifie, B. E., Gowda, M., Suresh, L. M., Olsen, M. S., Tongoona, P., Offei, S. K., Danquah, E., Prasanna, B. M., & Wegary, D. (2024). Agronomic Performance and Resistance to Maize Lethal Necrosis in Maize Hybrids Derived from Doubled Haploid Lines. Agronomy, 14(10), 2443. https://doi.org/10.3390/agronomy14102443








