Combining Image-Based Phenotyping and Multivariate Analysis to Estimate Fruit Fresh Weight in Segregation Lines of Lowland Tomatoes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growing Conditions
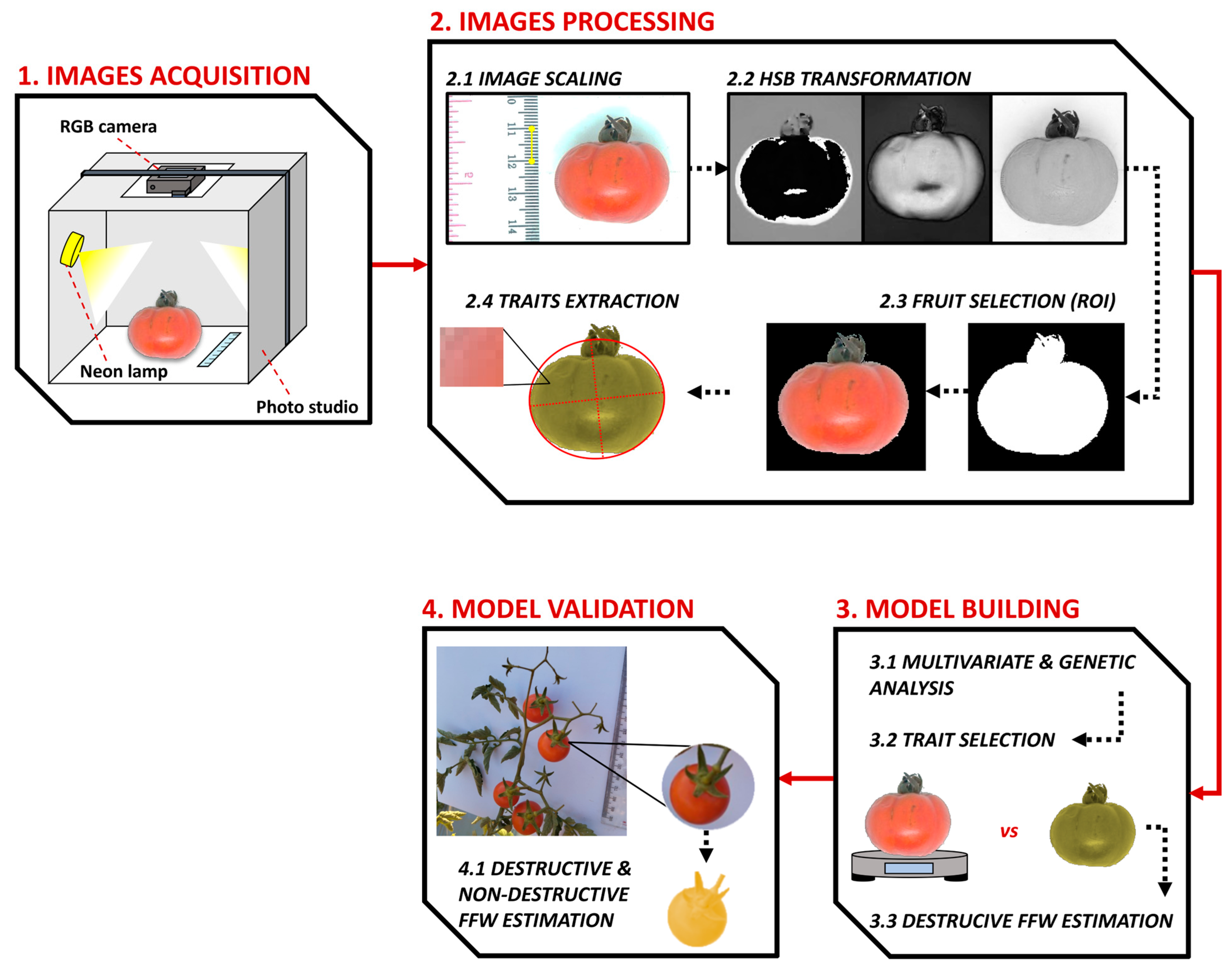
2.2. Fruits Imaging and Traits Extraction
2.3. Development of the Fruit Fresh Weight Estimation Model
2.3.1. Selection Intensity and Genetic Progress Percentage
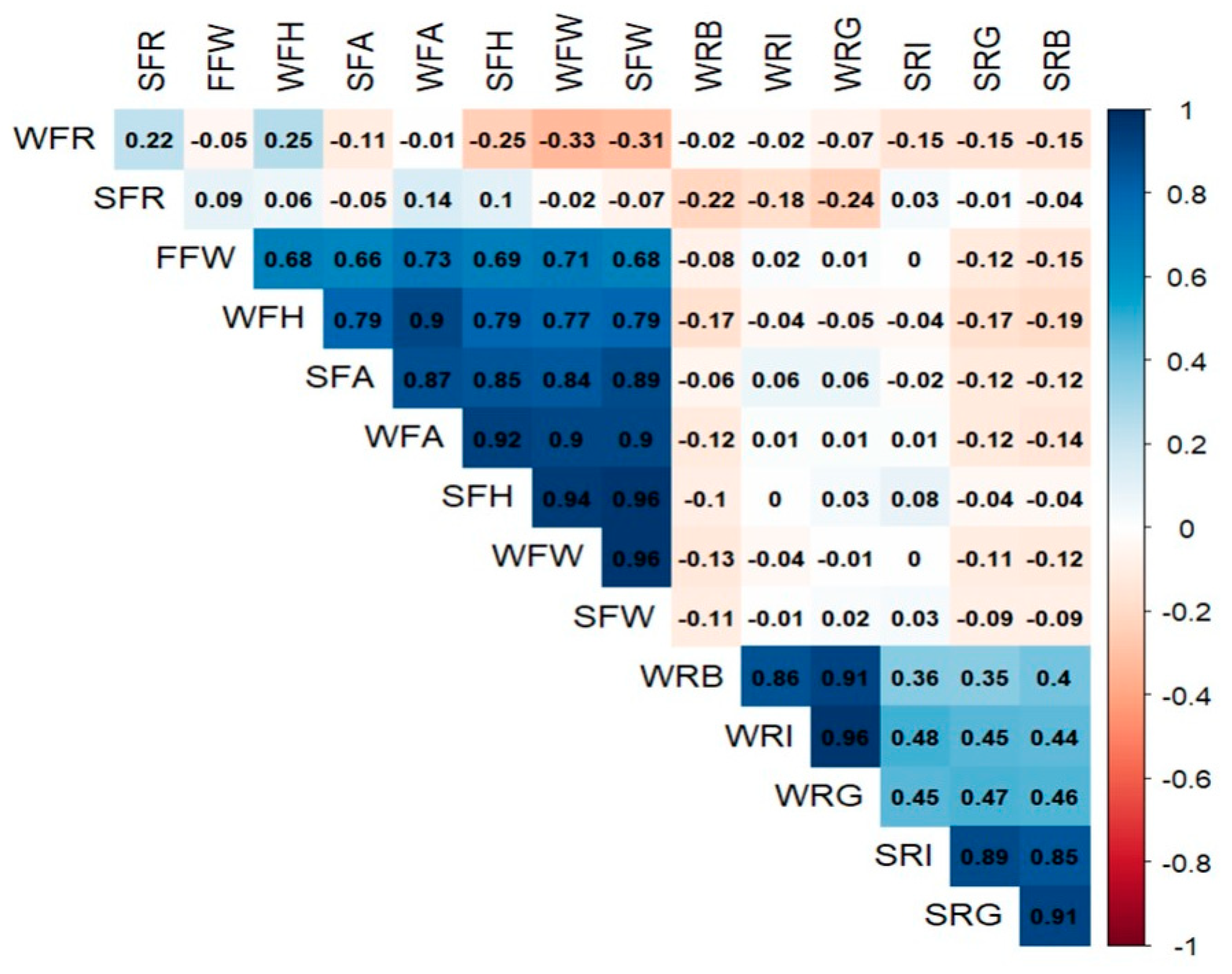
2.3.2. Effectiveness of Secondary Traits
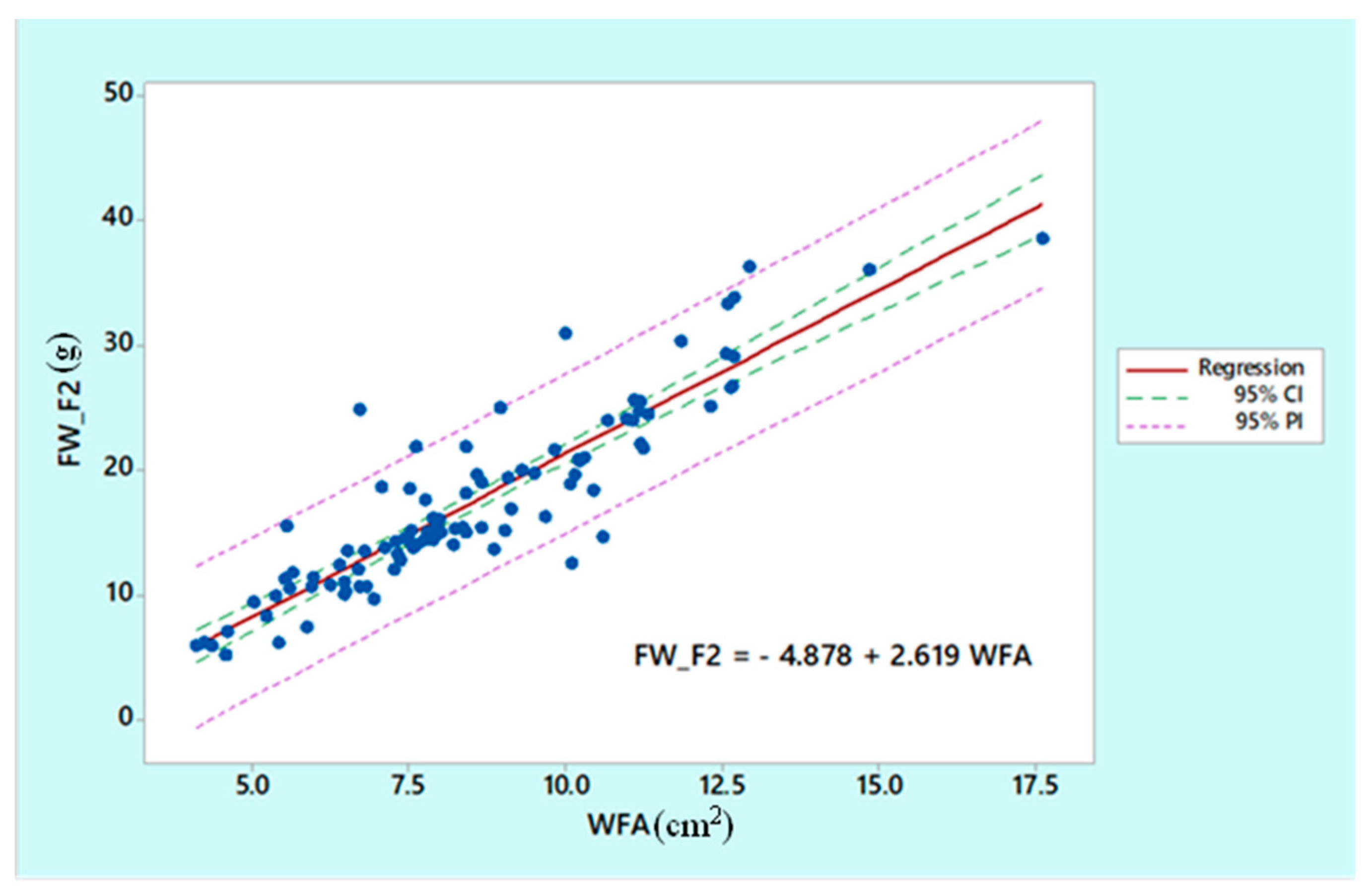
2.3.3. Model Sensitivity and Effectivness
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quinet, M.; Angosto, T.; Yuste-Lisbona, F.J.; Blanchard-Gros, R.; Bigot, S.; Martinez, J.P.; Lutts, S. Tomato Fruit Development and Metabolism. Front. Plant Sci. 2019, 10, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Martí, R.; Roselló, S.; Cebolla-Cornejo, J. Tomato as a Source of Carotenoids and Polyphenols Targeted to Cancer Prevention. Cancers 2016, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, H.; Zhang, Y.; Martin, C. Can the World’s Favorite Fruit, Tomato, Provide an Effective Biosynthetic Chassis for High-Value Metabolites? Plant Cell Rep. 2018, 37, 1443–1450. [Google Scholar] [CrossRef] [PubMed]
- Airoboman, F.A.; Onobhayedo, A.O. An Inquest into the Impacts of Population Pressure on the Natural Environment and Human Society. KIU J. Humanit. 2022, 7, 211–218. [Google Scholar]
- Rasheed, A.; Ilyas, M.; Khan, T.N.; Mahmood, A.; Riaz, U.; Chattha, M.B.; Al Kashgry, N.A.T.; Binothman, N.; Hassan, M.U.; Wu, Z.; et al. Study of Genetic Variability, Heritability, and Genetic Advance for Yield-Related Traits in Tomato (Solanum lycopersicon Mill.). Front. Genet. 2023, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mawasid, F.P.; Syukur, M. Trikoesoemaningtyas Epistatic Gene Control on the Yield of Tomato at Medium Elevation in the Tropical Agroecosystem. Biodiversitas 2019, 20, 1880–1886. [Google Scholar] [CrossRef]
- Skendžić, S.; Zovko, M.; Živković, I.P.; Lešić, V.; Lemić, D. The Impact of Climate Change on Agricultural Insect Pests. Insects 2021, 12, 440. [Google Scholar] [CrossRef]
- Schneider, P.; Asch, F. Rice Production and Food Security in Asian Mega Deltas—A Review on Characteristics, Vulnerabilities and Agricultural Adaptation Options to Cope with Climate Change. J. Agron. Crop Sci. 2020, 206, 491–503. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, M.; Song, Y. Abiotic and Biotic Stress Cascades in the Era of Climate Change Pose a Challenge to Genetic Improvements in Plants. Forests 2022, 13, 780. [Google Scholar] [CrossRef]
- Romadhon, M.R.; Sutjahjo, S.H.; Wirnas, D. Interaction Genetic x Environment Putatif Mutant Lines Tomato M5 on Two Agro Ecosystems. Int. J. Agron. Agric. Res. 2017, 10, 102–112. [Google Scholar]
- Yunandra, Y.; Deviona, D.; Zuhry, E.; Syukur, M.; Ardian, A.; Effendi, A.; Nurbaiti, N.; Yoseva, S.; Auliyanda, F.N. Selection Criteria for Lowland Tomatoes (Solanum lycopersicum L.). E3S Web Conf. 2023, 373, 03005. [Google Scholar] [CrossRef]
- Mustafa, M.; Syukur, M.; Sutjahjo, S.H. Sobir Inheritance Study for Fruit Characters of Tomato IPBT78 x IPBT73 Using Joint Scaling Test. IOP Conf. Ser. Earth Environ. Sci. 2019, 382, 012009. [Google Scholar] [CrossRef]
- Farid, M.; Anshori, M.F.; Ridwan, I. Tomato F3 Lines Development and Its Selection Index Based on Narrow-Sense Heritability and Factor Analysis. Biodiversitas 2022, 23, 5790–5797. [Google Scholar] [CrossRef]
- Fadhilah, A.N.; Farid, M.; Ridwan, I.; Anshori, M.F.; Yassi, A. Genetic Parameters and Selection Index of High-Yielding Tomato F2 Population. SABRAO J. Breed. Genet. 2022, 54, 1026–1036. [Google Scholar] [CrossRef]
- Farid, M.; Nasaruddin, N.; Musa, Y.; Anshori, M.F.; Ridwan, I.; Hendra, J.; Subroto, G. Genetic Parameters and Multivariate Analysis to Determine Secondary Traits in Selecting Wheat Mutant Adaptive on Tropical Lowlands. Plant Breed. Biotechnol. 2020, 8, 368–377. [Google Scholar] [CrossRef]
- Hernández-Bautista, A.; Lobato-Ortiz, R.; Cruz-Izquierdo, S.; García-Zavala, J.J.; Chávez-Servia, J.L.; Hernández-Leal, E.; Bonilla-Barrientos, O. Fruit Size QTLs Affect in a Major Proportion the Yield in Tomato. Chil. J. Agric. Res. 2015, 75, 402. [Google Scholar] [CrossRef]
- Cambiaso, V.; Gimenez, M.D.; Pereira da Costa, J.H.; Vazquez, D.V.; Picardi, L.A.; Pratta, G.R.; Rodríguez, G.R. Selected Genome Regions for Fruit Weight and Shelf Life in Tomato RILs Discernible by Markers Based on Genomic Sequence Information. Breed. Sci. 2019, 69, 447–454. [Google Scholar] [CrossRef]
- Ene, C.O.; Abtew, W.G.; Oselebe, H.O.; Ozi, F.U.; Ogah, O.; Okechukwu, E.C.; Chukwudi, U.P. Hybrid Vigor and Heritability Estimates in Tomato Crosses Involving Solanum lycopersicum × S. Pimpinellifolium under Cool Tropical Monsoon Climate. Int. J. Agron. 2023, 2023, 3003355. [Google Scholar] [CrossRef]
- Donoso, A.; Salazar, E. Yield Components and Development in Indeterminate Tomato Landraces: An Agromorphological Approach to Promoting Their Utilization. Agronomy 2023, 13, 434. [Google Scholar] [CrossRef]
- Skolik, P.; Morais, C.L.M.; Martin, F.L.; McAinsh, M.R. Determination of Developmental and Ripening Stages of Whole Tomato Fruit Using Portable Infrared Spectroscopy and Chemometrics. BMC Plant Biol. 2019, 19, 236. [Google Scholar] [CrossRef]
- da Silva, É.D.B.; Xavier, A.; Faria, M.V. Impact of Genomic Prediction Model, Selection Intensity, and Breeding Strategy on the Long-Term Genetic Gain and Genetic Erosion in Soybean Breeding. Front. Genet. 2021, 12, 637133. [Google Scholar] [CrossRef]
- Bucio, F.J.; Isaza, C.; Gonzalez, E.; De Paz, J.P.Z.; Sierra, J.A.R.; Rivera, E.K.A. Non-Destructive Post-Harvest Tomato Mass Estimation Model Based on Its Area via Computer Vision and Error Minimization Approaches. IEEE Access 2022, 10, 100247–100256. [Google Scholar] [CrossRef]
- Tolasa, M.; Gedamu, F.; Woldetsadik, K. Impacts of Harvesting Stages and Pre-Storage Treatments on Shelf Life and Quality of Tomato (Solanum lycopersicum L.). Cogent Food Agric. 2021, 7, 1863620. [Google Scholar] [CrossRef]
- Hairmansis, A.; Berger, B.; Tester, M.; Roy, S.J. Image-Based Phenotyping for Non-Destructive Screening of Different Salinity Tolerance Traits in Rice. Rice 2014, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Shi, R.; Pape, J.M.; Neumann, K.; Arend, D.; Graner, A.; Chen, M.; Klukas, C. Predicting Plant Biomass Accumulation from Image-Derived Parameters. Gigascience 2018, 7, 1–13. [Google Scholar] [CrossRef]
- Nyalala, I.; Okinda, C.; Chao, Q.; Mecha, P.; Korohou, T.; Yi, Z.; Nyalala, S.; Jiayu, Z.; Chao, L.; Kunjie, C. Weight and Volume Estimation of Single and Occluded Tomatoes Using Machine Vision. Int. J. Food Prop. 2021, 24, 818–832. [Google Scholar] [CrossRef]
- Anshori, M.F.; Farid, M.; Musa, Y.; Iswoyo, H.; Sakinah, A.I.; Arifuddin, M.; Laraswati, A.A. Development of Image-Based Phenotyping for Selection Characters of Rice Adaptability on the Seedling Salinity Screening. IOP Conf. Ser. Earth Environ. Sci. 2021, 807, 032022. [Google Scholar] [CrossRef]
- Al-Tamimi, N.; Langan, P.; Bernád, V.; Walsh, J.; Mangina, E.; Negrão, S. Capturing Crop Adaptation to Abiotic Stress Using Image-Based Technologies. Open Biol. 2022, 12, 210353. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.; Schurr, U.; Loreto, F.; Menesatti, P.; Carpentier, S. Plant Phenotyping Research Trends, a Science Mapping Approach. Front. Plant Sci. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, Q.; Huang, D. A Review of Imaging Techniques for Plant Phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, Camera, Action: High-Throughput Plant Phenotyping Is Ready for a Close-Up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef]
- Humplík, J.F.; Lazár, D.; Husičková, A.; Spíchal, L. Automated Phenotyping of Plant Shoots Using Imaging Methods for Analysis of Plant Stress Responses—A Review. Plant Methods 2015, 11, 29. [Google Scholar] [CrossRef]
- Araus, J.L.; Kefauver, S.C.; Zaman-Allah, M.; Olsen, M.S.; Cairns, J.E. Translating High-Throughput Phenotyping into Genetic Gain. Trends Plant Sci. 2018, 23, 451–466. [Google Scholar] [CrossRef] [PubMed]
- Jangra, S.; Chaudhary, V.; Yadav, R.C.; Yadav, N.R. High-Throughput Phenotyping: A Platform to Accelerate Crop Improvement. Phenomics 2021, 1, 31–53. [Google Scholar] [CrossRef]
- Anshori, M.F.; Dirpan, A.; Sitaresmi, T.; Rossi, R.; Farid, M.; Hairmansis, A.; Sapta Purwoko, B.; Suwarno, W.B.; Nugraha, Y. An Overview of Image-Based Phenotyping as an Adaptive 4.0 Technology for Studying Plant Abiotic Stress: A Bibliometric and Literature Review. Heliyon 2023, 9, e21650. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, H.; Zhou, J.; Fu, X.; Ye, H.; Nguyen, H.T. Development of an Automated Phenotyping Platform for Quantifying Soybean Dynamic Responses to Salinity Stress in Greenhouse Environment. Comput. Electron. Agric. 2018, 151, 319–330. [Google Scholar] [CrossRef]
- Laraswati, A.A.; Padjung, R.; Farid, M.; Nasaruddin, N.; Anshori, M.F.; Nur, A.; Sakinah, A.I. Image Based-Phenotyping and Selection Index Based on Multivariate Analysis for Rice Hydroponic Screening under Drought Stress. Plant Breed. Biotechnol. 2021, 9, 272–286. [Google Scholar] [CrossRef]
- Nyalala, I.; Okinda, C.; Nyalala, L.; Makange, N.; Chao, Q.; Chao, L.; Yousaf, K.; Chen, K. Tomato Volume and Mass Estimation Using Computer Vision and Machine Learning Algorithms: Cherry Tomato Model. J. Food Eng. 2019, 263, 288–298. [Google Scholar] [CrossRef]
- Quispe-Choque, G.; Rojas-Ledezma, S.; Maydana-Marca, A. Diversidad Morfológica de Fruto de Una Colección de Tomate (Solanum lycopersicum L.) Mediante Fenotipado Basado En Imágenes Digitales. J. Selva Andin. Res. Soc. 2022, 13, 51–68. [Google Scholar] [CrossRef]
- Zhu, Y.; Gu, Q.; Zhao, Y.; Wan, H.; Wang, R.; Zhang, X.; Cheng, Y. Quantitative Extraction and Evaluation of Tomato Fruit Phenotypes Based on Image Recognition. Front. Plant Sci. 2022, 13, 859290. [Google Scholar] [CrossRef]
- Rossi, R.; Costafreda-Aumedes, S.; Leolini, L.; Leolini, C.; Bindi, M.; Moriondo, M. Implementation of an Algorithm for Automated Phenotyping through Plant 3D-Modeling: A Practical Application on the Early Detection of Water Stress. Comput. Electron. Agric. 2022, 197, 106937. [Google Scholar] [CrossRef]
- Nankar, A.N.; Tringovska, I.; Grozeva, S.; Ganeva, D.; Kostova, D. Tomato Phenotypic Diversity Determined by Combined Approaches of Conventional and High-Throughput Tomato Analyzer Phenotyping. Plants 2020, 9, 197. [Google Scholar] [CrossRef] [PubMed]
- Taheri-Garavand, A.; Rafiee, S.; Keyhani, A. Study on Some Morphological and Physical Characteristics of Tomato Used in Mass Models to Characterize Best Post Harvesting Options. Aust. J. Crop Sci. 2011, 5, 433–438. [Google Scholar]
- de Luna, R.G.; Dadios, E.P.; Bandala, A.A.; Vicerra, R.R.P. Size Classification of Tomato Fruit Using Thresholding, Machine Learning and Deep Learning Techniques. Agrivita 2019, 41, 586–596. [Google Scholar] [CrossRef]
- Mahfud, M.; Murti, R.H. Inheritance Pattern of Fruit Color and Shape in Multi-Pistil and Purple Tomato Crossing. Agrivita 2020, 42, 572–583. [Google Scholar] [CrossRef]
- Bojarian, M.; Asadi-Gharneh, H.A.; Golabadi, M. Factor Analysis, Stepwise Regression and Path Coefficient Analyses of Yield, Yield-Associated Traits, and Fruit Quality in Tomato. Int. J. Veg. Sci. 2019, 25, 542–553. [Google Scholar] [CrossRef]
- Alsabah, R.; Purwoko, B.S.; Dewi, I.S.; Wahyu, Y. Selection Index for Selecting Promising Doubled Haploid Lines of Black Rice. Sabrao J. Breed. Genet. 2019, 51, 420–441. [Google Scholar]
- Mousavi, S.M.N.; Nagy, J. Evaluation of Plant Characteristics Related to Grain Yield of FAO410 and FAO340 Hybrids Using Regression Models. Cereal Res. Commun. 2021, 49, 161–169. [Google Scholar] [CrossRef]
- Padjung, R.; Farid, M.; Musa, Y.; Anshori, M.F.; Nur, A.; Masnenong, A. Drought-Adapted Maize Line Based on Morphophysiological Selection Index. Biodiversitas 2021, 22, 4028–4035. [Google Scholar] [CrossRef]
- Yeater, K.M.; Duke, S.E.; Riedell, W.E. Multivariate Analysis: Greater Insights into Complex Systems. Agron. J. 2015, 107, 799–810. [Google Scholar] [CrossRef]
- Barth, E.; de Resende, J.T.V.; Mariguele, K.H.; de Resende, M.D.V.; da Silva, A.L.B.R.; Ru, S. Multivariate Analysis Methods Improve the Selection of Strawberry Genotypes with Low Cold Requirement. Sci. Rep. 2022, 12, 11458. [Google Scholar] [CrossRef]
- Sakinah, A.I.; Musa, Y.; Farid, M.; Hairmansis, A.; Anshori, M.F.; Nasaruddin, N. Rice Selection Criteria Based on Morphological And Image-Based Phenotyping Under Drought-And Salinity-Stress Conditions. Sabrao J. Breed. Genet. 2022, 54, 686–699. [Google Scholar] [CrossRef]
- Duc, N.T.; Ramlal, A.; Rajendran, A.; Raju, D.; Lal, S.K.; Kumar, S.; Sahoo, R.N.; Chinnusamy, V. Image-Based Phenotyping of Seed Architectural Traits and Prediction of Seed Weight Using Machine Learning Models in Soybean. Front. Plant Sci. 2023, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Woolf, M.S.; Dignan, L.M.; Scott, A.T.; Landers, J.P. Digital Postprocessing and Image Segmentation for Objective Analysis of Colorimetric Reactions. Nat. Protoc. 2021, 16, 218–238. [Google Scholar] [CrossRef]
- Ayenan, M.A.T.; Danquah, A.; Ampomah-Dwamena, C.; Hanson, P.; Asante, I.K.; Danquah, E.Y. Optimizing Pollencounter for High Throughput Phenotyping of Pollen Quality in Tomatoes. MethodsX 2020, 7, 100977. [Google Scholar] [CrossRef]
- Barraj Barraj, R.; Segado, P.; Moreno-González, R.; Heredia, A.; Fernández-Muñoz, R.; Domínguez, E. Genome-Wide QTL Analysis of Tomato Fruit Cuticle Deposition and Composition. Hortic. Res. 2021, 8, 113. [Google Scholar] [CrossRef] [PubMed]
- Acquaah, G. Principles of Plant Genetics and Breeding; Blackwell Publishing: Oxford, UK, 2007; ISBN 1405136464. [Google Scholar]
- Anshori, M.F.; Purwoko, B.S.; Dewi, I.S.; Ardie, S.W.; Suwarno, W.B.; Safitri, H. Determination of Selection Criteria for Screening of Rice Genotypes for Salinity Tolerance. Sabrao J. Breed. Genet. 2018, 50, 279–294. [Google Scholar]
- Reflinur; Kim, B.; Jang, S.M.; Chu, S.H.; Bordiya, Y.; Akter, M.B.; Lee, J.; Chin, J.H.; Koh, H.J. Analysis of Segregation Distortion and Its Relationship to Hybrid Barriers in Rice. Rice 2014, 7, 3. [Google Scholar] [CrossRef]
- Nie, X.; Tu, J.; Wang, B.; Zhou, X.; Lin, Z. A BIL Population Derived from G. Hirsutum and G. Barbadense Provides a Resource for Cotton Genetics and Breeding. PLoS ONE 2015, 10, e0141064. [Google Scholar] [CrossRef]
- Dos Santos, V.O.; Viana, A.P.; da Costa Preisigke, S.; Santos, E.A. Backcrosses in a Segregating Population of Passiflora Mediated by Morphoagronomic and Resistance Traits. Bragantia 2019, 78, 542–552. [Google Scholar] [CrossRef]
- Merrick, L.F.; Herr, A.W.; Sandhu, K.S.; Lozada, D.N.; Carter, A.H. Optimizing Plant Breeding Programs for Genomic Selection. Agronomy 2022, 12, 714. [Google Scholar] [CrossRef]
- Schmidt, P.; Hartung, J.; Bennewitz, J.; Hans-Peter, P. Heritability in Plant Breeding on a Genotype-Difference Basis. Genetics 2019, 212, 991–1008. [Google Scholar] [CrossRef]
- Tesfaye, A. Genetic Variability, Heritability, and Genetic Advance Estimates in Garlic (Allium sativum) from the Gamo Highlands of Southern Ethiopia. Int. J. Agron. 2021, 2021, 3171642. [Google Scholar] [CrossRef]
- Diniz, R.P.; de Oliveira, E.J. Genetic Parameters, Path Analysis and Indirect Selection of Agronomic Traits of Cassava germplasm. An. Acad. Bras. Cienc. 2019, 91, 1–11. [Google Scholar] [CrossRef]
- Lozada, D.N.; Godoy, J.V.; Ward, B.P.; Carter, A.H. Genomic Prediction and Indirect Selection for Grain Yield in US Pacific Northwest Winter Wheat Using Spectral Reflectance Indices from High-Throughput Phenotyping. Int. J. Mol. Sci. 2020, 21, 165. [Google Scholar] [CrossRef]
- Pham, H. A New Criterion for Model Selection. Mathematics 2019, 7, 1215. [Google Scholar] [CrossRef]
- Chicco, D.; Warrens, M.J.; Jurman, G. The Coefficient of Determination R-Squared Is More Informative than SMAPE, MAE, MAPE, MSE and RMSE in Regression Analysis Evaluation. PeerJ Comput. Sci. 2021, 7, e623. [Google Scholar] [CrossRef] [PubMed]
- Schauer, N.; Semel, Y.; Balbo, I.; Steinfath, M.; Repsilber, D.; Selbig, J.; Pleban, T.; Zamir, D.; Fernie, A.R. Mode of Inheritance of Primary Metabolic Traits in Tomato. Plant Cell 2008, 20, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Acquaah, G. Principles of Plant Genetics and Breeding, 2nd ed.; Wiley: Oxford, UK, 2012; ISBN 9780470664766. [Google Scholar]
- Oliveira, G.H.; Amaral, C.B.; Silva, F.A.; Dutra, S.M.; Marconato, M.B.; Môro, G. V Mixed Models and Multivariate Analysis for Selection of Superior Maize Genotypes. Chil. J. Agric. Res. 2016, 76, 427–431. [Google Scholar] [CrossRef]
- Nardino, M.; Barros, W.S.; Olivoto, T.; Cruz, C.D.; Silva, F.F.E.; De Pelegrin, A.J.; De Souza, V.Q.; Carvalho, I.R.; Szareski, V.J.; De Oliveira, A.C.; et al. Multivariate Diallel Analysis by Factor Analysis for Establish Mega-Traits. An. Acad. Bras. Cienc. 2020, 92. [Google Scholar] [CrossRef] [PubMed]
- Mokhtar, A.; El-Ssawy, W.; He, H.; Al-Anasari, N.; Sammen, S.S.; Gyasi-Agyei, Y.; Abuarab, M. Using Machine Learning Models to Predict Hydroponically Grown Lettuce Yield. Front. Plant Sci. 2022, 13, 706042. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.-L.; Bao, W.-K. Statistical Analysis of Leaf Water Use Efficiency and Physiology Traits of Winter Wheat Under Drought Condition. J. Integr. Agric. 2012, 11, 82–89. [Google Scholar] [CrossRef]
- Bahmani, K.; Izadi-Darbandi, A.; Noori, S.A.S.; Jafari, A.A.; Moradi, N. Determination of Interrelationships among Phenotypic Traits of Iranian Fennel (Foeniculum vulgare Mill.) Using Correlation, Stepwise Regression and Path Analyses. J. Essent. Oil Bear. Plants 2012, 15, 424–444. [Google Scholar] [CrossRef]
- Rameeh, V. Multivariate Regression Analyses of Yield Associated Traits in Rapeseed (Brassica napus L.) Genotypes. Adv. Agric. 2014, 2014, 626434. [Google Scholar] [CrossRef]
- Smith, L.N.; Zhang, W.; Hansen, M.F.; Hales, I.J.; Smith, M.L. Innovative 3D and 2D Machine Vision Methods for Analysis of Plants and Crops in the Field. Comput. Ind. 2018, 97, 122–131. [Google Scholar] [CrossRef]
- Khan, M.M.H.; Rafii, M.Y.; Ramlee, S.I.; Jusoh, M.; Al Mamun, M. Path-Coefficient and Correlation Analysis in Bambara Groundnut (Vigna subterranea [L.] Verdc.) Accessions over Environments. Sci. Rep. 2022, 12, 245. [Google Scholar] [CrossRef]
- Olivoto, T.; de Souza, V.Q.; Nardino, M.; Carvalho, I.R.; Ferrari, M.; de Pelegrin, A.J.; Szareski, V.J.; Schmidt, D. Multicollinearity in Path Analysis: A Simple Method to Reduce Its Effects. Agron. J. 2017, 109, 131–142. [Google Scholar] [CrossRef]
- Del Conte, M.V.; Souza Carneiro, P.C.; De Resende, M.D.V.; Da Silva, F.L.; Peternelli, L.A. Overcoming Collinearity in Path Analysis of Soybean [Glycine max (L.) Merr.] Grain Oil Content. PLoS ONE 2020, 15, e0233290. [Google Scholar] [CrossRef]
- Patil, P.G.; Bohra, A.; Satheesh, N.S.J.; Dubey, J.; Pandey, P.; Dutta, D.; Singh, F.; Singh, I.P.; Singh, N.P. Validation of QTLs for Plant Ideotype, Earliness and Growth Habit Traits in Pigeonpea (Cajanus cajan Millsp.). Physiol. Mol. Biol. Plants 2018, 24, 1245–1259. [Google Scholar] [CrossRef]
- de Jesus Colwell, F.; Souter, J.; Bryan, G.J.; Compton, L.J.; Boonham, N.; Prashar, A. Development and Validation of Methodology for Estimating Potato Canopy Structure for Field Crop Phenotyping and Improved Breeding. Front. Plant Sci. 2021, 12, 612843. [Google Scholar] [CrossRef]
- Rahman, M.M.; Crain, J.; Haghighattalab, A.; Singh, R.P.; Poland, J. Improving Wheat Yield Prediction Using Secondary Traits and High-Density Phenotyping Under Heat-Stressed Environments. Front. Plant Sci. 2021, 12, 633651. [Google Scholar] [CrossRef] [PubMed]
- Ali, Y.; Raza, A.; Iqbal, S.; Khan, A.A.; Aatif, H.M.; Hassan, Z.; Hanif, C.M.S.; Ali, H.M.; Mosa, W.F.A.; Mubeen, I.; et al. Stepwise Regression Models-Based Prediction for Leaf Rust Severity and Yield Loss in Wheat. Sustainability 2022, 14, 13893. [Google Scholar] [CrossRef]
- Cobb, J.N.; Juma, R.U.; Biswas, P.S.; Arbelaez, J.D.; Rutkoski, J.; Atlin, G.; Hagen, T.; Quinn, M.; Ng, E.H. Enhancing the Rate of Genetic Gain in Public-Sector Plant Breeding Programs: Lessons from the Breeder’s Equation. Theor. Appl. Genet. 2019, 132, 627–645. [Google Scholar] [CrossRef] [PubMed]
- da Silva, T.R.; Costa, A.G.; Paes, J.L.; de Oliveira, M.V.M.; de Assis de Carvalho Pinto, F. Comparing Quality Parameters Obtained Using Destructive and Optical Methods in Grading Tomatoes. Rev. Cienc. Agron. 2021, 52, e20207475. [Google Scholar] [CrossRef]
- Zaki, H.E.M.; Radwan, K.S.A. Estimates of Genotypic and Phenotypic Variance, Heritability, and Genetic Advance of Horticultural Traits in Developed Crosses of Cowpea (Vigna unguiculata [L.] Walp). Front. Plant Sci. 2022, 13, 987985. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.A.; Rebetzke, G.J. Indirect Selection for Potential Yield in Early-Generation, Spaced Plantings of Wheat and Other Small-Grain Cereals: A Review. Crop Pasture Sci. 2018, 69, 439–459. [Google Scholar] [CrossRef]
- Del Medico, A.P.; Cabodevila, V.G.; Vitelleschi, M.S.; Pratta, G.R. Characterization of Tomato Generations According to a Three-Way Data Analysis. Bragantia 2020, 79, 8–18. [Google Scholar] [CrossRef]
- Mata-Nicolás, E.; Montero-Pau, J.; Gimeno-Paez, E.; Garcia-Carpintero, V.; Ziarsolo, P.; Menda, N.; Mueller, L.A.; Blanca, J.; Cañizares, J.; van der Knaap, E.; et al. Exploiting the Diversity of Tomato: The Development of a Phenotypically and Genetically Detailed Germplasm Collection. Hortic. Res. 2020, 7, 66. [Google Scholar] [CrossRef]
- Tausen, M.; Clausen, M.; Moeskjær, S.; Shihavuddin, A.S.M.; Dahl, A.B.; Janss, L.; Andersen, S.U. Greenotyper: Image-Based Plant Phenotyping Using Distributed Computing and Deep Learning. Front. Plant Sci. 2020, 11, 01181. [Google Scholar] [CrossRef]
- Hui, F.; Xie, Z.W.; Li, H.G.; Guo, Y.; Li, B.G.; Liu, Y.L.; Ma, Y.T. Image-Based Root Phenotyping for Field-Grown Crops: An Example under Maize/Soybean Intercropping. J. Integr. Agric. 2022, 21, 1606–1619. [Google Scholar] [CrossRef]
- Shrestha, S.; Deleuran, L.C.; Olesen, M.H.; Gislum, R. Use of Multispectral Imaging in Varietal Identification of Tomato. Sensors 2015, 15, 4496–4512. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Vinson, R.; Holmes, M.; Seibel, G.; Bechar, A.; Nof, S.; Tao, Y. Early Detection of Tomato Spotted Wilt Virus by Hyperspectral Imaging and Outlier Removal Auxiliary Classifier Generative Adversarial Nets (OR-AC-GAN). Sci. Rep. 2019, 9, 4377. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hu, R.; Zhang, M.; Zhai, Z.; Zhang, R. Identification of Tomatoes with Early Decay Using Visible and near Infrared Hyperspectral Imaging and Image-Spectrum Merging Technique. J. Food Process Eng. 2021, 44, e13654. [Google Scholar] [CrossRef]


| GB | S | FFW | WFA | WFR | WFW | WFH | WRI | WGI | WBI | SFA | SFR | SFW | SFH | SRI | SGI | SBI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K | 20.71 | 9.33 | 0.89 | 3.53 | 3.39 | 125.72 | 44.43 | 33.87 | 9.84 | 0.94 | 3.60 | 3.43 | 118.87 | 47.26 | 29.62 | |
| 5.71 | 1.98 | 0.04 | 0.48 | 0.27 | 26.35 | 14.59 | 11.74 | 2.69 | 0.02 | 0.54 | 0.46 | 25.16 | 15.86 | 12.59 | ||
| M | 18.26 | 8.90 | 0.71 | 3.94 | 2.97 | 114.24 | 44.09 | 30.24 | 6.95 | 0.92 | 3.94 | 3.75 | 114.24 | 44.09 | 30.24 | |
| 5.98 | 2.66 | 0.11 | 0.49 | 0.51 | 12.27 | 9.97 | 5.89 | 1.79 | 0.04 | 0.49 | 0.36 | 12.27 | 9.97 | 5.89 | ||
| F1 | 19.91 | 9.90 | 0.94 | 3.56 | 3.54 | 115.73 | 44.37 | 32.32 | 6.54 | 0.93 | 3.74 | 3.61 | 128.91 | 60.58 | 38.38 | |
| 3.91 | 0.96 | 0.02 | 0.25 | 0.16 | 27.34 | 14.96 | 12.23 | 0.88 | 0.02 | 0.33 | 0.22 | 25.48 | 24.45 | 11.79 | ||
| F2 | 17.32 | 8.49 | 0.84 | 3.56 | 3.12 | 114.35 | 40.49 | 31.75 | 5.93 | 0.93 | 3.53 | 3.41 | 121.95 | 50.99 | 33.25 | |
| 7.93 | 2.98 | 0.08 | 0.68 | 0.56 | 22.54 | 15.83 | 14.63 | 2.43 | 0.03 | 0.64 | 0.64 | 22.95 | 18.96 | 12.50 | ||
| VE | 5.20 | 1.87 | 0.06 | 0.41 | 0.31 | 21.99 | 13.17 | 9.95 | 1.79 | 0.03 | 0.45 | 0.35 | 20.97 | 16.76 | 10.09 | |
| VP | 7.93 | 2.98 | 0.08 | 0.68 | 0.56 | 22.54 | 15.83 | 14.63 | 2.43 | 0.03 | 0.64 | 0.64 | 22.95 | 18.96 | 12.50 | |
| VG | 2.73 | 1.12 | 0.02 | 0.27 | 0.24 | 0.55 | 2.66 | 4.68 | 0.64 | 0.01 | 0.19 | 0.29 | 1.98 | 2.20 | 2.41 | |
| H | 34.44 | 37.47 | 27.79 | 39.69 | 43.73 | 2.44 | 16.82 | 31.96 | 26.39 | 18.86 | 29.76 | 45.64 | 8.62 | 11.62 | 19.27 | |
| Trait | Direct Effect | Indirect Effect | Correlation | |||||
|---|---|---|---|---|---|---|---|---|
| WFA | WFW | WFH | SFA | SFW | SFH | |||
| WFA | 0.21 | 0.54 | 0.19 | 0.13 | −0.44 | 0.1 | 0.73 | |
| WFW | 0.60 | 0.19 | 0.16 | 0.13 | −0.47 | 0.1 | 0.71 | |
| WFH | 0.21 | 0.19 | 0.47 | 0.12 | −0.39 | 0.08 | 0.68 | |
| SFA | 0.15 | 0.18 | 0.51 | 0.17 | −0.44 | 0.09 | 0.66 | |
| SFW | −0.49 | 0.19 | 0.58 | 0.17 | 0.13 | 0.1 | 0.68 | |
| SFH | 0.11 | 0.19 | 0.57 | 0.17 | 0.13 | −0.47 | 0.69 | |
| total | 0.94 | 2.67 | 0.86 | 0.63 | −2.21 | 0.47 | ||
| Parameters | Estimate | Std. Error | t Value | Pr (>|t|) |
|---|---|---|---|---|
| (Intercept) | −4.9456 | 3.7527 | −1.318 | 0.19071 |
| WFA | 1.2911 | 0.4217 | 3.062 | 0.00286 ** |
| WFW | 3.2015 | 1.8520 | 1.729 | 0.08711 |
| Grouping | Traits | SG | S | i | (Cr/Rx) | ||
|---|---|---|---|---|---|---|---|
| Independent | WFW | 4.45 | 0.90 | 1.81 | 0.57 | 15.90 | |
| WFA | 12.56 | 4.06 | 2.35 | 2.49 | 29.29 | ||
| FFW | 28.85 | 11.53 | 4.09 | 6.76 | 39.05 | ||
| Dependent | WFA | 12.56 | 2.65 | 2.70 | 0.99 | 10.03 | 0.68 |
| FFW | 27.16 | 7.25 | 3.66 | 2.50 | 12.53 | ||
| WFW | 4.45 | 0.89 | 1.09 | 0.35 | 9.95 | 0.32 | |
| FFW | 26.89 | 6.98 | 3.53 | 2.40 | 12.06 |
| Data | n | p-Value | R2 | Adjusted-R2 | R2-Deviation | RMSE |
|---|---|---|---|---|---|---|
| Training | 96 | 0.000 | 81.5 | 81.3 | 0.2 | 3.14 |
| Destructive Validation | 30 | 0.000 | 70.7 | 69.8 | 0.9 | 4.46 |
| Non-destructive Validation | 45 | 0.000 | 80.7 | 80.2 | 0.5 | 2.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farid, M.; Anshori, M.F.; Rossi, R.; Haring, F.; Mantja, K.; Dirpan, A.; Larekeng, S.H.; Mustafa, M.; Adnan, A.; Tahara, S.A.M.; et al. Combining Image-Based Phenotyping and Multivariate Analysis to Estimate Fruit Fresh Weight in Segregation Lines of Lowland Tomatoes. Agronomy 2024, 14, 338. https://doi.org/10.3390/agronomy14020338
Farid M, Anshori MF, Rossi R, Haring F, Mantja K, Dirpan A, Larekeng SH, Mustafa M, Adnan A, Tahara SAM, et al. Combining Image-Based Phenotyping and Multivariate Analysis to Estimate Fruit Fresh Weight in Segregation Lines of Lowland Tomatoes. Agronomy. 2024; 14(2):338. https://doi.org/10.3390/agronomy14020338
Chicago/Turabian StyleFarid, Muh, Muhammad Fuad Anshori, Riccardo Rossi, Feranita Haring, Katriani Mantja, Andi Dirpan, Siti Halimah Larekeng, Marlina Mustafa, Adnan Adnan, Siti Antara Maedhani Tahara, and et al. 2024. "Combining Image-Based Phenotyping and Multivariate Analysis to Estimate Fruit Fresh Weight in Segregation Lines of Lowland Tomatoes" Agronomy 14, no. 2: 338. https://doi.org/10.3390/agronomy14020338
APA StyleFarid, M., Anshori, M. F., Rossi, R., Haring, F., Mantja, K., Dirpan, A., Larekeng, S. H., Mustafa, M., Adnan, A., Tahara, S. A. M., Amier, N., Amal, M. A. I., & Sakinah, A. I. (2024). Combining Image-Based Phenotyping and Multivariate Analysis to Estimate Fruit Fresh Weight in Segregation Lines of Lowland Tomatoes. Agronomy, 14(2), 338. https://doi.org/10.3390/agronomy14020338









