The Phenotypic Diversity of 232 Germplasm Accessions Identifies the Adverse Effects of Flowering Redundancy on Peanut Yield
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Field Trials
2.2. Phenotypic Data Collection
2.2.1. Flowering Time Traits
2.2.2. Yield and Its Related Traits
2.3. Data Analysis
2.3.1. Descriptive Statistics Analyses and Shannon–Weaver Index
2.3.2. Phenotypic Variation Partitioning and Heritability Estimation
2.3.3. Correlation Analysis and Principal Component Analysis (PCA)
2.3.4. Clustering Analysis
3. Results
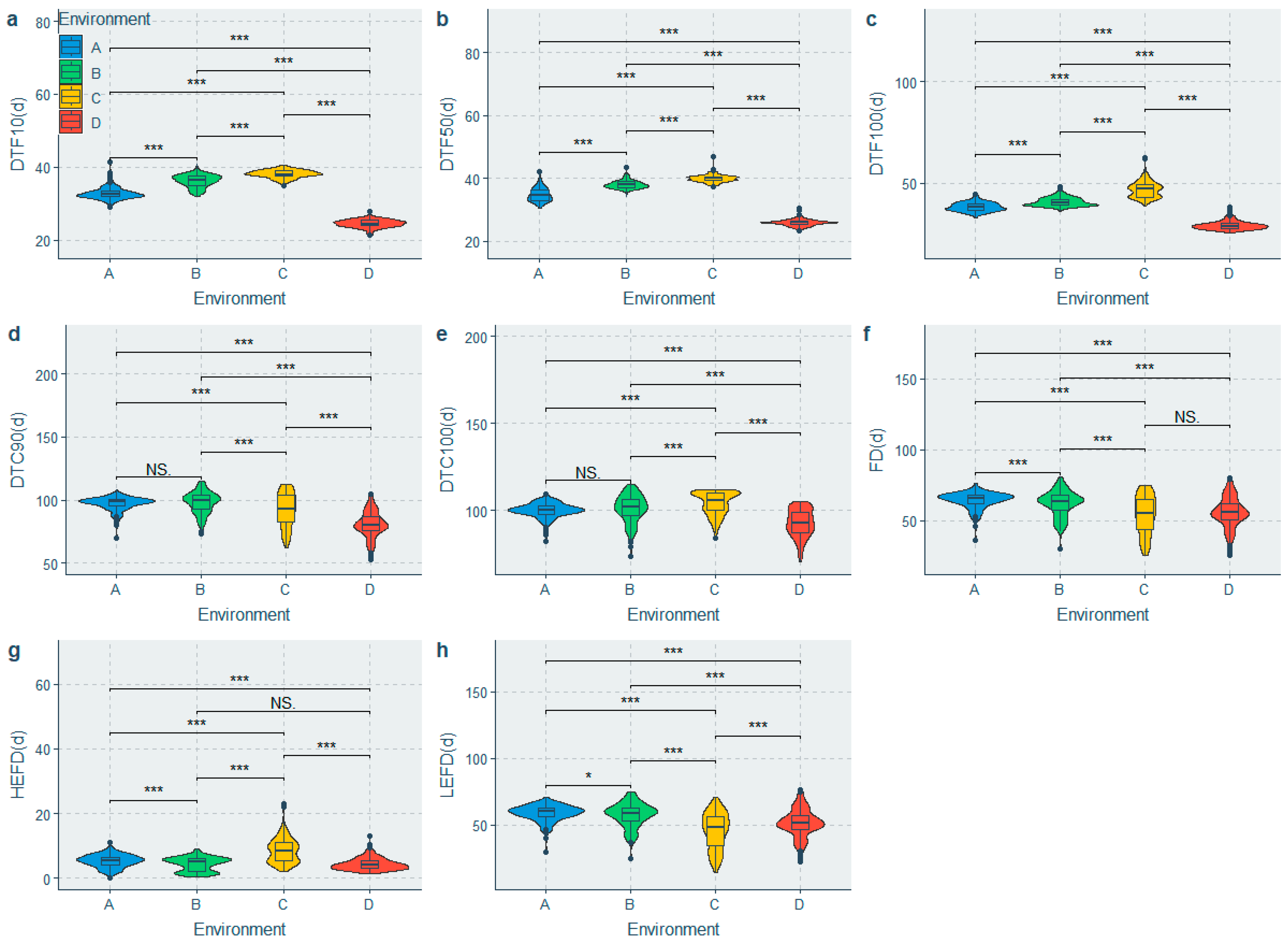
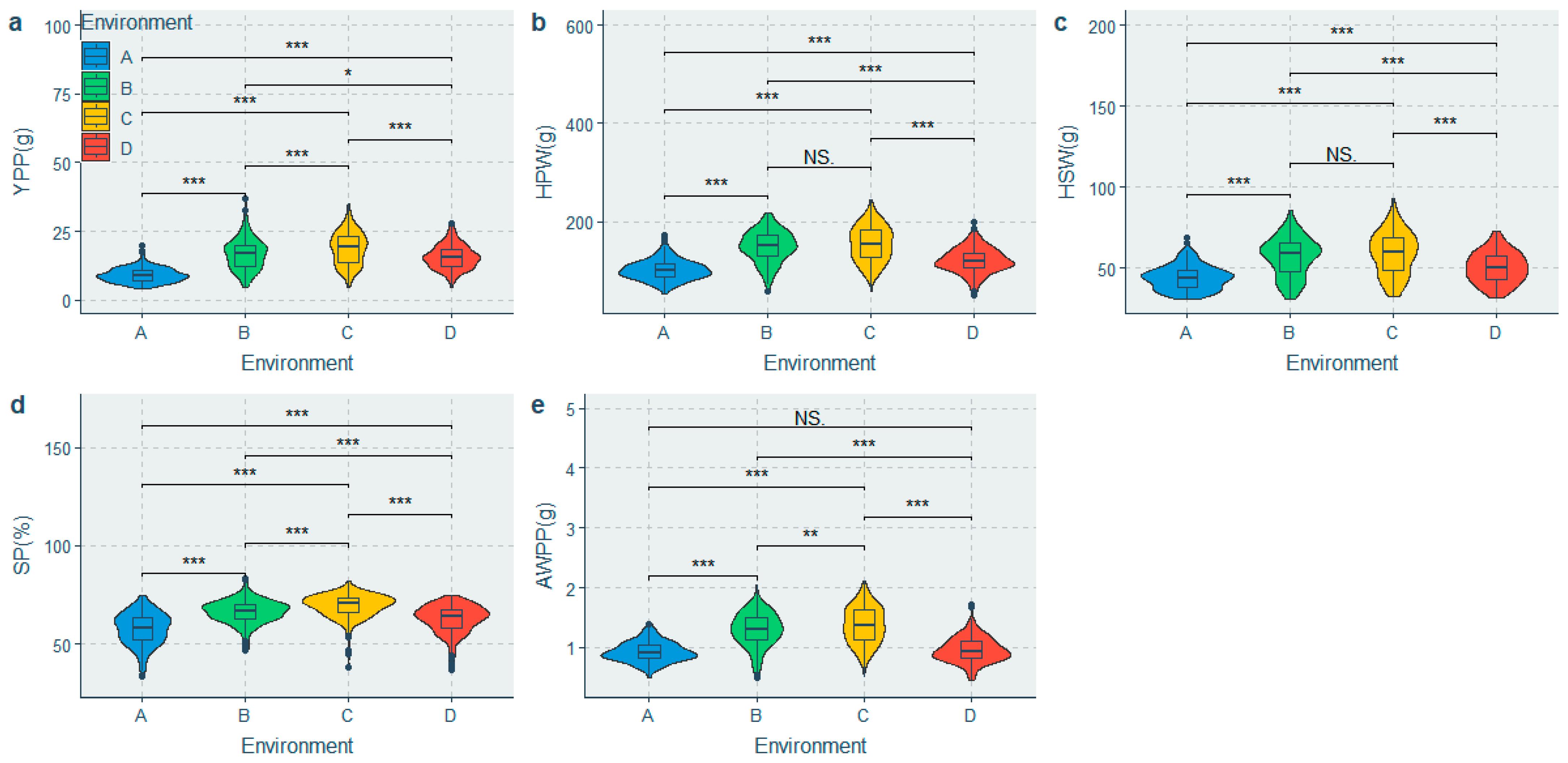
3.1. Phenotypic Diversity Analysis of Flowering and Yield-Related Traits
3.2. Phenotypic Variation Partitioning and Heritability Estimation
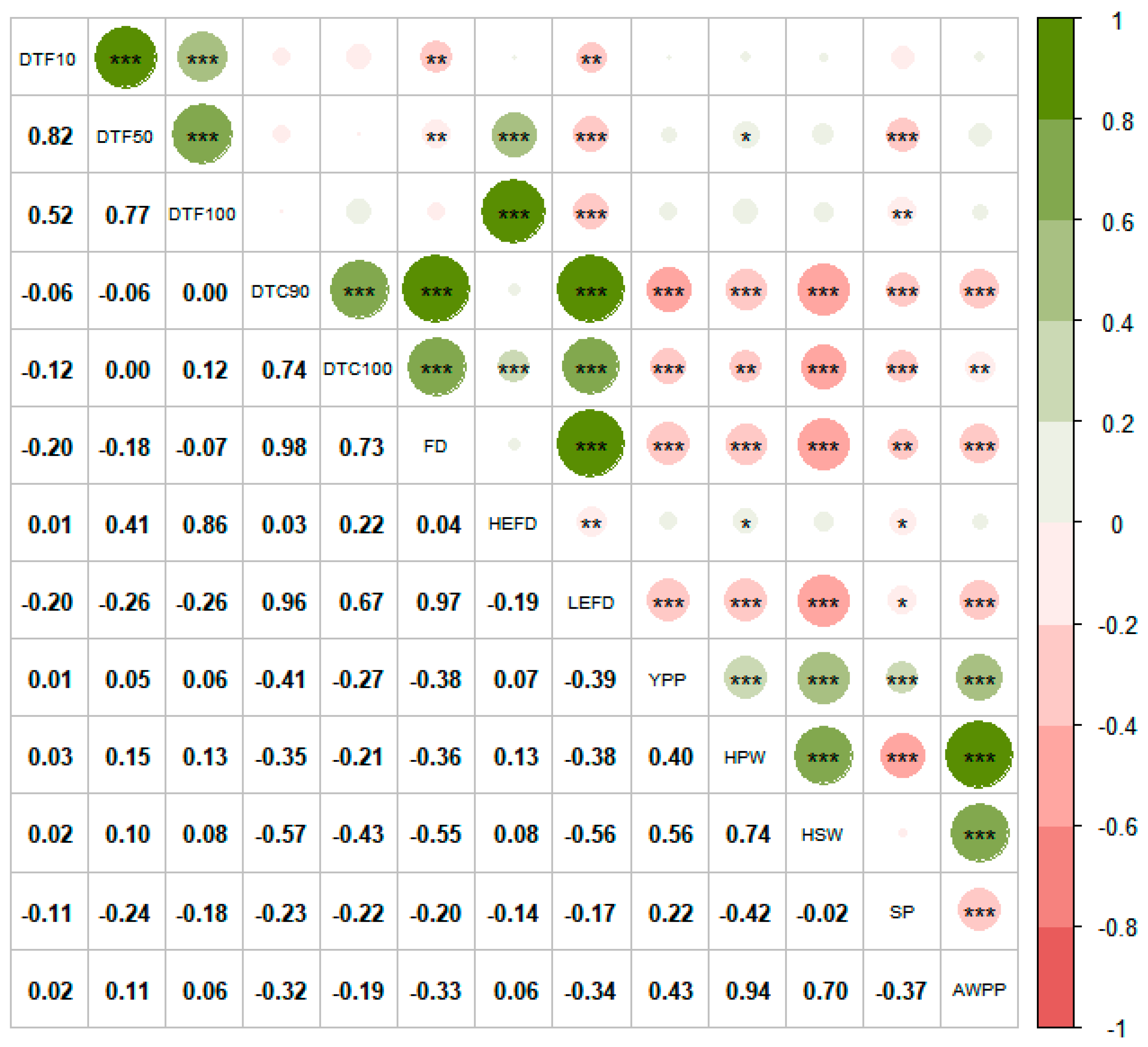
3.3. Correlation Analysis between Flowering Time and Yield—Related Traits
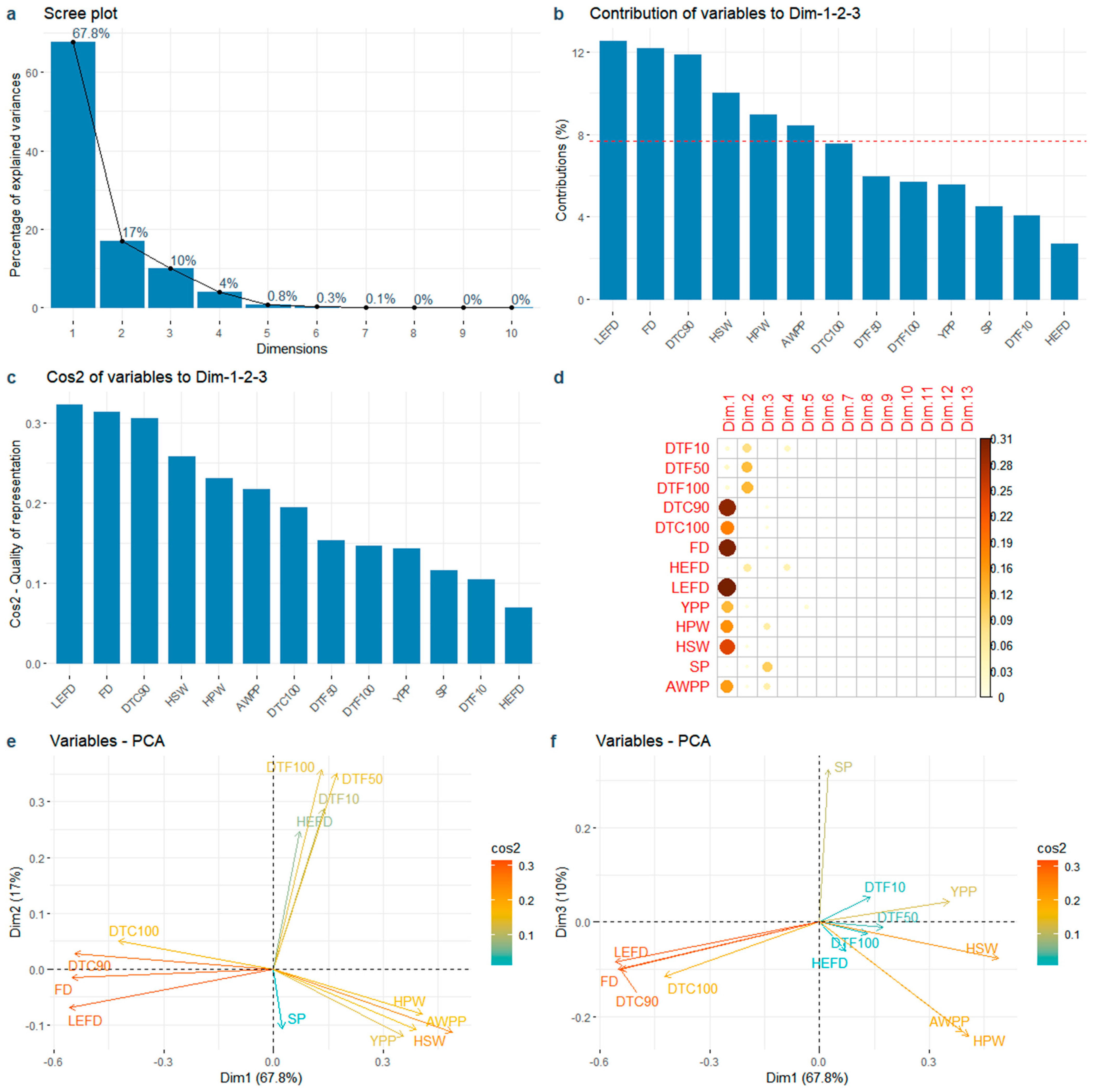
3.4. PCA of Flowering Time and Yield Related Traits
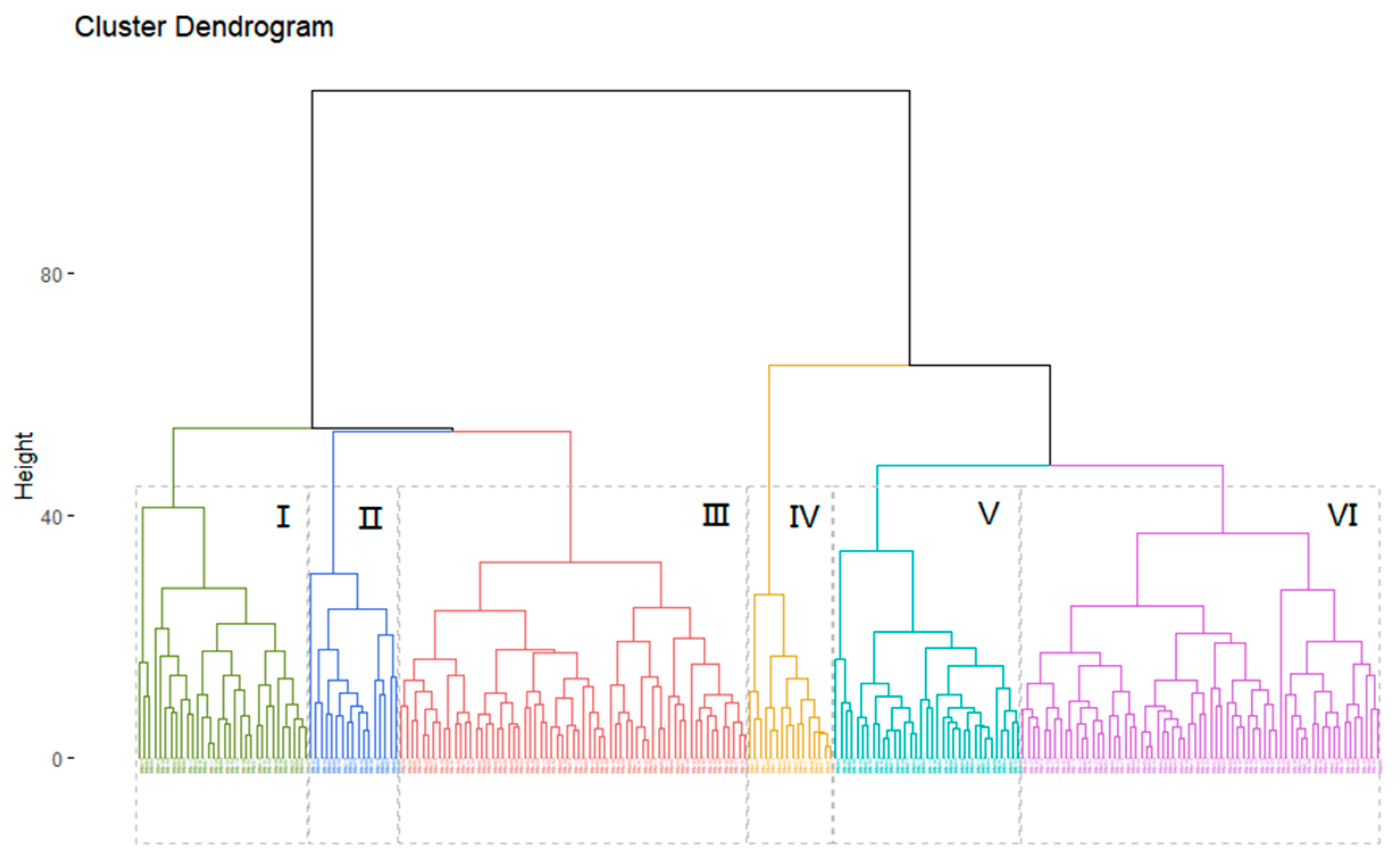
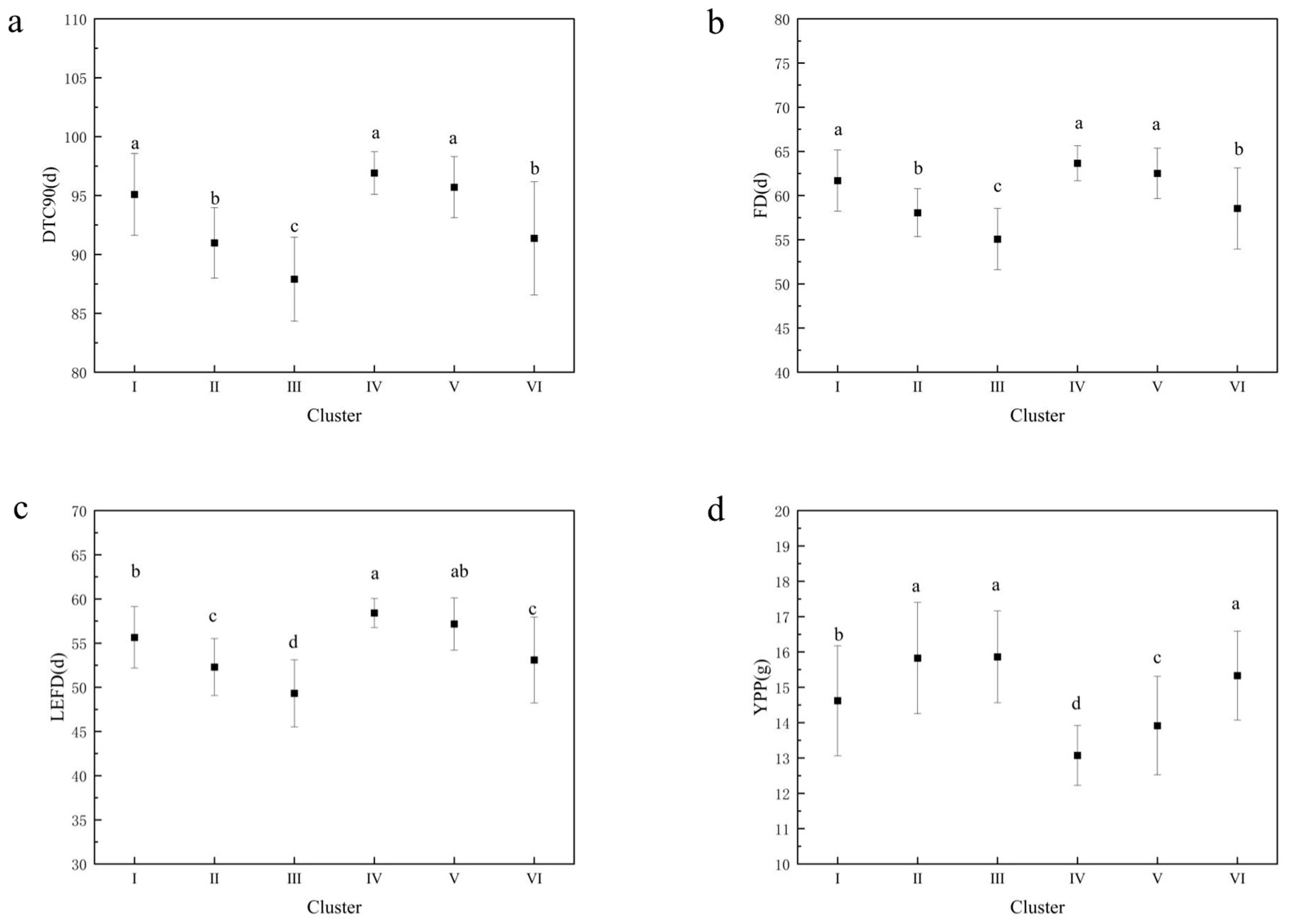
3.5. Cluster Analysis of Peanut Accessions
4. Discussion
4.1. Phenotypic Diversity Analysis of Flowering and Yield Traits in the Germplasm Panel
4.2. Genetic Diversity Analysis of Flowering and Yield Traits in the Germplasm Panel
4.3. Flowering Redundancy Has Significant Adverse Effects on Pod Yield
4.3.1. Correlation Analysis of Flowering Time and Yield Traits
4.3.2. PCA of Peanut Germplasm Resources
4.3.3. Cluster Analysis of Peanut Germplasm Resources
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Magassa, M. Improved Varieties Sow a Better Future in Mali’s Groundnut Hubs. ICRISAT. 2022. Available online: https://www.cgiar.org/news-events/news/improved-varieties-sow-a-better-future-in-malis-groundnut-hubs (accessed on 23 July 2023).
- Food and Agriculture Organization. 2021. Available online: https://www.fao.org/faostat/zh/#data/QCL/visualize (accessed on 10 January 2024).
- Huyghe, C. Genetics and genetic modifications of plant architecture in grain legumes: A review. Agronomie 1998, 18, 383–411. [Google Scholar] [CrossRef]
- Shin, K.; Kenichiro, F.; Setsuzo, Y.; Masao, I.; Tatsuhiko, S.; Takashi, S.; Akio, K.; Takeshi, N. Seed yield and its components of indeterminate and determinate lines in recombinant inbred lines of soybean. Breed. Sci. 2015, 65, 154–160. [Google Scholar] [CrossRef]
- Ashwini, K.V.R.; Ramesh, S.; Sunitha, N.C. Comparative BLUP, YREM-based performance and AMMI model-based stability of horse gram [Macrotyloma uniflorum (Lam.) Verdc.] genotypes differing in growth habit. Genet. Resour. Crop Evol. 2021, 68, 457–467. [Google Scholar] [CrossRef]
- Kaur, H.; Banga, S.S. Discovery and mapping of Brassica juncea Sdt1 gene associated with determinate plant growth habit. Theor. Appl. Genet. 2015, 128, 235–245. [Google Scholar] [CrossRef]
- Shin, K.; Takashi, S.; Fumio, T.; Akio, K.; Masao, I.; Elroy, C. Effect of change from a determinate to a semi-determinate growth habit on the yield and lodging resistance of soybeans in the northeast region of Japan. Breed. Sci. 2019, 69, 151–159. [Google Scholar] [CrossRef]
- Çalişkan, S.; Çalişkan, M.E.; Arslan, M. Genotypic differences for reproductive growth, yield, and yield components in groundnut (Arachis hypogaea L.). Turk. J. Agric. For. 2008, 32, 415–424. [Google Scholar]
- Kaba, J.S.; Kumaga, F.K.; Ofori, K. Effect of flower production and time of flowering on pod yield of peanut (Arachis hypogaea L.) genotypes. IOSR J. Agric. Vet. Sci. 2014, 7, 44–49. [Google Scholar] [CrossRef]
- Jung, C.; Müller, A.E. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009, 14, 563–573. [Google Scholar] [CrossRef]
- Patil, H.E.; Dikshit, N.; Aklade, S.A.; Vavdiya, P.A. Evaluation of vegetable type Pigeon pea (Cajanus cajan (L.) genotypes for yield and yield contributing traits. Ecol. Environ. Conserv. 2016, 22, 247–254. [Google Scholar]
- Gaur, P.M.; Samineni, S.; Tripathi, S.; Varshney, R.K.; Gowda, C.L. Allelic relationships of flowering time genes in chickpea. Euphytica 2015, 203, 295–308. [Google Scholar] [CrossRef]
- Raman, R.; Diffey, S.; Carling, J.; Cowley, R.B.; Kilian, A.; Luckett, D.J.; Raman, H. Quantitative genetic analysis of grain yield in an Australian Brassica napus doubled-haploid population. Crop Pasture Sci. 2016, 67, 298–307. [Google Scholar] [CrossRef]
- Raman, H.; Raman, R.; Qiu, Y.; Yadav, A.S.; Sureshkumar, S.; Borg, L.; Rohan, M.; Wheeler, D.; Owen, O.; Menz, I.; et al. GWAS hints at pleiotropic roles for FLOWERING LOCUS T in flowering time and yield-related traits in canola. BMC Genom. 2019, 20, 636. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Du, H.P.; Huang, Z.R.; He, M.L.; Kong, L.P.; Fang, C.; Chen, L.Y.; Yang, H.; Zhang, Y.H.; Liu, B.H.; et al. The AP2/ERF transcription factor TOE4b regulates photoperiodic flowering and grain yield of per plant in soybean. Plant Biotechnol. J. 2023, 21, 1682–1694. [Google Scholar] [CrossRef] [PubMed]
- Cockram, J.; Jones, H.; Leigh, F.J.; O’Sullivan, D.; Powell, W.; Laurie, D.A.; Greenland, A.J. Control of flowering time in temperate cereals: Genes, domestication, and sustainable productivity. J. Exp. Bot. 2007, 58, 1231–1244. [Google Scholar] [CrossRef] [PubMed]
- Upadhyaya, H.D.; Dwivedi, S.L.; Vadez, V.; Hamidou, F.; Singh, S.; Varshney, R.K.; Liao, B. Multiple resistant and nutritionally dense germplasm identified from mini core collection in peanut. Crop Sci. 2014, 54, 679–693. [Google Scholar] [CrossRef]
- Patil, R.; Viswanatha, K.P.; Upadhyaya, H.D.; Lokesha, R.; Khan, H.; Gururaj, S. Genetic diversity, association and principle component analyses for agronomical and quality traits in genomic selection training population of groundnut (Arachis hypogaea L.). Indian J. Genet. Plant Breed. 2020, 80, 282–290. [Google Scholar] [CrossRef]
- Ali, M.A.; Pal, A.K.; Baidya, A.; Gunri, S.K. Variation in dry matter production, partitioning, yield and its correlation in groundnut (Arachis hypogaea L.) genotypes. Legume Res. 2021, 44, 706–711. [Google Scholar] [CrossRef]
- Fonceka, D.; Tossim, H.A.; Rivallan, R.; Vignes, H.; Faye, I.; Ndoye, O.; Moretzsohn, M.C.; Bertioli, D.J.; Glaszmann, J.C.; Courtois, B.; et al. Fostered and left behind alleles in peanut: Interspecific QTL mapping reveals footprints of domestication and useful natural variation for breeding. BMC Plant Biol. 2012, 12, 26. [Google Scholar] [CrossRef]
- Wang, L.; Yang, X.L.; Cui, S.L.; Wang, J.H.; Hou, M.Y.; Mu, G.J.; Li, Z.C.; Liu, L.F. Identification of main effect and epistatic QTLs controlling initial flowering date in cultivated peanut (Arachis hypogaea L.). J. Integr. Agric. 2020, 19, 2383–2393. [Google Scholar] [CrossRef]
- Shirasawa, K.; Koilkonda, P.; Aoki, K.; Hirakawa, H.; Tabata, S.; Watanabe, M.; Hasegawa, M.; Kiyoshima, H.; Suzuki, S.; Kuwata, C.; et al. In silico polymorphism analysis for the development of simple sequence repeat and transposon markers and construction of linkage map in cultivated peanut. BMC Plant Biol. 2012, 12, 80. [Google Scholar] [CrossRef]
- Wang, L.; Yang, X.; Cui, S.; Zhao, N.N.; Li, L.; Hou, M.Y.; Mu, G.J.; Liu, L.F.; Li, Z.C. High-density genetic map development and QTL mapping for concentration degree of floret flowering date in cultivated peanut (Arachis hypogaea L.). Mol. Breed. 2020, 40, 17. [Google Scholar] [CrossRef]
- Vasanthi, R.P.; Suneetha, N.; Sudhakar, P. Genetic variability and correlation studies for morphological, yield and yield attributes in groundnut (Arachis hypogaea L.). Legume Res. 2015, 38, 9–15. [Google Scholar] [CrossRef]
- Yol, E.; Furat, S.; Upadhyaya, H.D.; Uzun, B. Characterization of groundnut (Arachis hypogaea L.) collection using quantitative and qualitative traits in the Mediterranean Basin. J. Integr. Agric. 2018, 17, 63–75. [Google Scholar] [CrossRef]
- Banla, E.M.; Dzidzienyo, D.K.; Diangar, M.M.; Melomey, L.D.; Offei, S.K.; Tongoona, P.; Desmae, H. Molecular and phenotypic diversity of groundnut (Arachis hypogaea L.) cultivars in Togo. Physiol. Mol. Biol. Plants 2020, 26, 1489–1504. [Google Scholar] [CrossRef]
- Swamy, B.M.; Upadhyaya, H.D.; Goudar, P.K.; Kullaiswamy, B.Y.; Singh, S. Phenotypic variation for agronomic characteristics in a groundnut core collection for Asia. Field Crops Res. 2003, 84, 359–371. [Google Scholar] [CrossRef][Green Version]
- Gan, Y.T.; Harker, K.N.; Kutcher, H.R.; Gulden, R.H.; Irvine, B.; May, W.E.; O’Donovan, J.T. Canola seed yield and phenological responses to plant density. Can. J. Plant Sci. 2016, 96, 151–159. [Google Scholar] [CrossRef]
- Jiang, H.F.; Duan, N.X. Descriptors and Data Standard for Peanut (Arachis spp.); China Agriculture Press: Beijing, China, 2006; pp. 26–29. [Google Scholar]
- Pan, J.; Zhang, L.; Wang, L.; Fu, S. Effects of long-term fertilization treatments on the weed seed bank in a wheat-soybean rotation system. Glob. Ecol. Conserv. 2020, 21, e00870. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Piepho, H.P.; Mohring, J. Computing heritability and selection response from unbalanced plant breeding trials. Genetics 2007, 177, 1881–1888. [Google Scholar] [CrossRef]
- Friendly, M. Corrgrams: Exploratory displays for correlation matrices. Am. Stat. 2002, 56, 316–324. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A. Practical Guide to Principal Component Methods in R; STHDA: Los Angeles, CA, USA, 2017; Volume 2. [Google Scholar]
- Steel, R.G.D.; Torrie, J.H.; Dicey, D.A. Principles and Procedures of Statistics: A Biometrical Approach; McGraw-Hill Co. Inc.: New York, NY, USA, 1997. [Google Scholar]
- Chen, T.T.; Zhang, J.L.; Wang, X.Y.; Zeng, R.E.; Zhang, L. Improving Peanut Growth and Yield Responses with Monoseeding and Paclobutrazol Applications in Southern China. Legume Res. 2022, 45, 327–333. [Google Scholar] [CrossRef]
- Liu, Z.; Nan, Z.W.; Lin, S.M.; Meng, W.W.; Xie, L.Y.; Yu, H.Q.; Zhang, Z.; Wan, S.B. Organ removal of maize increases peanut canopy photosynthetic capacity, dry matter accumulation, and yield in maize/peanut intercropping. Front. Plant Sci. 2023, 24, 1266969. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.Y.; Dai, J.; Li, N.; Zhao, H.T.; Chang, H.B.; Liao, X.; Sheng, F.; Lu, Q. Comparative evaluation of microbially-produced biostimulants on peanut growth. Sustainability 2023, 15, 8025. [Google Scholar] [CrossRef]
- Zaman, M.A.; Tuhina-Khatun, M.; Ullah, M.Z.; Moniruzzamn1, M.; Alam, K.H. Genetic variability and path analysis of groundnut (Arachis hypogaea L.). Agriculturists 2011, 9, 29–36. [Google Scholar] [CrossRef]
- Mukri, G.; Nadaf, H.L.; Bhat, R.S.; Gowda, M.V.C.; Upadhyaya, H.D.; Sujay, V. Phenotypic and molecular dissection of ICRISAT mini core collection of peanut (Arachis hypogaea L.) for high oleic acid. Plant Breed. 2012, 131, 418–422. [Google Scholar] [CrossRef]
- Sudini, H.; Upadhyaya, H.D.; Reddy, S.V.; Mangala1, U.N.; Rathore1, A.; Kumar, K.V.K. Resistance to late leaf spot and rust diseases in ICRISAT’s mini core collection of peanut (Arachis hypogaea L.). Australas. Plant Pathol. 2015, 44, 557–566. [Google Scholar] [CrossRef]
- Chaudhari, S.; Khare, D.; Patil, S.C.; Sundravadana, S.; Variath, M.T.; Sudini, H.K.; Manohar, S.S.; Bhat, R.S.; Pasupuleti, J. Genotype × Environment Studies on Resistance to Late Leaf Spot and Rust in Genomic Selection Training Population of Peanut (Arachis hypogaea L.). Front. Plant Sci. 2019, 10, 1338. [Google Scholar] [CrossRef]
- Guclu, V.; Aydogdu, M.; Basak, M.; Kizil, S.; Uzun, B.; Yol, E. Characterization of a groundnut collection to stem rot disease caused by Sclerotium rolfsii. Australas. Plant Pathol. 2020, 49, 691–700. [Google Scholar] [CrossRef]
- Hamidou, F.; Halilou, O.; Vadez, V. Assessment of groundnut under combined heat and drought stress. J. Agron. Crop Sci. 2013, 199, 1–11. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Reddy, L.J.; Dwivedi, S.L.; Gowda, C.L.L.; Singh, S. Phenotypic diversity in cold-tolerant peanut (Arachis hypogaea L.) germplasm. Euphytica 2009, 165, 279–291. [Google Scholar] [CrossRef]
- Upadhyaya, H.D. Variability for drought resistance related traits in the mini core collection of peanut. Crop Sci. 2005, 45, 1432–1440. [Google Scholar] [CrossRef]
- Lal, C.; Hariprasanna, K.; Rathnakumar, A.L.; Misra, J.B.; Samdur, M.Y.; Gor, H.K.; Chikani, B.M.; Jain, V.K. Response of peanut genotypes to mid-season moisture stress: Phenological, morpho-physiological, and yield traits. Crop Pasture Sci. 2009, 60, 339–347. [Google Scholar] [CrossRef]
- Holbrook, C.C.; Anderson, W.F.; Pittman, R.N. Selection of a core collection from the US germplasm collection of peanut. Crop Sci. 1993, 33, 859–861. [Google Scholar] [CrossRef]
- Upadhyaya, H.D. Phenotypic diversity in groundnut (Arachis hypogaea L.) core collection assessed by morphological and agronomical evaluations. Genet. Resour. Crop Evol. 2003, 50, 539–550. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Bramel, P.J.; Ortiz, R.; Singh, S. Developing a mini core of peanut for utilization of genetic resources. Crop Sci. 2002, 42, 2150–2156. [Google Scholar] [CrossRef]
- Cheong, Y.K.; Park, K.H.; Doo, H.S.; Ryu, J.H.; Choi, S.Y.; Suh, D.Y. Flowering and Fruiting of Characteristics of Short Flowering Period Lines in Peanut. Korean J. Crop Sci. 2002, 47, 437–442. [Google Scholar]
- Mazzani, C.E.; Segovia, V.; Marín, C.; Pacheco, W. Peanut (Arachis hypogaea L.) cultivars classification based on quantitative traits to establish core collections of the genebank of Centro Nacional de Investigaciones Agropecuarias, Venezuela. Rev. UDO Agrícola 2009, 9, 756–763. [Google Scholar]
- Kumar, N.; Ajay, B.C.; Rathanakumar, A.L.; Radhakrishnan, T.; Jadon, K.S.; Chikani, B.M. Genetic variability analyses for yield and physiological traits in groundnut (Arachis hypogaea L.) genotypes. J. Oilseeds Res. 2019, 36, 1–7. [Google Scholar] [CrossRef]
- Jiang, H.F.; Huang, L.; Ren, X.P.; Chen, Y.N.; Zhou, X.J.; Xia, Y.L.; Huang, J.Q.; Lei, Y.; Yan, L.Y.; Wan, L.Y.; et al. Diversity characterization and association analysis of agronomic traits in a Chinese peanut (Arachis hypogaea L.) mini-core collection. J. Integr. Plant Biol. 2014, 56, 159–169. [Google Scholar] [CrossRef]
- Kiranmai, S.M.; Venkataravana, P.; Pushpa, H.D. Correlation and path analysis studies in groundnut under different environment. Legume Res. 2016, 39, 1048–1050. [Google Scholar] [CrossRef]
- Mubai, N.; Sibiya, J.; Mwololo, J.; Musvosvi, C.; Charlie, H.; Munthali, W.; Kachulu, L.; Okori, P. Phenotypic correlation, path coefficient and multivariate analysis for yield and yield-associated traits in groundnut accessions. Cogent Food Agric. 2020, 6, 1823591. [Google Scholar] [CrossRef]
- Jannat, S.; Hassan, M.U.; Ortiz, G.T.; Shah, M.K.N.; Ahmed, M.; Shah, A.H.; Qayyum, A. Genetic characterization of flowering and phytochrome genes in peanut (Arachis hypogaea L.) for early maturity. Mol. Biol. Rep. 2022, 49, 5495–5504. [Google Scholar] [CrossRef] [PubMed]
- Monpara, B.A.; Dhameliya, H.R. Genetic behaviour of earliness related traits and seed yield in chickpea (Cicer arietinum L.). Pak. J. Biol. Sci. 2013, 16, 955–959. [Google Scholar] [CrossRef] [PubMed]
- Mallikarjuna, B.P.; Viswanatha, K.P.; Samineni, S.; Gaur, P.M. Association of flowering time with phenological and productivity traits in chickpea. Euphytica 2019, 215, 77. [Google Scholar] [CrossRef]
- Endres, L.; da Cruz, S.J.S.; Vilela, R.D.; dos Santos, J.M.; Barbosa, G.V.D.S.; Silva, J.A.C. Foliar applications of calcium reduce and delay sugarcane flowering. BioEnergy Res. 2016, 9, 98–108. [Google Scholar] [CrossRef]
- Khokhar, J.S.; Jamwal, N.S.; Sanghera, G.S.; Singh, P. Evaluation of sugarcane (Saccharum officinarum) germplasm for quality, yield traits and effects of flowering on cane traits. Indian J. Agric. Sci. 2022, 92, 842–846. [Google Scholar] [CrossRef]
- Vinothini, N.; Vijayan, R.; Umarani, R. Studies on flowering pattern in relation to seed filling and seed multiplication rate in groundnut (Arachis hypogaea L.). Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 3321–3328. [Google Scholar] [CrossRef]
- Hariprasanna, K.; Lal, C.; Radhakrishnan, T. Relationship between flowering duration and physical quality traits as well as pod yield in groundnut (Arachis hypogaea). Indian J. Agric. Sci. 2008, 78, 180–182. [Google Scholar]
- Khatoon, B.; Sharma, V.R.; Yadav, H.K. Phenotypic diversity and trait association in cluster bean (Cyamopsis tetragonoloba). Vegetos 2023, 37, 329–340. [Google Scholar] [CrossRef]
- Rana, J.C.; Sharma, T.R.; Tyagi, R.K.; Chahota, R.K.; Gautam, N.K.; Singh, M.; Sharma, P.N.; Ojha, S.N. Characterisation of 4274 accessions of common bean (Phaseolus vulgaris L.) germplasm conserved in the Indian gene bank for phenological, morphological and agricultural traits. Euphytica 2015, 205, 441–457. [Google Scholar] [CrossRef]
- Gerrano, A.S.; Van Rensburg, W.S.J.; Kutu, F.R. Agronomic evaluation and identification of potential cowpea (Vigna unguiculata L. Walp) genotypes in South Africa. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2019, 69, 295–303. [Google Scholar] [CrossRef]
- Nyirenda Yohane, E.; Shimelis, H.; Laing, M.; Mathew, I.; Shayanowako, A. Phenotypic divergence analysis in pigeonpea [Cajanus cajan (L.) Millspaugh] germplasm accessions. Agronomy 2020, 10, 1682. [Google Scholar] [CrossRef]
- Ullah, A.; Akram, Z.; Malik, S.I.; Khan, K.S.U. Assessment of phenotypic and molecular diversity in soybean [Glycine max (L.) Merr.] germplasm using morpho-biochemical attributes and SSR markers. Genet. Resour. Crop Evol. 2021, 68, 2827–2847. [Google Scholar] [CrossRef]
- Yimram, T.; Somta, P.; Srinives, P. Genetic variation in cultivated mungbean germplasm and its implication in breeding for high yield. Field Crops Res. 2009, 112, 260–266. [Google Scholar] [CrossRef]
- Adu, M.O.; Asare, P.A.; Yawson, D.O.; Dzidzienyo, D.K.; Nyadanu, D.; Asare-Bediako, E.; Afutu, E.; Tachie-Menson, J.W.; Amoah, M.N. Identifying key contributing root system traits to genetic diversity in field-grown cowpea (Vigna unguiculata L. Walp.) genotypes. Field Crops Res. 2019, 232, 106–118. [Google Scholar] [CrossRef]
- Kwak, M.; Gepts, P. Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae). Theor. Appl. Genet. 2009, 118, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.A.; Naidu, G.K.; Nadaf, H.L.; Tippannavar, P.S. Genetic diversity in groundnut (Arachis hypogaea L.) based on reaction to biotic stresses and productivity parameters. Legume Res. 2021, 44, 131–137. [Google Scholar] [CrossRef]
- Luo, X.; Liu, Z. Plant development: Unveiling cytokinin’s role in the end of flowering. Curr. Biol. 2022, 32, R168–R170. [Google Scholar] [CrossRef]
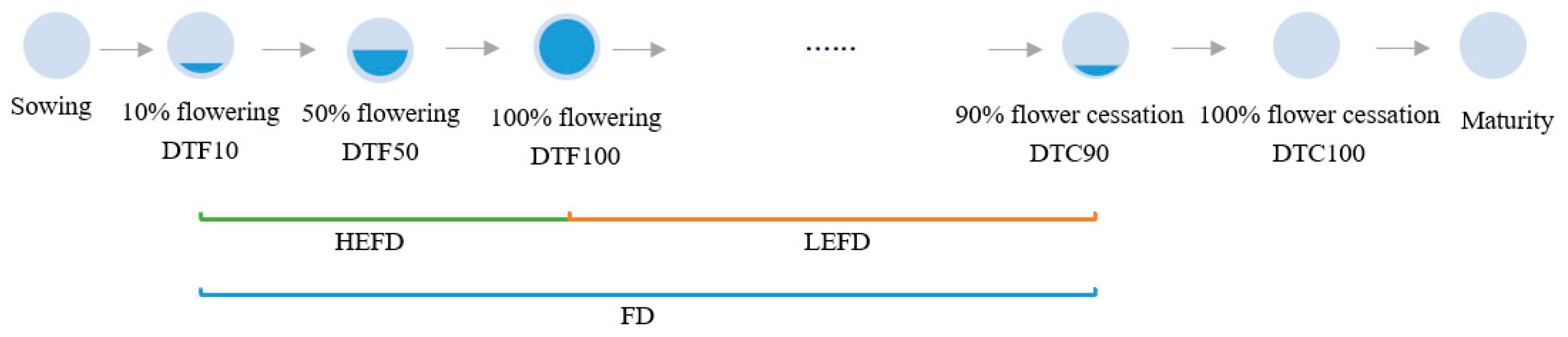
| Environment | Soil Type | pH | Hydrolyzable N mg/kg | Available P mg/kg | Rapidly Available K mg/kg | Organic Matter g/kg | Exchangeable Ca cmol/kg | Water-Soluble Ca cmol/kg |
|---|---|---|---|---|---|---|---|---|
| Yongzhou-spring | red soil | 4.48 | 95 | 0.5 | 35 | 17.1 | 0.95 | 0.21 |
| Yueyang-spring | red soil | 4.98 | 78 | 0.5 | 79 | 8.41 | 4.03 | 0.19 |
| Changsha-spring | alluvial soil | 5.62 | 166 | 17.2 | 231 | 25.6 | 3.91 | 0.03 |
| Changsha-summer | alluvial soil | 6.14 | 205 | 17.2 | 74 | 23.8 | 4.50 | 0.03 |
| Phenotypic Trait | Minimum | Maximum | Range | Median | Mean ± SE | SD | CV (%) | H′ |
|---|---|---|---|---|---|---|---|---|
| DTF10 (d) | 21.50 | 41.50 | 20.00 | 34.00 | 32.96 ± 0.18 | 5.39 | 16.35 | 1.807 |
| DTF50 (d) | 23.50 | 47.00 | 23.50 | 37.00 | 34.69 ± 0.18 | 5.57 | 16.07 | 1.710 |
| DTF100 (d) | 25.50 | 62.50 | 37.00 | 39.00 | 38.61 ± 0.23 | 6.92 | 17.91 | 1.948 |
| DTC90 (d) | 49.00 | 115.00 | 66.00 | 94.50 | 91.92 ± 0.4 | 12.09 | 13.15 | 1.964 |
| DTC100 (d) | 69.00 | 115.00 | 46.00 | 100.50 | 99.52 ± 0.27 | 8.16 | 8.20 | 1.932 |
| FD (d) | 25.00 | 81.00 | 56.00 | 62.00 | 59.05 ± 0.36 | 10.96 | 18.56 | 1.982 |
| HEFD (d) | 0.00 | 23.00 | 23.00 | 5.50 | 5.65 ± 0.1 | 3.02 | 53.43 | 1.825 |
| LEFD (d) | 14.00 | 76.50 | 62.50 | 56.00 | 53.4 ± 0.38 | 11.47 | 21.47 | 1.978 |
| YPP (g) | 4.23 | 37.09 | 32.87 | 14.59 | 15.11 ± 0.2 | 6.01 | 39.74 | 0.862 |
| HPW (g) | 49.08 | 243.37 | 194.29 | 128.36 | 131.49 ± 1.2 | 36.23 | 27.55 | 0.919 |
| HSW (g) | 30.36 | 92.49 | 62.14 | 51.34 | 52.44 ± 0.41 | 12.34 | 23.53 | 0.861 |
| SP (%) | 33.47 | 83.51 | 50.04 | 65.17 | 63.95 ± 0.27 | 8.30 | 12.98 | 0.918 |
| AWPP (g) | 0.44 | 2.11 | 1.66 | 1.09 | 1.14 ± 0.01 | 0.32 | 27.80 | 0.841 |
| Phenotypic Trait | G | E | G × E | PCV (%) | GCV (%) | H2 (%) |
|---|---|---|---|---|---|---|
| DTF10 (d) | 0.682 *** | 36.913 *** | 0.449 *** | 2.915 | 2.505 | 73.837 |
| DTF50 (d) | 0.949 *** | 38.287 *** | 0.324 *** | 3.209 | 2.807 | 76.516 |
| DTF100 (d) | 2.311 *** | 54.021 *** | 1.236 *** | 4.855 | 3.937 | 65.779 |
| DTC90 (d) | 34.308 *** | 64.858 ** | 24.733 *** | 7.686 | 6.372 | 68.728 |
| DTC100 (d) | 10.659 *** | 25.016 *** | 7.932 *** | 4.486 | 3.281 | 53.476 |
| FD (d) | 33.308 *** | 22.002 * | 29.492 *** | 11.988 | 9.774 | 66.482 |
| HEFD (d) | 1.325 *** | 3.878 ** | 1.551 *** | 28.292 | 20.370 | 51.838 |
| LEFD (d) | 35.591 *** | 35.299 * | 25.572 *** | 13.503 | 11.171 | 68.447 |
| YPP (g) | 4.752 *** | 16.534 ** | 5.258 *** | 19.535 | 14.427 | 54.543 |
| HPW (g) | 413.310 *** | 606.090 ** | 152.580 *** | 17.211 | 15.462 | 80.704 |
| HSW (g) | 75.077 *** | 49.475 ** | 18.975 *** | 17.638 | 16.523 | 87.755 |
| SP (%) | 28.592 *** | 25.578 *** | 9.901 *** | 9.066 | 8.362 | 85.060 |
| AWPP (g) | 0.028 *** | 0.050 ** | 0.013 *** | 16.562 | 14.643 | 78.166 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, N.; Liu, D.; Tang, K.; Lu, X.; Tao, Y.; Yan, X.; Zeng, N.; Li, L.; Luo, Z. The Phenotypic Diversity of 232 Germplasm Accessions Identifies the Adverse Effects of Flowering Redundancy on Peanut Yield. Agronomy 2024, 14, 434. https://doi.org/10.3390/agronomy14030434
Liu N, Liu D, Tang K, Lu X, Tao Y, Yan X, Zeng N, Li L, Luo Z. The Phenotypic Diversity of 232 Germplasm Accessions Identifies the Adverse Effects of Flowering Redundancy on Peanut Yield. Agronomy. 2024; 14(3):434. https://doi.org/10.3390/agronomy14030434
Chicago/Turabian StyleLiu, Na, Dengwang Liu, Kang Tang, Xuankang Lu, Yu Tao, Xin Yan, Ningbo Zeng, Lin Li, and Zinan Luo. 2024. "The Phenotypic Diversity of 232 Germplasm Accessions Identifies the Adverse Effects of Flowering Redundancy on Peanut Yield" Agronomy 14, no. 3: 434. https://doi.org/10.3390/agronomy14030434
APA StyleLiu, N., Liu, D., Tang, K., Lu, X., Tao, Y., Yan, X., Zeng, N., Li, L., & Luo, Z. (2024). The Phenotypic Diversity of 232 Germplasm Accessions Identifies the Adverse Effects of Flowering Redundancy on Peanut Yield. Agronomy, 14(3), 434. https://doi.org/10.3390/agronomy14030434





