Abstract
This study explores spectroscopy in the 350 to 2500 nm range for detecting powdery mildew (Erysiphe necator) in grapevine leaves, crucial for precision agriculture and sustainable vineyard management. In a controlled experimental vineyard setting, the spectral reflectance on leaves with varying infestation levels was measured using a FieldSpec 4 spectroradiometer during July and September. A detailed assessment was conducted following the guidelines recommended by the European and Mediterranean Plant Protection Organization (EPPO) to quantify the level of infestation; categorising leaves into five distinct grades based on the percentage of leaf surface area affected. Subsequently, spectral data were collected using a contact probe with a tungsten halogen bulb connected to the spectroradiometer, taking three measurements across different areas of each leaf. Partial Least Squares Regression (PLSR) analysis yielded coefficients of determination R2 = 0.74 and 0.71, and Root Mean Square Errors (RMSEs) of 12.1% and 12.9% for calibration and validation datasets, indicating high accuracy for early disease detection. Significant spectral differences were noted between healthy and infected leaves, especially around 450 nm and 700 nm for visible light, and 1050 nm, 1425 nm, 1650 nm, and 2250 nm for the near-infrared spectrum, likely due to tissue damage, chlorophyll degradation and water loss. Finally, the Powdery Mildew Vegetation Index (PMVI) was introduced, calculated as PMVI = (R755 − R675)/(R755 + R675), where R755 and R675 are the reflectances at 755 nm (NIR) and 675 nm (red), effectively estimating disease severity (R2 = 0.7). The study demonstrates that spectroscopy, combined with PMVI, provides a reliable, non-invasive method for managing powdery mildew and promoting healthier vineyards through precision agriculture practices.
1. Introduction
Crop diseases and pests significantly impact global agricultural production, quality, and economic outcomes [1]. In the field of viticulture, several diseases significantly impact grapevine health, such as grapevine trunk diseases like Esca [2], Grapevine Red Blotch Disease and Grapevine Leafroll Disease [3] or Botrytis, which leads to botrytis bunch rot or grey mould [4]. Each disease presents unique challenges, requiring specific management and control strategies. Among these diseases, powdery mildew (PM) caused by the fungus Erysiphe necator (aka Uncinula necator) stands out as particularly harmful, especially with the challenges posed by climate change [5]. PM is a major economic threat to grape cultivation as it can alter both yield and grape quality, thereby impacting wine quality [6,7]. The epidemiology of the fungus, as well as its ecology, morphology, and reproductive biology, are well-documented [8]. This extensive research has revealed that the frequent use of intensive spraying programs to control PM contributes to resistance across various fungicide classes, thus necessitating the invention of novel, effective molecular control strategies [9]. Concurrently, research into the molecular dynamics between Vitis vinifera and Erysiphe necator offers insights into resistance mechanisms that hold promise for breeding PM-resistant strains [10]. Therefore, early disease detection and monitoring are crucial for improving productivity and promoting sustainable agriculture by facilitating targeted pesticide and fungicide applications [11], highlighting the urgency in developing rapid detection and surveillance techniques for PM.
PM severity is typically carried out through visual inspections, yet recent advancements offer more precise methods to diagnose, measure, and manage the disease by leveraging core plant pathology techniques [8]. The primary molecular tools for disease detection currently include ELISA and PCR (including real-time PCR), alongside methods based on immunofluorescence (IF), flow cytometry (FCM), fluorescence in situ hybridisation (FISH), and DNA microarrays [12]. PCR techniques, for instance, determine the quantity of pathogen DNA or the percentage of surface area affected by powdery mildew through infrared (IR) spectroscopy or by identifying disease biomarkers like volatile organic compounds [11]. These objective methods demand meticulous sampling and processing to identify plant diseases accurately [11]. Despite these technological advances, manual scouting remains prevalent due to its simplicity, albeit being laborious, time-consuming, and costly [13]. Hence, developing a reliable, non-destructive, fast and real-time disease monitoring system would significantly advance disease control and management in sustainable viticulture. Current trends in plant disease detection increasingly rely on advanced imaging techniques [1,14], with a notable emphasis on remote sensing, especially multispectral and hyperspectral (HSI) imagery captured by unmanned aerial vehicles (UAVs). This approach has become a key tool for monitoring grapevine health and identifying diseases [15,16,17,18,19]. These techniques often combine near-infrared (NIR) and red reflectance to gather data, exploiting biophysical parameters detectable via sensors equipped on UAVs. Notably, in the context of detecting diseases and environmental safety, HSI has been extensively utilised [20,21]. Moreover, HSI is adept at reducing constants and linear functions, thereby refining the precision of remote sensing in measuring crop parameters [22]. Specifically in this field, spectroscopy has shown considerable promise for detecting plant diseases [23] and in analysing compositional parameters in wine grapes, grape juice, and grapevine tissues critical for assessing quality and ripeness [24]. Techniques such as Vis–NIR [25], MIR [26], and Raman [27] spectroscopy offer rapid, cost-effective, and efficient approaches for disease detection and diagnosis in plants, applicable both in laboratory settings and in the field [28]. These methods can identify diseases before symptoms become visually apparent [29,30,31,32], and they can complement broad detection methods like UAVs due to the portability and ease of use of handheld spectroscopy devices for targeted, detailed analysis of specific areas. Additionally, spectroscopy has the potential to identify specific wavelengths that correlate directly with plant traits of interest, aiding in crafting new vegetation indices from specific spectral bands to improve the precision of agricultural assessment. Vegetation indices are useful for estimating plant and fruit health [33], biomass [34], water stress [35], quality traits [36], and chlorophyll content [37], and they could also enhance disease assessment capabilities by accurately detecting early signs of plant diseases through refined spectral analysis.
Particularly, in the field of viticulture, several researchers have effectively utilised spectroscopy for diverse applications, highlighting the method’s capability to capture critical biophysical parameters essential for enhancing grape production quality and efficiency. De Bei et al. [38] showcased the potential of near-infrared spectroscopy for the non-destructive determination of grapevine water potential, which is crucial for precise irrigation management. Likewise, Cuq et al. [39] employed NIR spectroscopy to swiftly determine the macro-element composition in vine leaves and grape berries, affirming its utility in vineyard management and ensuring grape quality. González-Caballero et al. [40] and González-Fernández et al. [41] explored field spectroscopy for tracking grape maturation and assessing vine water status, respectively. In the realm of vineyard management, significant progress has been made in identifying diseases through the application of spectroscopy techniques. Through Raman Spectroscopy, Baratto et al. [42] distinguished between healthy and Esca-infected vines, including those without symptoms. Junges et al. [43] identified unique spectral signatures between healthy and infected grapevine leaves, demonstrating hyperspectral reflectance as an effective method for prompt disease identification. In terms of quantifying disease severity, Hill et al. [44] applied NIR and mid-IR spectroscopy to objectively quantify Botrytis Bunch Rot in grapes. AL-Saddik et al. [45] created specific spectral disease indices for ‘Flavescence Dorée’ detection, reaching a classification accuracy of over 90%. Galvan et al. [46] utilised airborne imaging spectroscopy for the widespread early detection of grapevine viral diseases with an accuracy of up to 87%.
The main goal of this research is to enhance the early detection of powdery mildew infection levels in grapevines through spectroscopy. By utilising this technique, this research aims to identify specific wavelengths useful for this purpose and measure the disease’s early impact, thereby facilitating more prompt and effective management measures. This approach aims to support healthier vineyards and promote more sustainable agricultural practices.
2. Materials and Methods
2.1. Field Site and Sampling
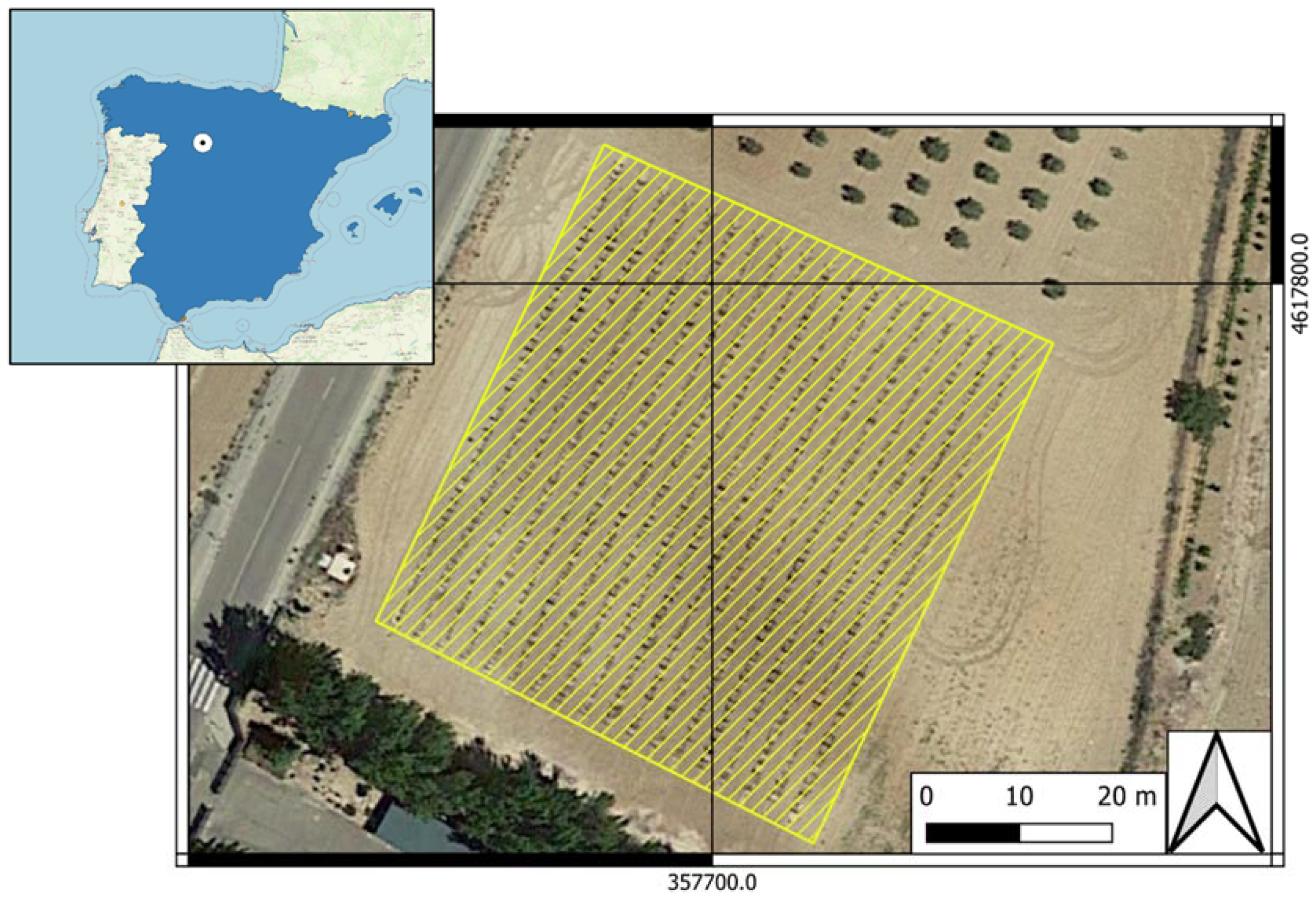
The study was conducted during the growing season of 2020 in an experimental vineyard (Figure 1) located in Zamadueñas estate (Valladolid, Spain; coordinates X:357700.6; Y: 4617774.6, ETRS89/UTM zone 30N, EPSG: 25830).
Figure 1.
Location of the experimental vineyard. Zamadueñas estate (Valladolid, Spain; coordinates X: 357700.6; Y: 4617774.6, ETRS89/UTM zone 30N, EPSG: 25830).
The grapevines, Vitis vinifera L. cv Tempranillo, under organic management, were treated with standard agricultural inputs and practices compatible with this system. The application of plant protection products was minimised, involving just two treatments: the first occurred before flowering on 23 June, utilising products compatible with organic agriculture (a combination of sulphur and copper-based products along with a binder, specifically horsetail), and the second took place after budding, employing wettable sulphur. This approach encouraged the natural progression of diseases, allowing for a range of infestation levels to be observed and studied within the experiment. As a result, the vineyard exhibited various degrees of natural powdery mildew infestation, which was endemic to the area due to the existing climatic conditions.
The fieldwork was organised into two phases: one for collecting a calibration dataset (D1) and another for a validation dataset (D2). The first dataset was collected on 31 July, comprising 68 leaf samples. The second dataset, consisting of 54 leaf samples, was gathered on 10 September. The guidelines recommended by the European and Mediterranean Plant Protection Organization (2002) [47] were followed to perform the samplings, focusing on the percentage of leaf surface affected by disease for sample assessment (Figure 2).

Figure 2.
Damage symptoms due to powdery mildew on leaf vine at different levels of infestation expressed on the percentage of leaf surface area affected: Grade 1 (no disease), Grade 2 (1–5%), Grade 3 (5–25%), Grade 4 (25–50%) and Grade 5 (>50%).
Based on these guidelines, the sampled leaves were graded into five levels of infestation: Grade 1 (no disease), Grade 2 (1–5%), Grade 3 (5–25%), Grade 4 (25–50%) and Grade 5 (>50%). Five to seven grapevines were selected for each grade, and one representative leaf from each grapevine was collected for spectral measurements.
2.2. Spectral Measurements
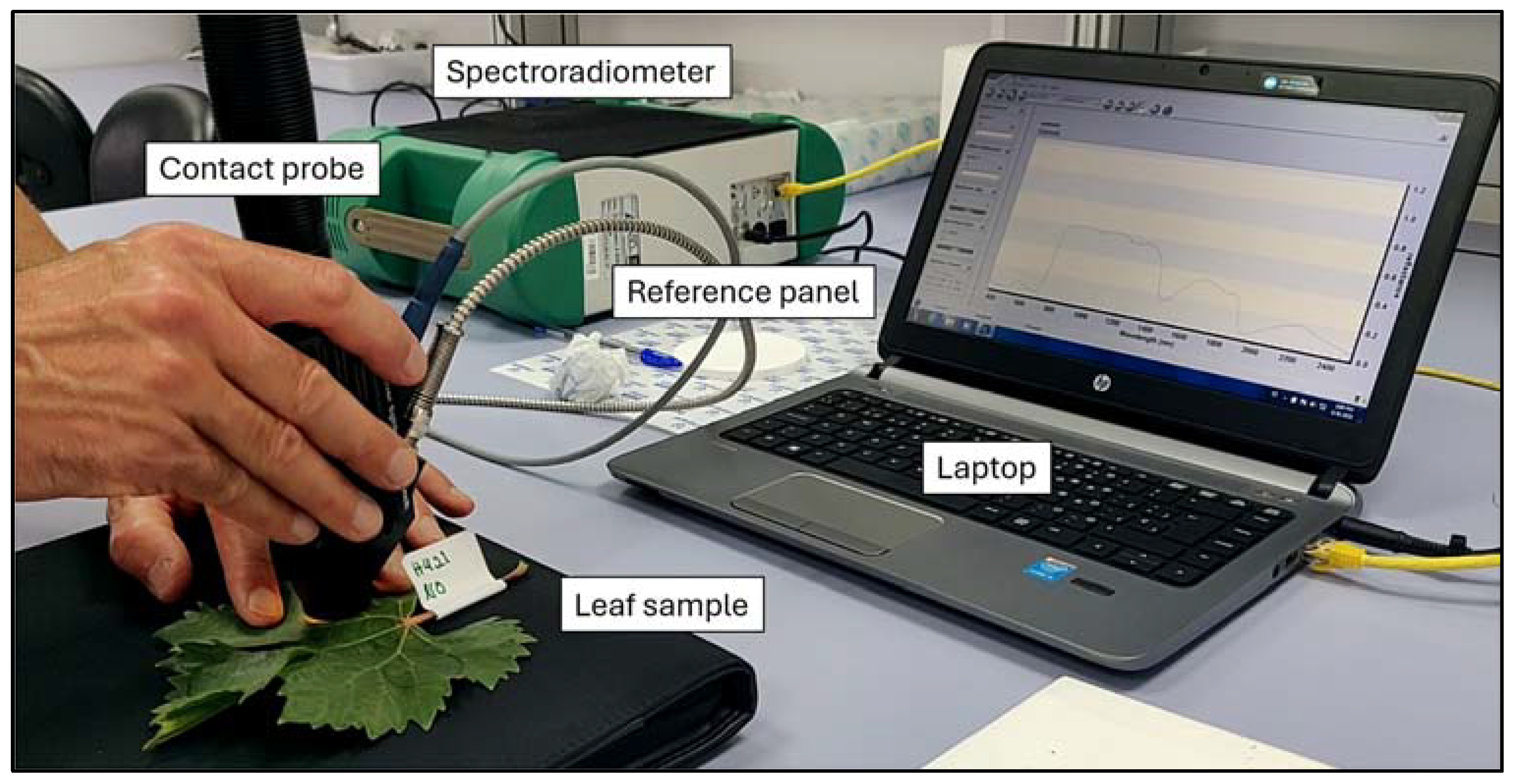
Spectra were obtained using a FieldSpec 4 spectroradiometer (Analytical Spectral Devices, Boulder, CO, USA). A contact probe with a tungsten halogen bulb was used to take leaf reflectance data (Figure 3), ensuring homogeneity in measurements. Three spectral measurements at different locations on each leaf were taken, and the damage level was assessed in order to obtain the spectral signature of detailed infestation levels. An average of 15 successive scans over the wavelength, ranging from 350 to 2500 nm, were done to calculate each spectrum of the leaf sample.
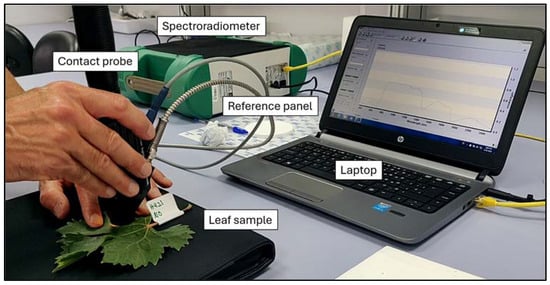
Figure 3.
Leaf under analysis using the contact probe connected to the FieldSpec 4 spectroradiometer. The data are visualised in real-time on the laptop screen.
The calibration and validation datasets were collected in the field during leaf sampling activities in two separate collection periods: the first on July 31st, at the beginning of veraison, and the second on September 10th, when the berries were ready to be harvested, corresponding to BBCH81 and BBCH89 stages, respectively, according to the BBCH scale for the growth stages of the grapevine [48]. In each of these sessions, every leaf sampled was measured, resulting in 68 samples in the first session and 54 in the second (Table 1). The reflectance was processed using ASD View Spec Pro (ASD, Boulder, CO, USA), Spectral Analysis and Management System -SAMS V2 (CSTARS, UC Davis, CA, USA) and the Unscrambler V11 (CAMO, Oslo, Norway).

Table 1.
Number of leaves sampled for each dataset and infestation level (severity degree).
2.3. PLS Regressions
The performance of the regression model was initially evaluated through Leave-One-Out Cross-Validation (LOOCV) using the calibration dataset. Subsequently, its applicability was further tested via an independent validation employing the validation dataset. PLS calibrations were performed based on raw reflectance from the first dataset (31 July), and, using the second dataset (10 September), an independent validation was conducted to assess the model’s predictive ability. The model’s performance was evaluated by calculating the coefficient of determination (R2) and the root mean squared error (RMSE) values from the validation data. Notably, two types of RMSE were considered: one for the LOOCV assessment (RMSECV) and another for the independent validation (RMSEV). The standard deviation (SD) to RMSE ratio, known as RPD, was also utilised to evaluate the model’s predictive capability. The R2 is calculated as:
where RSS is the residual sum of squares and TSS is the total sum of squares.
The RMSE is defined as:
where yi and yp represent the observed and predicted level of infestation for sample i, respectively, and n is the sample size.
The RPD is calculated as:
R2 quantifies the proportion of variance in the dependent variable (PM infestation level) that is predictable from the independent variable (reflectance). It is presented as the percentage of the variation explained by a best-fit regression line. RMSE indicates the total prediction error of the model. In general, a high R2 and low RMSE, along with an RPD > 2.5, indicate the adequacy of the prediction model.
3. Results
3.1. Severity of Infestation
Both data sets showed similar characteristics and distributions of descriptive statistics (Table 2). The calibration dataset had a mean and SD of 32.1 and 23.7 (range of 0–80%), while the validation dataset had 34.8 and 24.1 (range of 0–90%), respectively. Infestation levels were right-skewed, with a median of 30% in both datasets. The CVs were 73.8% for calibration and 69.4% for validation, indicating that both datasets feature diverse distributions and substantial variation.

Table 2.
Descriptive statistics of the level of infestation (as a percentage of leaf area) for calibration (July 31st) and validation (September 10th) data sets.
3.2. Spectral Reflectances at Different Levels of Infestation
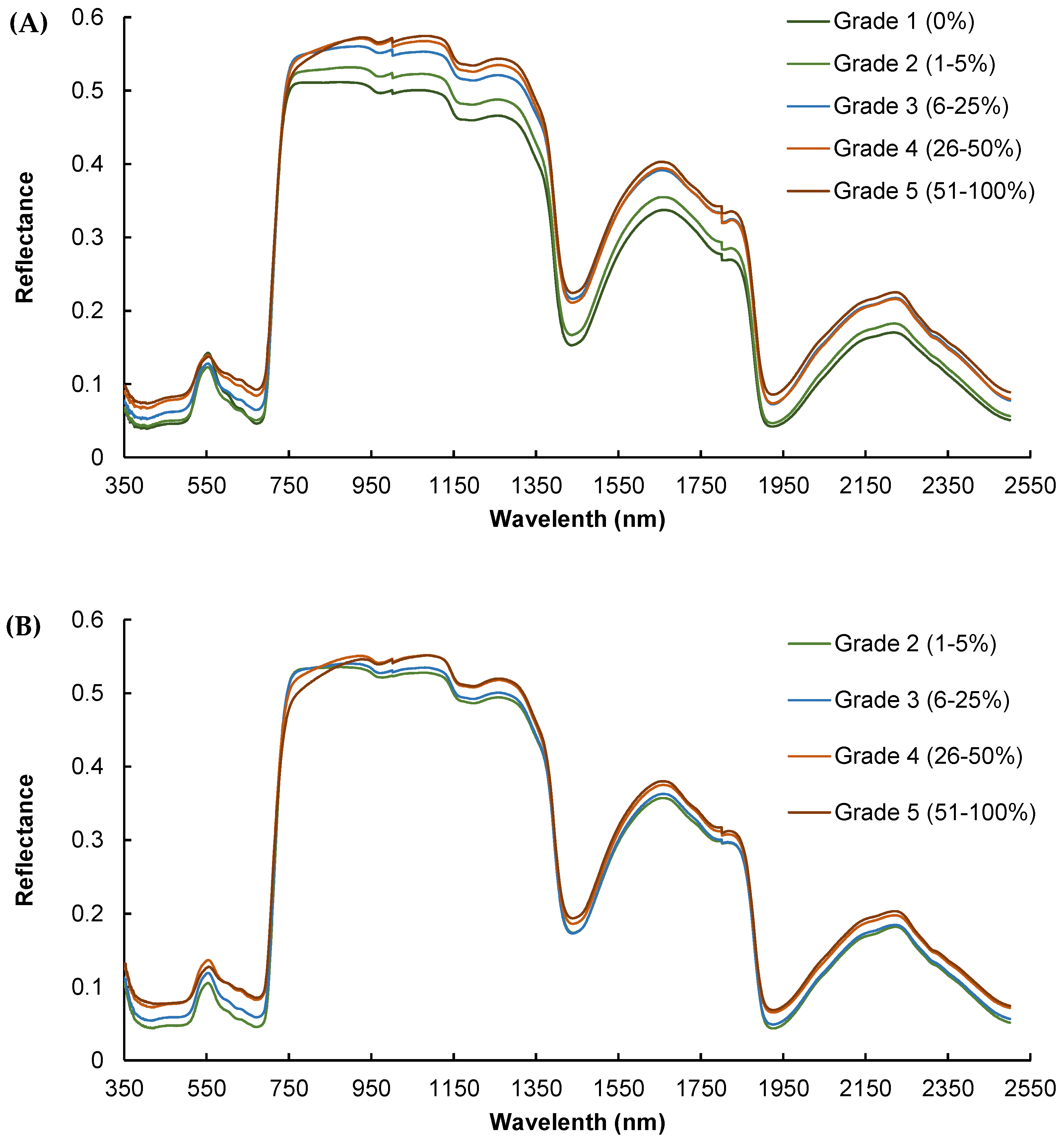
Significant differences were noted in the mean reflectance spectra across various infestation grades, particularly in the visible and near-infrared spectra (Figure 4). This figure contains five series corresponding to different levels of infection, each with a distinct “Grade” of reflectance. The x-axis represents the wavelength of light, and the y-axis represents the reflectance, measuring how much light is being reflected off a surface at each wavelength. As the level of infestation increases, there is a general trend of increased reflectance.
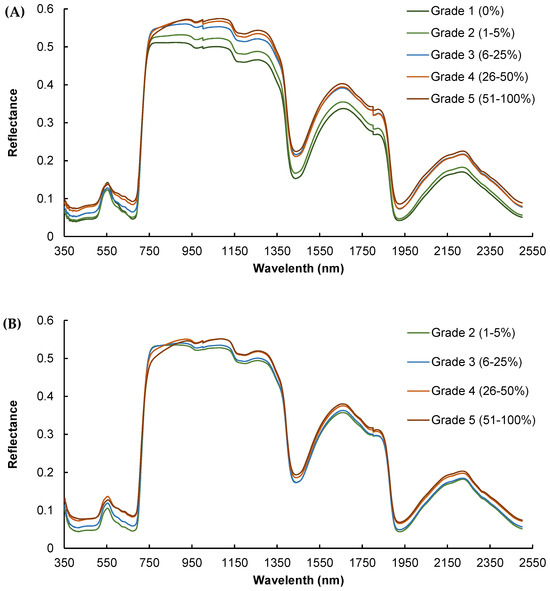
Figure 4.
Mean reflectance spectra of leaves at different grades/levels of infestation by powdery mildew for (A) calibration dataset in July and (B) validation dataset in September.
Generally, the higher the degree of infection, the greater the difference in the spectral signature compared to non-infected leaves, with infested leaves exhibiting higher mean reflectance than healthy ones, which can be attributed to chlorophyll degradation (visible spectrum) and water loss (near-infrared spectrum) due to tissue damage. Furthermore, the separation between the curves is not uniform across all wavelengths, especially around 450 nm and 700 nm for visible and 1050 nm, 1425 nm, 1650 nm and 2250 nm for infrared, indicating that the fungus has a more pronounced effect on reflectance at specific wavelengths. The reflectance measurements for calibration recorded in July, marking the onset of disease detection, showed clear differences in spectral signatures, especially between severity levels 1 and 5 within the 750–1300 nm range (Figure 4A).
The spectral dataset for validation, collected in September when the disease was more widespread, also demonstrated clear differences in spectral signatures, although these differences were not as pronounced as those observed in July (Figure 4B). It can be noted that there was no “Grade 1” (infestation level: 0%) in September because all the leaves were infected due to the aforementioned vineyard management. It can be observed that the separation between the curves is smaller in September than in July, with lower mean reference values for all wavelengths. In addition, the slope of the spectral signature between 750 and 850 nm was notably different, being steeper for grades 4 and 5 compared to grades 1, 2, and 3. This pattern was even more pronounced in the validation dataset (Figure 4A).
3.3. Predictive Ability of the Level of Infestation (PLS Model)
The results of LOOCV yielded a high coefficient of determination value (R2 = 0.74) and a root mean square error of cross-validation (RMSECV) of 12.1%, while the values for the independent validation were R2 = 0.71 and an RMSEP of 12.9% (Table 3). Moreover, the high RPD values for LOOCV and independent validation (2.6 and 2.7, respectively) underscored the robustness of the prediction.

Table 3.
Performance of PLSR model.
3.4. Predicted vs. Observed Level of Infestation
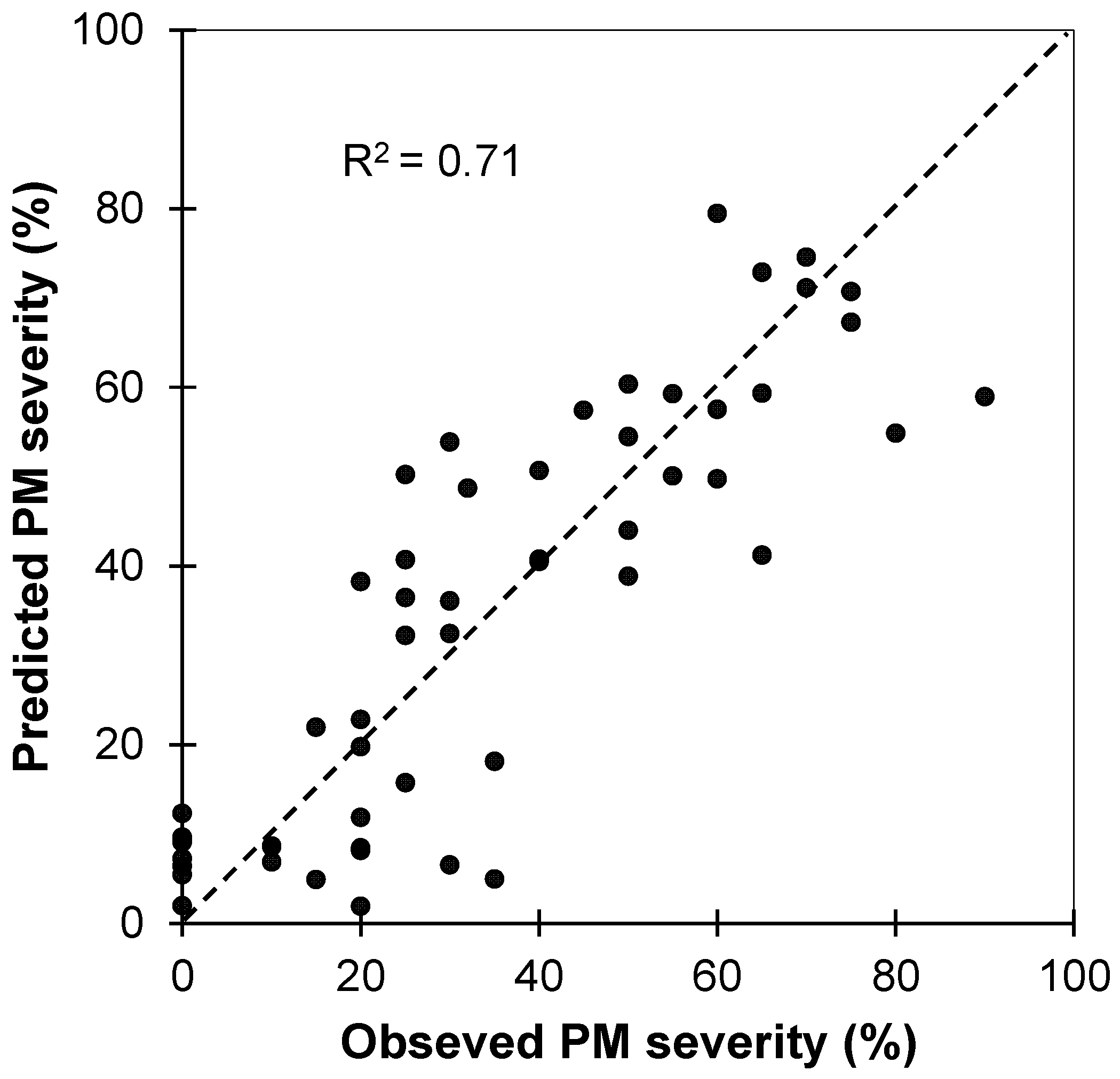
The relationship between observed and predicted levels of infestation for powdery mildew in the test dataset (n = 54) from the PLSR model fitted using the calibration spectra dataset (Figure 5) showed a satisfactory accuracy for predictions (R2 = 0.71 and a RMSEP of 12.9%), corroborated by a RPD = 2.7, confirming the high quality and future applicability of the results as it can be considered a reflection of excellent predictive ability [49].
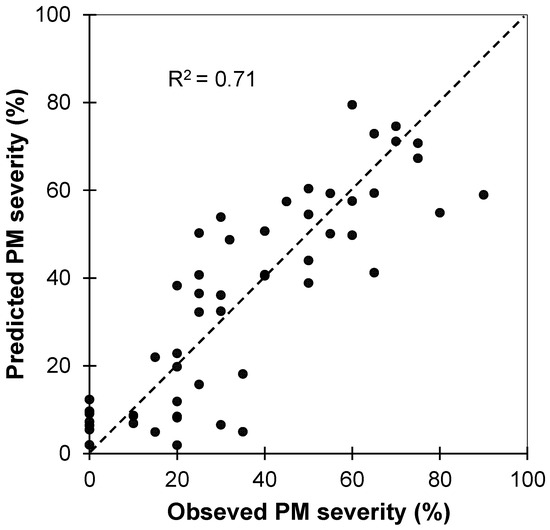
Figure 5.
Observed versus predicted severity degree/levels of powdery mildew (PM) infestation derived from the PLS regression. These results correspond to the independent validation dataset. The dotted line is 1:1.
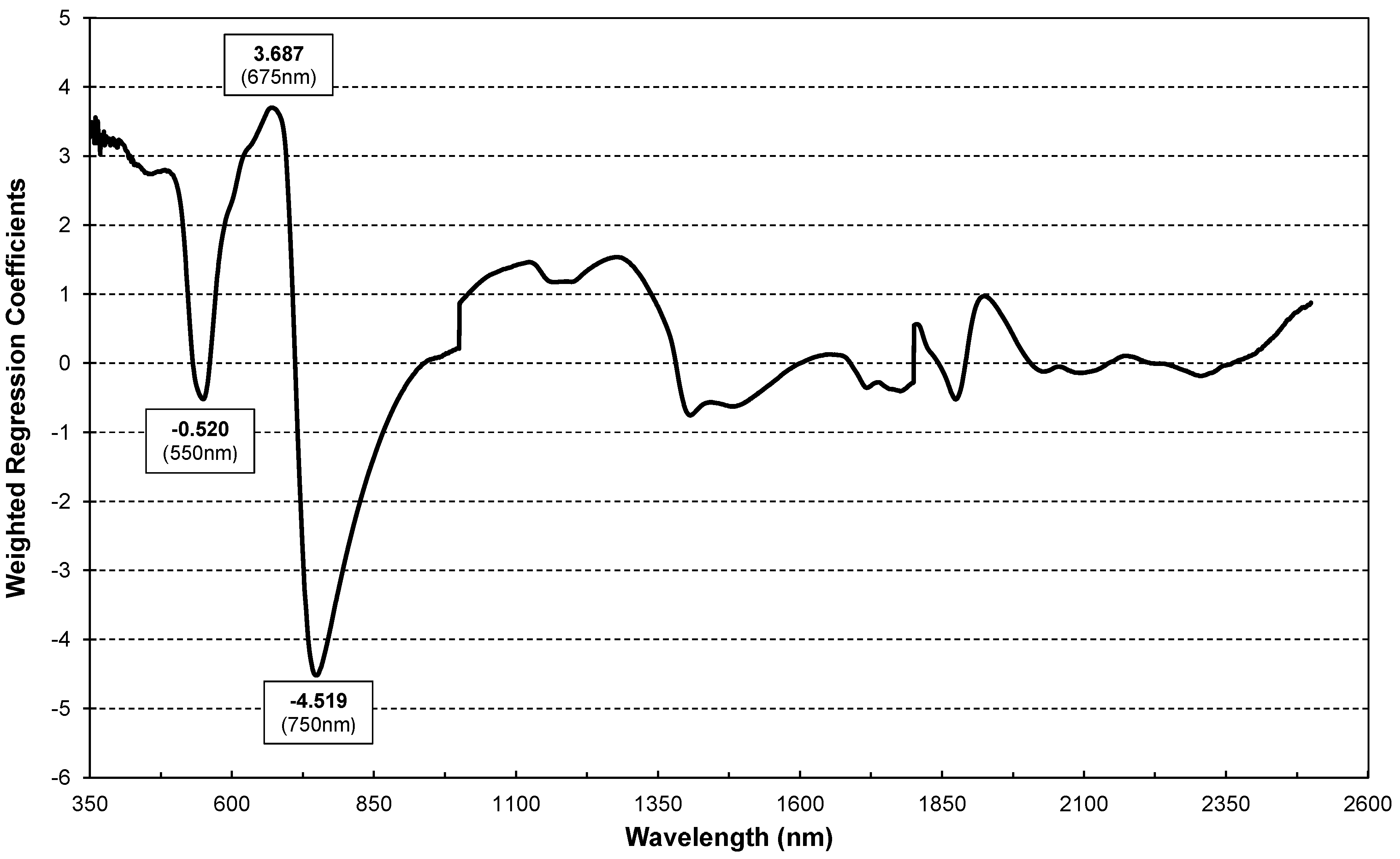
Figure 6 shows the Weighted Regression Coefficients of the PLS regression modes derived from the calibration dataset (10 July). Higher regression coefficients (Figure 6) are centred at 550 (green), 675nm (red) and 750 (red edge) nm, suggesting that the disease impacts chlorophyll assimilation [50]. Peaks at 1400 and 1900 nm highlight water metabolism disturbances due to powdery mildew.
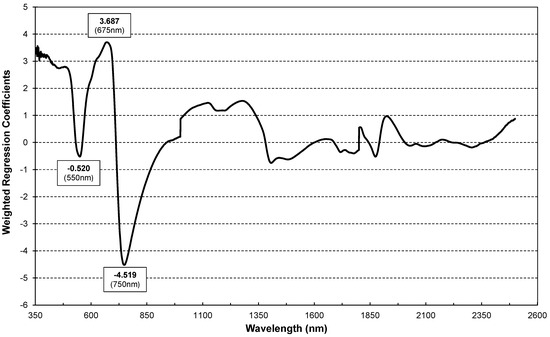
Figure 6.
Weighted Regression Coefficients of PLS regression for estimation of infestation levels using reflectance values of dataset 1 (31 July) as prediction variables.
3.5. Powdery Mildew Vegetation Index (PMVI)
The sensitivity of the 675 nm and 755 nm bands (Figure 6) to disease severity suggests their potential for early detecting and estimating powdery mildew damage, leading to the formulation of the Powdery Mildew Vegetation Index (PMVI) as follows:
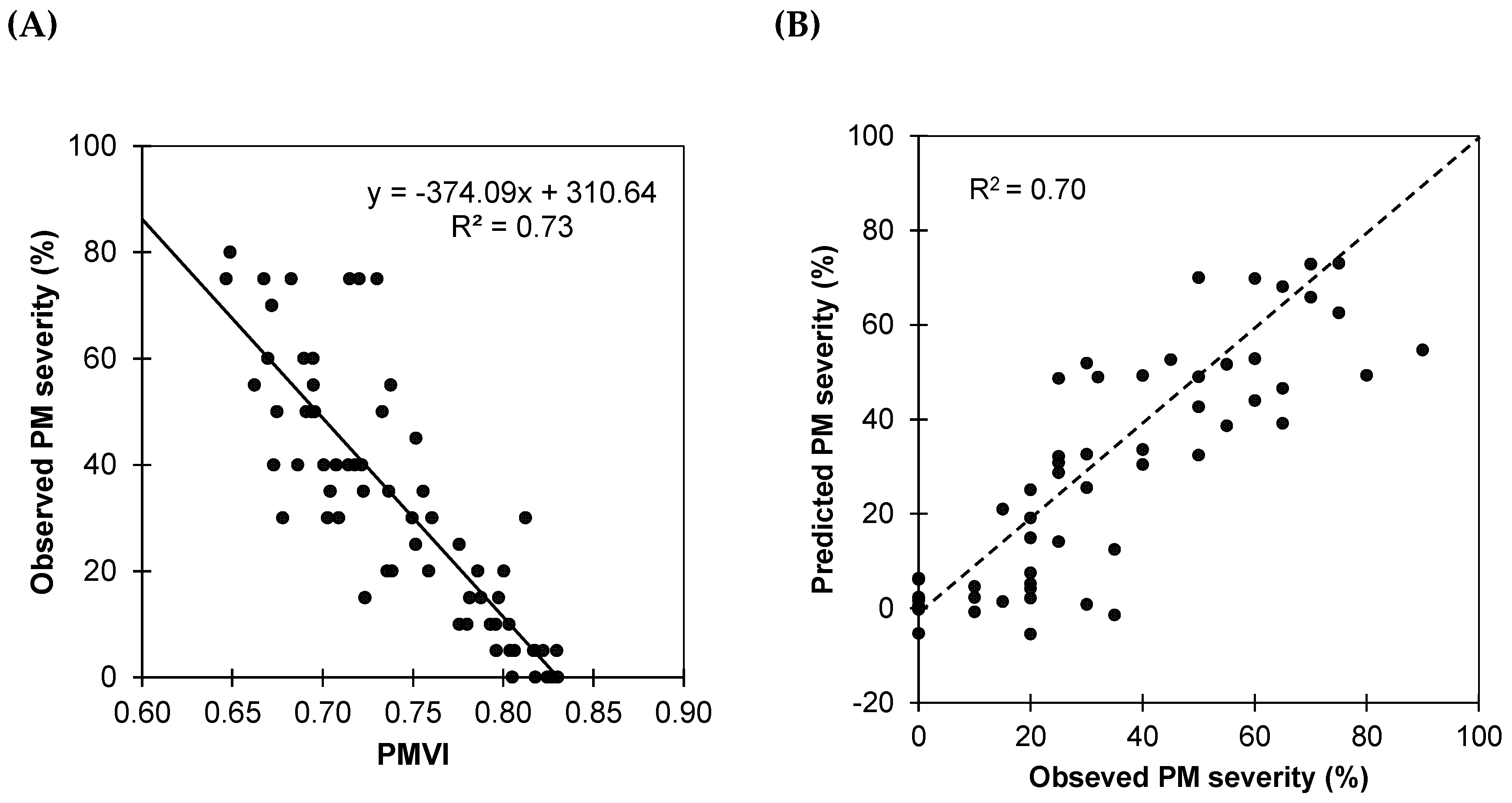
where R755 and R675 are reflectance at 755 and 675 nm, respectively. Reflectances R755 and R675 were calculated as average reflectances of ranges 740–770 nm and 660–690 nm, respectively. An ordinary linear regression analysis (OLS) using the PMVI as the predictor (Figure 7A) showed a significant correlation between PM severity and PMVI (R2 = 0.73), demonstrating the potential of the new index to detect disease severity in vineyards. The negative linear relationship between PMVI and observed severity indicates that as PMVI increases, the observed severity of infestation decreases.
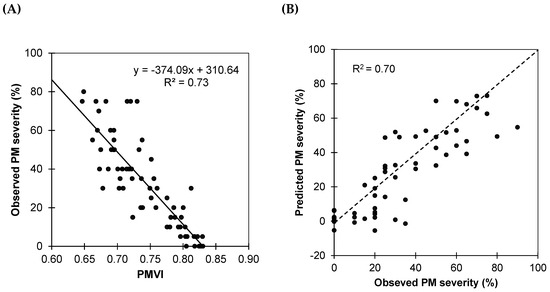
Figure 7.
(A) Ordinary linear regression (OLS) between PMVI and severity of infestation of powdery mildew. Data correspond to calibration dataset D1 (31 July). (B) Observed versus predicted severity of powdery mildew derived from the OLS model. Results correspond to validation dataset D2 (10 September). The dotted line is 1:1.
In order to test the predictive accuracy of PMVI, the fitted model was validated against the second dataset (Figure 7B). Although there is some dispersion, the general trend indicates that the model has an acceptable accuracy, as most of the points are clustered close to the line of equality, and the RMSE of 14.63% confirms PMVI’s utility in the prediction of the severity.
4. Discussion
This research highlights the potential of spectroscopy in the spectral range of 350 to 2500 nm as a rapid and non-invasive disease detection technique. This approach is well aligned with the current needs of precision agriculture and sustainable wine-growing practices. The comparative analysis between the spectral data collected in July and September offers a critical insight into the temporal dynamics of PM infestation. In July, the spectroscopy data marked the onset of PM detection, with clear spectral differences noted in the reflectance data between the infected and non-infected leaves. This finding is of particular importance as it suggests the ability of spectroscopic methods to detect subtle changes in the vine leaves that precede clear visible symptoms of infestation.
Consequently, this early detection could enable viticulturists to implement preventative rather than reactive measures. This shift can potentially improve sustainability and reduce the extent of infection and associated costs. The spectral signatures in July, especially the significant peaks at wavelengths corresponding to chlorophyll absorption and water content (Figure 4A), indicate the initial physiological responses of the vines to the pathogen. In the progression towards September, these spectral differences become less pronounced (Figure 4B), with the separation between the curves being smaller in September than in July, with lower mean reference values for all wavelengths. This shift in the spectral characteristics could be influenced by the phenological stage of the vines, which is pivotal when considering the application of spectral indices for monitoring over an entire growing season [51].
Moreover, the reflectance of mature leaves may be lower due to several factors, including structural, chemical and photochemical changes. As the leaf ages, there is a variation in the ratio of chlorophyll to carotenoids, changes in the leaf cuticle and adaptations to light exposure that can affect how the leaf reflects light [52,53,54]. In addition, the consistency in the Weighted Regression Coefficients of PLS across both datasets underscores the reliability of spectral reflectance as an indicator of PM infestation, irrespective of the temporal development of the disease. There are similarities in the spectral patterns between July and September. These similarities imply that the spectral signature of PM infestation, once established, can be a reliable metric and could serve as a continuous monitoring tool throughout the growing season. Although the subtle variations in the slope changes in the spectral signature between 750 and 850 nm highlight the importance of continuous monitoring to capture the full spectrum of the impact of the disease, the reflectance centred at the red (R675) and NIR (R755) suggest that this spectral region remains a robust indicator of PM severity over time. Furthermore, the consistency in the PLS Weighted Regression Coefficients across datasets highlights the reliability of the spectral reflectance as a PM infestation indicator, regardless of the progression. The similarities in spectral patterns between July and September indicate that the disease can be spectrally monitored throughout the season. In addition, it is important to note that the red (R675) and NIR (R755) reflectance suggest these regions as strong indicators of PM severity over time.
Therefore, introducing the Powdery Mildew Vegetation Index (PMVI) aims to harness these spectral regions for early detection and estimation of the severity of downy mildew in vineyards. This index has a standardised equation that is easy to apply, similar to other well-known and helpful indices in agriculture, such as the Normalised Difference Red Edge Index (NDRE) [55] and Normalised Difference Vegetation Index (NDVI) [56]. In addition, the PMVI bands are located in an intermediate region between those used for NDVI and NDRE, highlighting the importance of this region in vegetation assessment. This finding is not surprising, as this region concentrates on indicators for estimating plant health, photosynthetic capacity, changes in chlorophyll content and the nutritional status of plants [57,58]. Nevertheless, PMVI specifically uses the 675 nm and 755 nm wavelengths, as opposed to those traditionally used for NDRE, which has a red-edge band (700 nm to 730 nm) and a band in the near-infrared (NIR) (often near 780 nm or more), and for NDVI, which uses one band in the red (approximately 640 nm to 680 nm) and one band in the NIR (approximately 800 nm to 900 nm). Thus, the bands chosen for PMVI lie between the specific spectral sensitivities of NDRE and NDVI, taking advantage of the transition between strong chlorophyll absorption in the red and increased reflectance in the NIR.
Concerning PM, Pithan et al. [59] identified distinct spectral shifts in grapevine leaves associated with various fungal diseases, and Atanassova et al. [60] identified spectral differences between healthy and infected plants, noting the most significant disparities occurred in a distinct spectral region, specifically between 540 and 680 nm, diverging from the range examined in our study. Oberti et al. [61] investigated the enhancement of grapevine powdery mildew detection via multispectral imaging from various angles, finding that the detection sensitivity increased significantly, from 9% at 0° to 73% at 60° for symptoms in early to middle stages, indicating that angle-based imaging could provide a more focused method for disease management. Bierman et al. [62] developed an automated imaging and analysis system to evaluate the severity of Erysiphe necator on grape leaf disks, employing a 46-megapixel RGB camera and a convolutional neural network (CNN) with over 94% accuracy and, as in this study, they successfully differentiated between susceptible, moderately resistant and highly resistant samples. Nevertheless, these investigations primarily concentrated on the visible (VIS) or near-infrared (NIR) spectral range rather than exploring the complete spectral signature range from 350 to 2500 nm. Some studies have embraced a broader spectral range, such as Knauer et al. [63], who introduced an advanced spatial–spectral segmentation approach using hyperspectral imaging to detect powdery mildew infection levels, effectively distinguishing between healthy, infected, and severely diseased levels. Pérez-Roncal et al. [64] also utilised hyperspectral imaging, achieving 85.33% accuracy in disease identification through image feature extraction and Partial Least Squares Discriminant Analysis (PLS-DA), adeptly differentiating between healthy and infected samples. Nevertheless, these authors focused on grapevine bunches rather than leaves.
This research thus makes a novel contribution to the fields of plant pathology and agronomy, marking the first study to examine the impact of powdery mildew on the spectral signature range from 350 to 2500 nm of grapevine leaves across different infection levels and throughout various periods of the year. The results of the present study reveal that spectral reflectance data underscore pronounced differences among various infestation grades, with a notable increase in reflectance as the infestation severity escalated. The results are in harmony with those of Pérez-Roncal et al. [64], who observed that infected pixels exhibited higher reflectance values than those from healthy samples, although their research was conducted on bunches, not leaves. This trend is further corroborated by other studies [65], which highlighted the effectiveness of spectral and textural data in accurately identifying grapevine diseases. The present research extends these insights, introducing the PMVI and demonstrating that specific wavelengths especially indicate powdery mildew infestation, likely attributable to chlorophyll degradation and water loss in infected leaves. Thus, these findings not only enrich the existing knowledge on spectral responses to varying PM levels using spectrometry from 350 to 2500 nm but also pinpoint specific wavelengths associated with PM infestation, augmenting the existing literature on the non-destructive evaluation of vineyard health.
Future research should focus on developing comparative studies across grape varieties. Resistance to PM depends on the cultivar [66], and these experiments could uncover variety-specific spectral disease indicators, potentially enabling more precise monitoring strategies. In addition, the temporal dynamics of the disease should be further explored to understand how spectral reflectance evolves over vine growth stages and seasons, potentially refining disease management timing. Finally, integrating spectral analysis with precision agriculture technologies like drone-based monitoring and machine learning could improve scalable, real-time disease detection, significantly boosting vineyard management efficiency. Portable sensors designed to compute the PMVI could play a crucial role in achieving this goal.
5. Conclusions
This research validates the efficacy of spectroscopy in the spectral range of 350 to 2500 as a fast and non-invasive disease detection technique for early detection of powdery mildew infestation levels in grapevines, matching the current needs in precision farming and sustainable wine-growing practices. This study underscores its reliability through statistical, spectral, and predictive analyses. Spectral signatures of non-infected and infected leaves were obtained using a FieldSpec 4 spectroradiometer equipped with a contact probe with a tungsten halogen bulb to collect reflectance data at wavelengths between 350 and 2500 nm. Spectral reflectance analysis revealed significant differences in the mean spectra between various infestation grades, particularly in the visible and near-infrared regions, with these discrepancies becoming more marked with increasing infection severity. The Partial Least Squares Regression (PLSR) model demonstrated substantial predictive capability, with a coefficient of determination R2 = 0.74 and a Root Mean Square Error of Cross-Validation (RMSECV) of 12.1% for the calibration set, and an R2 = 0.71 and a Root Mean Square Error of Prediction (RMSEP) of 12.9% for independent validation. The model’s robustness was further evidenced by Relative Percent Difference (RPD) values of 2.6 and 2.7 for Leave-One-Out Cross-Validation and independent validation, respectively.
In addition, key spectral regions indicative of disease impact, particularly on chlorophyll assimilation and water metabolism, were identified, highlighting the importance of green (550 nm) and red edge (750 nm) bands, as well as wavelengths at 1400 and 1900 nm. A new vegetation index was introduced using this information: the Powdery Mildew Vegetation Index (PMVI), derived from reflectance at 675 and 755 nm, potentially facilitating the disease’s rapid and non-destructive prediction. A significant linear relationship between PM severity and PMVI was confirmed, with potential practical application in vineyard management. These findings suggest that spectroscopy, enhanced by PLS regression and PMVI, offers a promising tool for early disease detection and management in viticulture. This approach not only supports farmers in timely treatment application but also holds the potential for developing portable sensors for continuous vineyard monitoring. Future work will extend to different grape cultivars and management practices, aiming to generalise the calibration’s applicability, ultimately aiding in improved grape production and contributing to food security.
Author Contributions
Conceptualisation, S.V., E.B. and J.R.R.-P.; methodology, S.V., E.B. and J.R.R.-P.; software, S.V., J.R.R.-P. and D.P.-O.; validation, S.V., E.B., J.R.R.-P. and D.P.-O.; formal analysis, S.V. and J.R.R.-P.; investigation, S.V. and J.R.R.-P.; resources, S.V., E.B., J.A.R. and J.R.R.-P.; data curation, S.V. and J.R.R.-P.; writing—original draft preparation, S.V. and J.R.R.-P.; writing—review and editing, S.V., E.B. and J.R.R.-P.; visualisation, S.V., J.R.R.-P. and D.P.-O.; supervision, E.B., J.A.R. and J.R.R.-P.; project administration, E.B. and J.A.R.; funding acquisition, E.B., J.A.R. and J.R.R.-P. All authors have read and agreed to the published version of the manuscript.
Funding
The research was part of the EVARRES project Evaluación de variedades de vid resistentes a enfermedades (Assessment of Disease-Resistant Grape Varieties, code 2018/879) and VITICOM project Alternativas para una viticultura competitiva y sostenible (Alternatives for Competitive and Sustainable Viticulture, code 2021/1168), both financially supported by the Centre for the Development of Industrial Technology of Spain (CDTI), and it was supported by the company VITICAMPO, SL (grant id: 2021/00009/001; T132). Dr. Sergio Vélez’s contract has been supported by the Iberdrola Foundation and the European Commission under the Marie Skłodowska-Curie Actions (MSCA)—E4F, part of the Horizon 2020 program (Grant Agreement No 101034297, https://doi.org/10.3030/101034297, accessed on 20 March 2024).
Data Availability Statement
Data are available upon reasonable request.
Acknowledgments
We would like to thank “Agromillora” for their collaboration.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Singh, V.; Sharma, N.; Singh, S. A Review of Imaging Techniques for Plant Disease Detection. Artif. Intell. Agric. 2020, 4, 229–242. [Google Scholar] [CrossRef]
- Lorrain, B.; Ky, I.; Pasquier, G.; Jourdes, M.; Dubrana, L.G.; Gény, L.; Rey, P.; Donèche, B.; Teissedre, P.-L. Effect of Esca Disease on the Phenolic and Sensory Attributes of Cabernet Sauvignon Grapes, Musts and Wines. Aust. J. Grape Wine Res. 2012, 18, 64–72. [Google Scholar] [CrossRef]
- Sawyer, E.; Laroche-Pinel, E.; Flasco, M.; Cooper, M.L.; Corrales, B.; Fuchs, M.; Brillante, L. Phenotyping Grapevine Red Blotch Virus and Grapevine Leafroll-Associated Viruses before and after Symptom Expression through Machine-Learning Analysis of Hyperspectral Images. Front. Plant Sci. 2023, 14, 1117869. [Google Scholar] [CrossRef] [PubMed]
- Lopez Pinar, A.; Rauhut, D.; Ruehl, E.; Buettner, A. Effects of Bunch Rot (Botrytis cinerea) and Powdery Mildew (Erysiphe necator) Fungal Diseases on Wine Aroma. Front. Chem. 2017, 5, 20. [Google Scholar] [CrossRef]
- Bois, B.; Zito, S.; Calonnec, A. Climate v.s. Grapevine Pests and Diseases Worldwide: The First Results of a Global Survey. OENO One 2017, 51, 133. [Google Scholar] [CrossRef]
- Gadoury, D.M.; Cadle-Davidson, L.; Wilcox, W.F.; Dry, I.B.; Seem, R.C.; Milgroom, M.G. Grapevine Powdery Mildew (Erysiphe necator): A Fascinating System for the Study of the Biology, Ecology and Epidemiology of an Obligate Biotroph: Grapevine Powdery Mildew. Mol. Plant Pathol. 2012, 13, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Stummer, B.E.; Francis, I.L.; Markides, A.J.; Scott, E.S. The Effect of Powdery Mildew Infection of Grape Berries on Juice and Wine Composition and on Sensory Properties of Chardonnay Wines. Aust. J. Grape Wine Res. 2003, 9, 28–39. [Google Scholar] [CrossRef]
- Scott, E.S. 2019 Daniel McAlpine Memorial Lecture. Grapevine Powdery Mildew: From Fundamental Plant Pathology to New and Future Technologies. Australas. Plant Pathol. 2021, 50, 1–6. [Google Scholar] [CrossRef]
- Kunova, A.; Pizzatti, C.; Saracchi, M.; Pasquali, M.; Cortesi, P. Grapevine Powdery Mildew: Fungicides for Its Management and Advances in Molecular Detection of Markers Associated with Resistance. Microorganisms 2021, 9, 1541. [Google Scholar] [CrossRef]
- Qiu, W.; Feechan, A.; Dry, I. Current Understanding of Grapevine Defense Mechanisms against the Biotrophic Fungus (Erysiphe necator), the Causal Agent of Powdery Mildew Disease. Hortic. Res. 2015, 2, 15020. [Google Scholar] [CrossRef]
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A Review of Advanced Techniques for Detecting Plant Diseases. Comput. Electron. Agric. 2010, 72, 1–13. [Google Scholar] [CrossRef]
- Fang, Y.; Ramasamy, R.P. Current and Prospective Methods for Plant Disease Detection. Biosensors 2015, 5, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Istiak, M.d.A.; Syeed, M.M.M.; Hossain, M.S.; Uddin, M.F.; Hasan, M.; Khan, R.H.; Azad, N.S. Adoption of Unmanned Aerial Vehicle (UAV) Imagery in Agricultural Management: A Systematic Literature Review. Ecol. Inform. 2023, 78, 102305. [Google Scholar] [CrossRef]
- Ariza-Sentís, M.; Vélez, S.; Martínez-Peña, R.; Baja, H.; Valente, J. Object Detection and Tracking in Precision Farming: A Systematic Review. Comput. Electron. Agric. 2024, 219, 108757. [Google Scholar] [CrossRef]
- Vélez, S.; Ariza-Sentís, M.; Valente, J. Mapping the Spatial Variability of Botrytis Bunch Rot Risk in Vineyards Using UAV Multispectral Imagery. Eur. J. Agron. 2023, 142, 126691. [Google Scholar] [CrossRef]
- Albetis, J.; Duthoit, S.; Guttler, F.; Jacquin, A.; Goulard, M.; Poilvé, H.; Féret, J.-B.; Dedieu, G. Detection of Flavescence Dorée Grapevine Disease Using Unmanned Aerial Vehicle (UAV) Multispectral Imagery. Remote Sens. 2017, 9, 308. [Google Scholar] [CrossRef]
- MacDonald, S.L.; Staid, M.; Staid, M.; Cooper, M.L. Remote Hyperspectral Imaging of Grapevine Leafroll-Associated Virus 3 in Cabernet Sauvignon Vineyards. Comput. Electron. Agric. 2016, 130, 109–117. [Google Scholar] [CrossRef]
- Pérez-Roncal, C.; Arazuri, S.; Lopez-Molina, C.; Jarén, C.; Santesteban, L.G.; López-Maestresalas, A. Exploring the Potential of Hyperspectral Imaging to Detect Esca Disease Complex in Asymptomatic Grapevine Leaves. Comput. Electron. Agric. 2022, 196, 106863. [Google Scholar] [CrossRef]
- Ariza-Sentís, M.; Vélez, S.; Valente, J. BBR: An Open-Source Standard Workflow Based on Biophysical Crop Parameters for Automatic Botrytis cinerea Assessment in Vineyards. SoftwareX 2023, 24, 101542. [Google Scholar] [CrossRef]
- Lu, G.; Fei, B. Medical Hyperspectral Imaging: A Review. J. Biomed. Opt. 2014, 19, 010901. [Google Scholar] [CrossRef]
- Kendler, S.; Mano, Z.; Aharoni, R.; Raich, R.; Fishbain, B. Hyperspectral Imaging for Chemicals Identification: A Human-Inspired Machine Learning Approach. Sci. Rep. 2022, 12, 17580. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Feng, Y.; Guo, A.; Huang, W.; Cai, Z.; Dong, Y.; Guo, J.; Qian, B.; Hao, Z.; Chen, G.; et al. Detection of Rubber Tree Powdery Mildew from Leaf Level Hyperspectral Data Using Continuous Wavelet Transform and Machine Learning. Remote Sens. 2023, 16, 105. [Google Scholar] [CrossRef]
- Ali, M.M.; Bachik, N.A.; Muhadi, N.A.; Tuan Yusof, T.N.; Gomes, C. Non-Destructive Techniques of Detecting Plant Diseases: A Review. Physiol. Mol. Plant Pathol. 2019, 108, 101426. [Google Scholar] [CrossRef]
- Dambergs, R.; Gishen, M.; Cozzolino, D. A Review of the State of the Art, Limitations, and Perspectives of Infrared Spectroscopy for the Analysis of Wine Grapes, Must, and Grapevine Tissue. Appl. Spectrosc. Rev. 2015, 50, 261–278. [Google Scholar] [CrossRef]
- Conrad, A.O.; Li, W.; Lee, D.-Y.; Wang, G.-L.; Rodriguez-Saona, L.; Bonello, P. Machine Learning-Based Presymptomatic Detection of Rice Sheath Blight Using Spectral Profiles. Plant Phenomics 2020, 2020, 8954085. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Feng, X.; Wang, J.; Liu, F.; He, Y.; Zhou, W. Mid-Infrared Spectroscopy Combined with Chemometrics to Detect Sclerotinia Stem Rot on Oilseed Rape (Brassica napus L.) Leaves. Plant Methods 2017, 13, 39. [Google Scholar] [CrossRef] [PubMed]
- Vallejo-Pérez, M.R.; Sosa-Herrera, J.A.; Navarro-Contreras, H.R.; Álvarez-Preciado, L.G.; Rodríguez-Vázquez, Á.G.; Lara-Ávila, J.P. Raman Spectroscopy and Machine-Learning for Early Detection of Bacterial Canker of Tomato: The Asymptomatic Disease Condition. Plants 2021, 10, 1542. [Google Scholar] [CrossRef]
- Sylvain, T.; Cecile, L.-G. Disease Identification: A Review of Vibrational Spectroscopy Applications. In Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2018; Volume 80, pp. 195–225. ISBN 978-0-444-64048-2. [Google Scholar]
- Prechsl, U.E.; Mejia-Aguilar, A.; Cullinan, C.B. In Vivo Spectroscopy and Machine Learning for the Early Detection and Classification of Different Stresses in Apple Trees. Sci. Rep. 2023, 13, 15857. [Google Scholar] [CrossRef]
- Khaled, A.Y.; Abd Aziz, S.; Bejo, S.K.; Nawi, N.M.; Seman, I.A.; Onwude, D.I. Early Detection of Diseases in Plant Tissue Using Spectroscopy—Applications and Limitations. Appl. Spectrosc. Rev. 2018, 53, 36–64. [Google Scholar] [CrossRef]
- Altangerel, N.; Ariunbold, G.O.; Gorman, C.; Alkahtani, M.H.; Borrego, E.J.; Bohlmeyer, D.; Hemmer, P.; Kolomiets, M.V.; Yuan, J.S.; Scully, M.O. In Vivo Diagnostics of Early Abiotic Plant Stress Response via Raman Spectroscopy. Proc. Natl. Acad. Sci. USA 2017, 114, 3393–3396. [Google Scholar] [CrossRef]
- Maimaitiyiming, M.; Ghulam, A.; Bozzolo, A.; Wilkins, J.L.; Kwasniewski, M.T. Early Detection of Plant Physiological Responses to Different Levels of Water Stress Using Reflectance Spectroscopy. Remote Sens. 2017, 9, 745. [Google Scholar] [CrossRef]
- Eh Teet, S.; Hashim, N. Recent Advances of Application of Optical Imaging Techniques for Disease Detection in Fruits and Vegetables: A Review. Food Control 2023, 152, 109849. [Google Scholar] [CrossRef]
- Vélez, S.; Barajas, E.; Rubio, J.A.; Vacas, R.; Poblete-Echeverría, C. Effect of Missing Vines on Total Leaf Area Determined by NDVI Calculated from Sentinel Satellite Data: Progressive Vine Removal Experiments. Appl. Sci. 2020, 10, 3612. [Google Scholar] [CrossRef]
- Poblete-Echeverría, C.; Espinace, D.; Sepúlveda-Reyes, D.; Zúñiga, M.; Sanchez, M. Analysis of Crop Water Stress Index (CWSI) for Estimating Stem Water Potential in Grapevines: Comparison between Natural Reference and Baseline Approaches. Acta Hortic. 2017, 1150, 189–194. [Google Scholar] [CrossRef]
- Martínez-Peña, R.; Vélez, S.; Vacas, R.; Martín, H.; Álvarez, S. Remote Sensing for Sustainable Pistachio Cultivation and Improved Quality Traits Evaluation through Thermal and Non-Thermal UAV Vegetation Indices. Appl. Sci. 2023, 13, 7716. [Google Scholar] [CrossRef]
- De Grave, C.; Pipia, L.; Siegmann, B.; Morcillo-Pallarés, P.; Rivera-Caicedo, J.P.; Moreno, J.; Verrelst, J. Retrieving and Validating Leaf and Canopy Chlorophyll Content at Moderate Resolution: A Multiscale Analysis with the Sentinel-3 OLCI Sensor. Remote Sens. 2021, 13, 1419. [Google Scholar] [CrossRef] [PubMed]
- De Bei, R.; Cozzolino, D.; Sullivan, W.; Cynkar, W.; Fuentes, S.; Dambergs, R.; Pech, J.; Tyerman, S. Non-Destructive Measurement of Grapevine Water Potential Using near Infrared Spectroscopy: Measure of Grapevine Water Potential Using NIR. Aust. J. Grape Wine Res. 2011, 17, 62–71. [Google Scholar] [CrossRef]
- Cuq, S.; Lemetter, V.; Kleiber, D.; Levasseur-Garcia, C. Assessing Macro-Element Content in Vine Leaves and Grape Berries of Vitis vinifera by Using near-Infrared Spectroscopy and Chemometrics. Int. J. Environ. Anal. Chem. 2020, 100, 1179–1195. [Google Scholar] [CrossRef]
- González-Caballero, V.; Sánchez, M.-T.; Fernández-Novales, J.; López, M.-I.; Pérez-Marín, D. On-Vine Monitoring of Grape Ripening Using Near-Infrared Spectroscopy. Food Anal. Methods 2012, 5, 1377–1385. [Google Scholar] [CrossRef]
- González-Fernández, A.B.; Sanz-Ablanedo, E.; Gabella, V.M.; García-Fernández, M.; Rodríguez-Pérez, J.R. Field Spectroscopy: A Non-Destructive Technique for Estimating Water Status in Vineyards. Agronomy 2019, 9, 427. [Google Scholar] [CrossRef]
- Baratto, C.; Ambrosio, G.; Faglia, G.; Turina, M. Early Detection of Esca Disease in Asymptomatic Vines by Raman Spectroscopy. IEEE Sens. J. 2022, 22, 23286–23292. [Google Scholar] [CrossRef]
- Junges, A.H.; Almança, M.A.K.; Fajardo, T.V.M.; Ducati, J.R. Leaf Hyperspectral Reflectance as a Potential Tool to Detect Diseases Associated with Vineyard Decline. Trop. Plant Pathol. 2020, 45, 522–533. [Google Scholar] [CrossRef]
- Hill, G.N.; Evans, K.J.; Beresford, R.M.; Dambergs, R.G. Near and Mid-Infrared Spectroscopy for the Quantification of Botrytis Bunch Rot in White Wine Grapes. J. Near Infrared Spectrosc. 2013, 21, 467–475. [Google Scholar] [CrossRef]
- AL-Saddik, H.; Simon, J.-C.; Cointault, F. Development of Spectral Disease Indices for ‘Flavescence Dorée’ Grapevine Disease Identification. Sensors 2017, 17, 2772. [Google Scholar] [CrossRef] [PubMed]
- Galvan, F.E.R.; Pavlick, R.; Trolley, G.; Aggarwal, S.; Sousa, D.; Starr, C.; Forrestel, E.; Bolton, S.; Alsina, M.D.M.; Dokoozlian, N.; et al. Scalable Early Detection of Grapevine Viral Infection with Airborne Imaging Spectroscopy. Phytopathology® 2023, 113, 1439–1446. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization. Efficacy Evaluation of Fungicides: Uncinula necator. EPPO Bull. 2002, 32, 315–318. [Google Scholar] [CrossRef]
- Lorenz, D.H.; Eichhorn, K.W.; Bleiholder, H.; Klose, R.; Meier, U.; Weber, E. Growth Stages of the Grapevine: Phenological Growth Stages of the Grapevine (Vitis vinifera L. Ssp. Vinifera)—Codes and Descriptions According to the Extended BBCH Scale. Aust. J. Grape Wine Res. 1995, 1, 100–103. [Google Scholar] [CrossRef]
- Kawamura, K.; Nishigaki, T.; Tsujimoto, Y.; Andriamananjara, A.; Rabenaribo, M.; Asai, H.; Rakotoson, T.; Razafimbelo, T. Exploring Relevant Wavelength Regions for Estimating Soil Total Carbon Contents of Rice Fields in Madagascar from Vis-NIR Spectra with Sequential Application of Backward Interval PLS. Plant Prod. Sci. 2021, 24, 1–14. [Google Scholar] [CrossRef]
- Baranoski, G.V.G.; Rokne, J.G. A Practical Approach for Estimating the Red Edge Position of Plant Leaf Reflectance. Int. J. Remote Sens. 2005, 26, 503–521. [Google Scholar] [CrossRef]
- Vélez, S.; Rançon, F.; Barajas, E.; Brunel, G.; Rubio, J.A.; Tisseyre, B. Potential of Functional Analysis Applied to Sentinel-2 Time-Series to Assess Relevant Agronomic Parameters at the within-Field Level in Viticulture. Comput. Electron. Agric. 2022, 194, 106726. [Google Scholar] [CrossRef]
- Moncholi-Estornell, A.; Van Wittenberghe, S.; Cendrero-Mateo, M.P.; Alonso, L.; Malenovský, Z.; Moreno, J. Impact of Structural, Photochemical and Instrumental Effects on Leaf and Canopy Reflectance Variability in the 500–600 Nm Range. Remote Sens. 2021, 14, 56. [Google Scholar] [CrossRef]
- Neuwirthová, E.; Kuusk, A.; Lhotáková, Z.; Kuusk, J.; Albrechtová, J.; Hallik, L. Leaf Age Matters in Remote Sensing: Taking Ground Truth for Spectroscopic Studies in Hemiboreal Deciduous Trees with Continuous Leaf Formation. Remote Sens. 2021, 13, 1353. [Google Scholar] [CrossRef]
- Villa, P.; Bolpagni, R.; Pinardi, M.; Tóth, V.R. Leaf Reflectance Can Surrogate Foliar Economics Better than Physiological Traits across Macrophyte Species. Plant Methods 2021, 17, 115. [Google Scholar] [CrossRef] [PubMed]
- Gitelson, A.A.; Merzlyak, M.N. Remote Estimation of Chlorophyll Content in Higher Plant Leaves. Int. J. Remote Sens. 1997, 18, 2691–2697. [Google Scholar] [CrossRef]
- Rouse, W.; Haas, R.H.; Welland, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with Erts. In Proceedings of the 3rd ERTS Symposium, Washington, DC, USA, 10–14 December 1973; NASA: Washington, DC, USA; Volume 10–14, pp. 309–317. [Google Scholar]
- Huang, S.; Tang, L.; Hupy, J.P.; Wang, Y.; Shao, G. A Commentary Review on the Use of Normalised Difference Vegetation Index (NDVI) in the Era of Popular Remote Sensing. J. For. Res. 2021, 32, 1–6. [Google Scholar] [CrossRef]
- Vélez, S.; Martínez-Peña, R.; Castrillo, D. Beyond Vegetation: A Review Unveiling Additional Insights into Agriculture and Forestry through the Application of Vegetation Indices. J 2023, 6, 421–436. [Google Scholar] [CrossRef]
- Pithan, P.A.; Ducati, J.R.; Garrido, L.R.; Arruda, D.C.; Thum, A.B.; Hoff, R. Spectral Characterization of Fungal Diseases Downy Mildew, Powdery Mildew, Black-Foot and Petri Disease on Vitis vinifera Leaves. Int. J. Remote Sens. 2021, 42, 5680–5697. [Google Scholar] [CrossRef]
- Atanassova, S.; Nikolov, P.; Valchev, N.; Masheva, S.; Yorgov, D. Early Detection of Powdery Mildew (Podosphaera xanthii) on Cucumber Leaves Based on Visible and Near-Infrared Spectroscopy; AIP Publishing: Sofia, Bulgaria, 2019; p. 160014. [Google Scholar]
- Oberti, R.; Marchi, M.; Tirelli, P.; Calcante, A.; Iriti, M.; Borghese, A.N. Automatic Detection of Powdery Mildew on Grapevine Leaves by Image Analysis: Optimal View-Angle Range to Increase the Sensitivity. Comput. Electron. Agric. 2014, 104, 1–8. [Google Scholar] [CrossRef]
- Bierman, A.; LaPlumm, T.; Cadle-Davidson, L.; Gadoury, D.; Martinez, D.; Sapkota, S.; Rea, M. A High-Throughput Phenotyping System Using Machine Vision to Quantify Severity of Grapevine Powdery Mildew. Plant Phenomics 2019, 2019, 9209727. [Google Scholar] [CrossRef]
- Knauer, U.; Matros, A.; Petrovic, T.; Zanker, T.; Scott, E.S.; Seiffert, U. Improved Classification Accuracy of Powdery Mildew Infection Levels of Wine Grapes by Spatial-Spectral Analysis of Hyperspectral Images. Plant Methods 2017, 13, 47. [Google Scholar] [CrossRef]
- Pérez-Roncal, C.; López-Maestresalas, A.; Lopez-Molina, C.; Jarén, C.; Urrestarazu, J.; Santesteban, L.G.; Arazuri, S. Hyperspectral Imaging to Assess the Presence of Powdery Mildew (Erysiphe necator) in Cv. Carignan Noir Grapevine Bunches. Agronomy 2020, 10, 88. [Google Scholar] [CrossRef]
- Al-Saddik, H.; Laybros, A.; Billiot, B.; Cointault, F. Using Image Texture and Spectral Reflectance Analysis to Detect Yellowness and Esca in Grapevines at Leaf-Level. Remote Sens. 2018, 10, 618. [Google Scholar] [CrossRef]
- Cadle-Davidson, L.; Chicoine, D.R.; Consolie, N.H. Variation Within and Among Vitis Spp. for Foliar Resistance to the Powdery Mildew Pathogen Erysiphe necator. Plant Dis. 2011, 95, 202–211. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).