Effects of Intercropping and Nitrogen Application on Soil Fertility and Microbial Communities in Peanut Rhizosphere Soil
Abstract
:1. Introduction
2. Materials and Methods
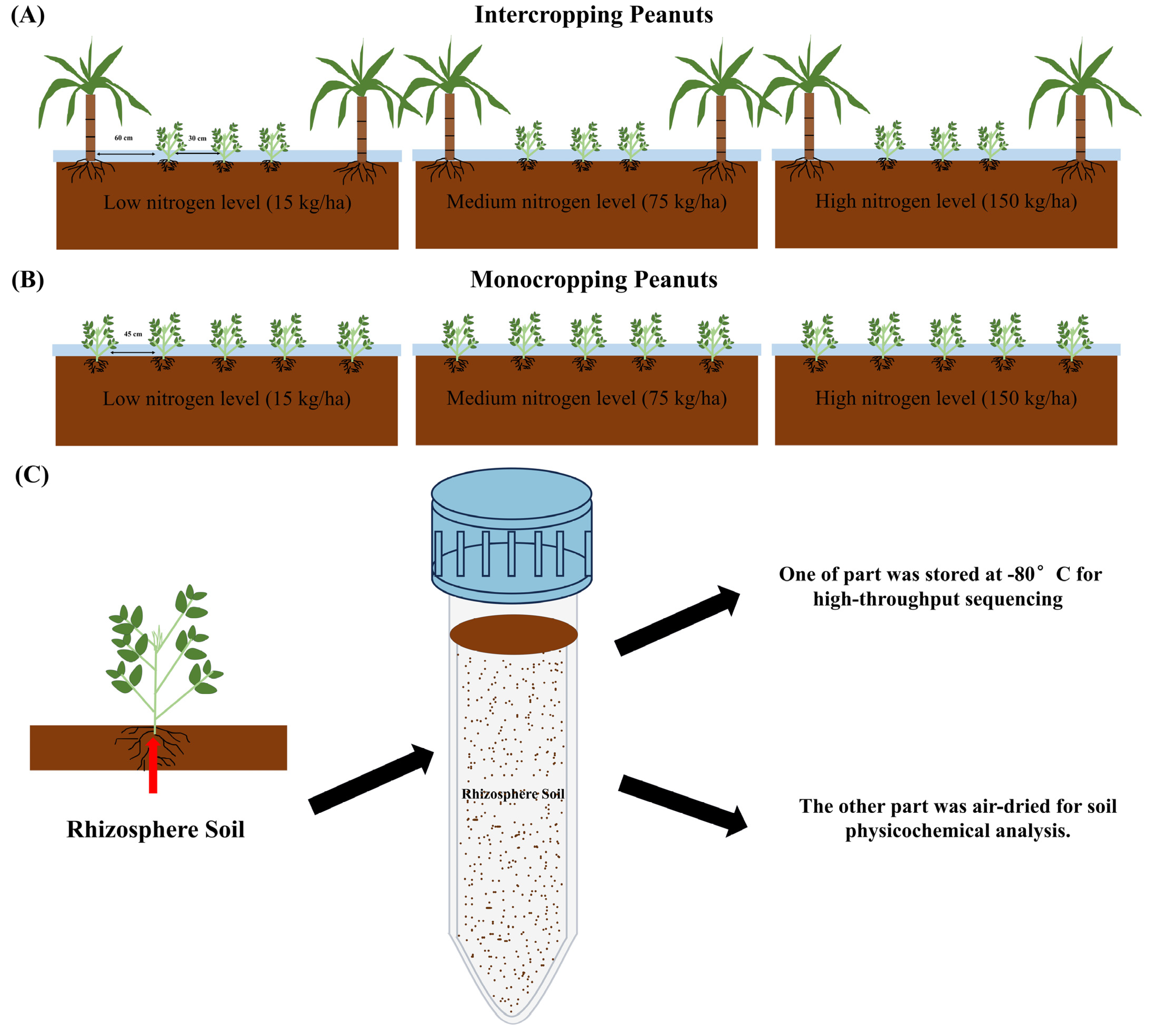
2.1. Study Site and Soil Collection
2.2. Physicochemical Characterization of Soil
2.3. Soil Metagenomic Sequencing
2.4. Illumina MiSeq Sequencing and Data Analysis
3. Results
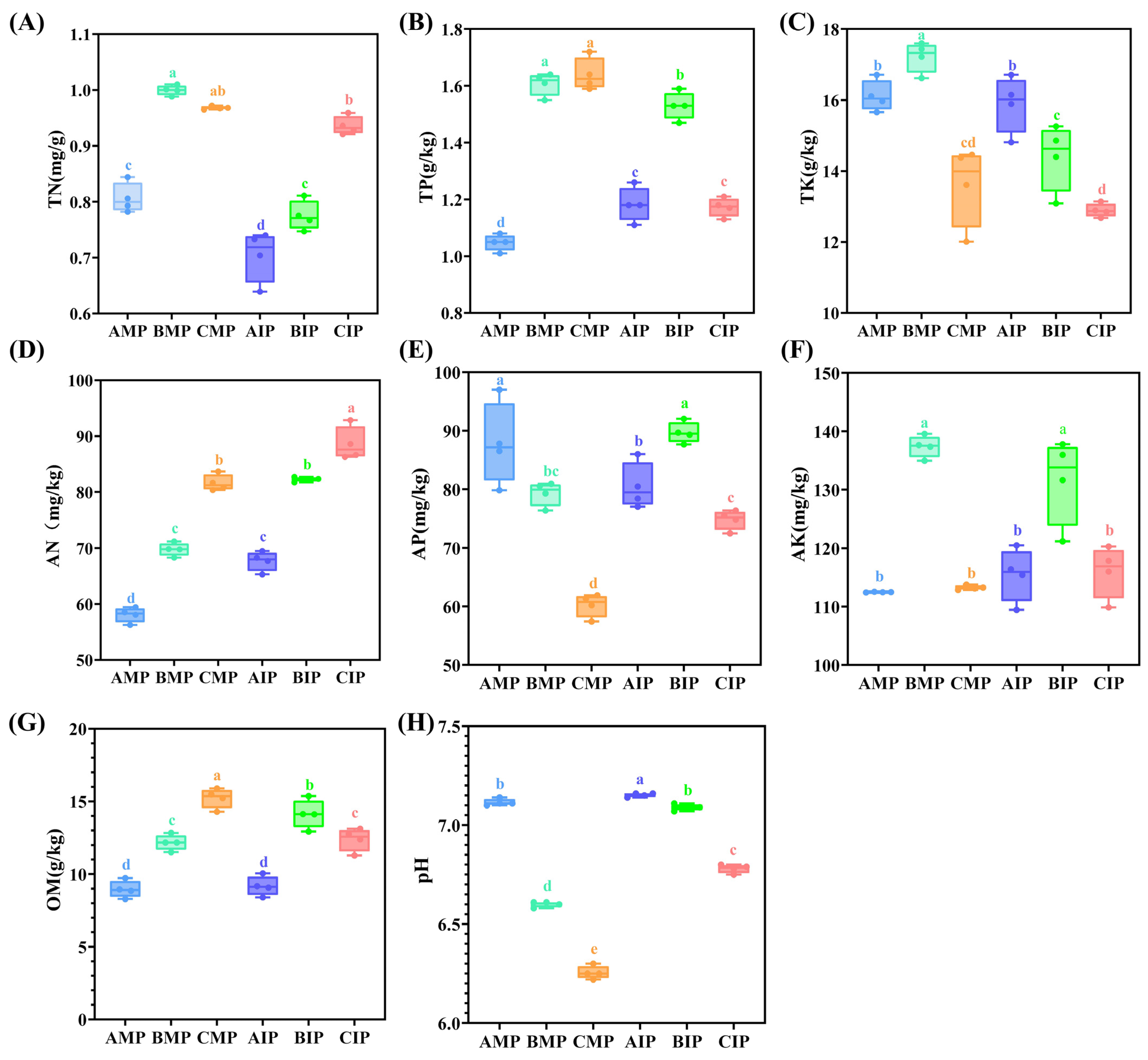
3.1. Physicochemical Characteristics of Soil
3.2. High Throughput Sequencing Analyses
3.3. Analyses of the Rhizosphere Soil Microbial Composition and Diversity
3.4. Functional Pathways of Rhizosphere Soil Microbes
3.5. The Impact of Environmental Factors on Rhizosphere Soil Microbial Community Structure and Function
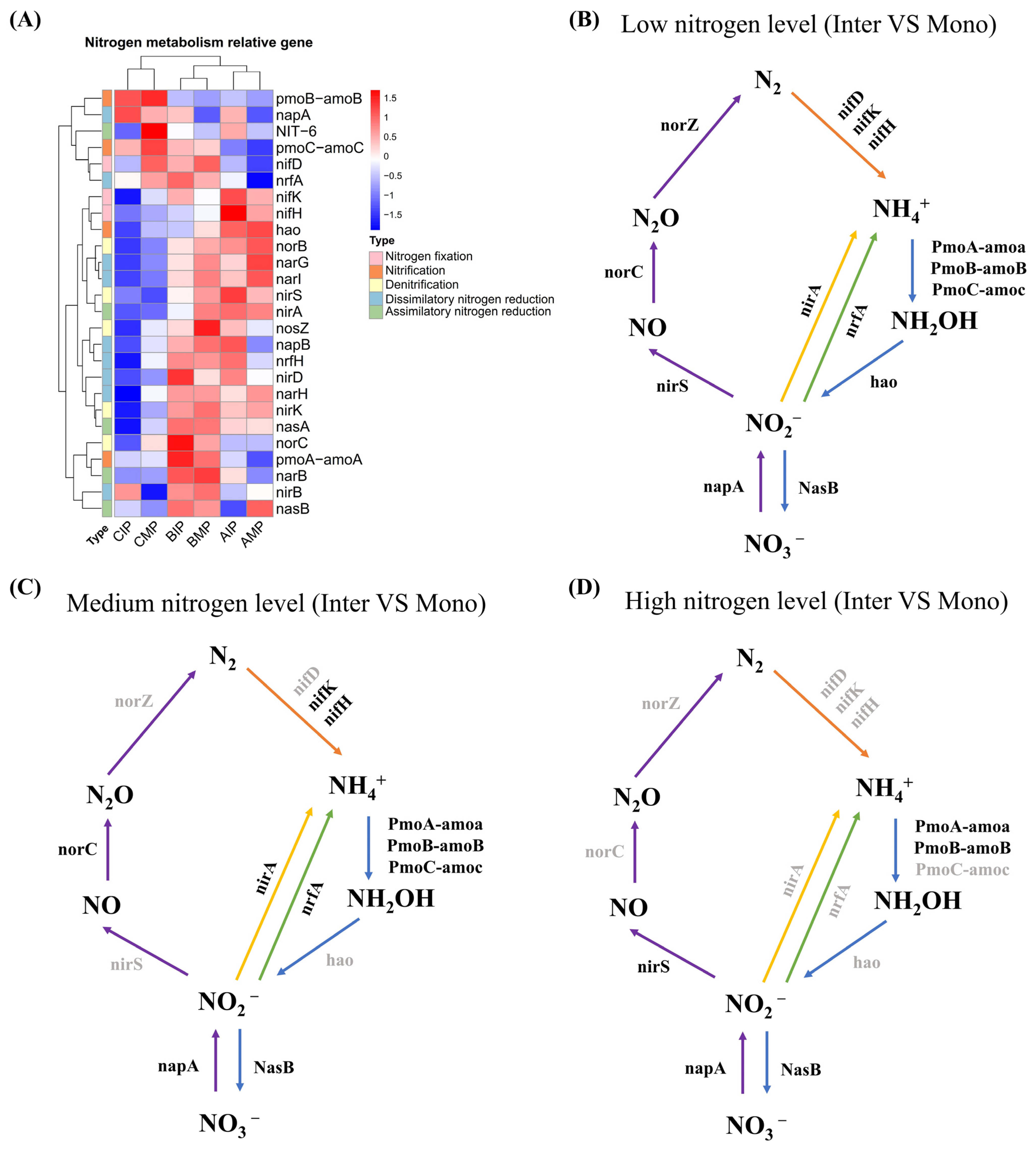
3.6. Analyses of the Nitrogen Cycle in Peanut Rhizosphere Soil Microbes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clough, T.J.; Mueller, C. Advances in understanding nitrogen flows and transformations: Gaps and research pathways. J. Agric. Sci. 2014, 152, 34–44. [Google Scholar] [CrossRef]
- Yao, H.; Bowman, D.; Shi, W. Seasonal variations of soil microbial biomass and activity in warm- and cool-season turfgrass systems. Soil Biol. Biochem. 2011, 43, 1536–1543. [Google Scholar] [CrossRef]
- Creamer, C.A.; Menezes, A.B.D.; Krull, E.S.; Sanderman, J.; Farrell, M. Microbial community structure mediates response of soil c decomposition to litter addition and warming. Soil Biol. Biochem. 2014, 80, 175–188. [Google Scholar] [CrossRef]
- Panke-Buisse, K.; Poole, A.C.; Goodrich, J.K.; Ley, R.E.; Kao-Kniffin, J. Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J. 2014, 9, 980–989. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.W.; Thomas, S.M. Microbial nitrogen cycles: Physiology, genomics and applications. Curr. Opin. Microbiol. 2001, 4, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.L.; Hao, Y.; Kou, L.; Song, G.L. Effects of nitrogen deposition and fertilization on n transformations in forest soils: A review. J. Soils Sediments 2015, 15, 863–879. [Google Scholar] [CrossRef]
- Kakirde, K.S.; Parsley, L.C.; Liles, M.R. Size does matter: Application-driven approaches for soil metagenomics. Soil Biol. Biochem. 2010, 42, 1911–1923. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Evans, S.E.; Friesen, M.L.; Tiemann, L.K. Effect of nitrogen fertilization on the abundance of nitrogen cycling genes in agricultural soils: A meta-analysis of field studies. Soil Biol. Biochem. 2018, S0038-0717, 30286–30292. [Google Scholar] [CrossRef]
- Scarlett, K.; Denman, S.; Clark, D.R.; Forster, J.; Vanguelova, E.; Brown, N.; Whitby, C. Relationships between nitrogen cycling microbial community abundance and composition reveal the indirect effect of soil pH on oak decline. ISME J. 2021, 15, 623–635. [Google Scholar] [CrossRef]
- Mooshammer, M.; Wanek, W.; Zechmeister-Boltenstern, S.; Richter, A. Stoichiometric imbalances between terrestrial decomposer communities and their resources: Mechanisms and implications of microbial adaptations to their resources. Front. Microbiol. 2014, 5, 22. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Smith, A.P.; Marín-Spiotta, E.; de Graaff, M.A.; Balser, T.C. Microbial community structure varies across soil organic matter aggregate pools during tropical land cover change. Soil Biol. Biochem. 2014, 77, 292–303. [Google Scholar] [CrossRef]
- Chu, H.; Lin, X.; Fujii, T.; Morimoto, S.; Yagi, K.; Hu, J.; Zhang, J. Soil microbial biomass, dehydrogenase activity, bacterial community structure in response to long-term fertilizer management. Soil Biol. Biochem. 2007, 39, 2971–2976. [Google Scholar] [CrossRef]
- Philippot, L.; Chenu, C.; Kappler, A.; Fierer, N. The interplay between microbial communities and soil properties. Nat. Rev. Microbiol. 2023, 22, 226–239. [Google Scholar] [CrossRef]
- Wu, M.; Li, G.; Li, W.; Liu, J.; Liu, M.; Jiang, C.; Li, Z. Nitrogen fertilizer deep placement for increased grain yield and nitrogen recovery efficiency in rice grown in subtropical china. Front. Plant Sci. 2017, 8, 1227. [Google Scholar] [CrossRef]
- Belay, A.; Claassens, A.S.; Wehner, F.C. Effect of direct nitrogen and potassium and residual phosphorus fertilizers on soil chemical properties, microbial components and maize yield under long-term crop rotation. Biol. Fertil. Soils 2002, 35, 420–427. [Google Scholar] [CrossRef]
- Du, Y.; Wang, T.; Wang, C.; Anane, P.; Liu, S.; Paz-Ferreiro, J. Nitrogen fertilizer is a key factor affecting the soil chemical and microbial communities in a mollisol. Can. J. Microbiol. 2019, 65, 510–521. [Google Scholar] [CrossRef]
- Huang, L.; Riggins, C.W.; Rodríguez-Zas, S.; Zabaloy, M.C.; Villamil, M.B. Long-term N fertilization imbalances potential N acquisition and transformations by soil microbes. Sci. Total Environ. 2019, 691, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, K.S.; Craine, J.M.; Fierer, N. Nitrogen fertilization inhibits soil microbial respiration regardless of the form of nitrogen applied. Soil Biol. Biochem. 2010, 42, 2336–2338. [Google Scholar] [CrossRef]
- Zheng, B.; Hao, X.; Ding, K.; Zhou, G.; Chen, Q.; Zhang, J.; Zhu, Y.G. Long-term nitrogen fertilization decreased the abundance of inorganic phosphate solubilizing bacteria in an alkaline soil. Sci. Rep. 2017, 7, 42284. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wang, C.; Zheng, M.; Jiang, L.; Luo, Y. Patterns and mechanisms of responses by soil microbial communities to nitrogen addition. Soil Biol. Biochem. 2017, 115, 433–441. [Google Scholar] [CrossRef]
- Willey, R.W. Resource use in intercropping systems. Agric. Water Manag. 1990, 17, 215–231. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, X.; Li, X.; Shen, J.; Sun, L.; Zaman, S.; Wang, Y.; Ding, Z. Different changes of bacterial diversity and soil metabolites in tea plants-legume intercropping systems. Front. Microbiol. 2023, 14, 1110623. [Google Scholar] [CrossRef] [PubMed]
- Dang, K.; Gong, X.; Zhao, G.; Wang, H.; Ivanistau, A.; Feng, B. Intercropping Alters the Soil Microbial Diversity and Community to Facilitate Nitrogen Assimilation: A Potential Mechanism for Increasing Proso Millet Grain Yield. Front. Microbiol. 2020, 11, 601054. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Arafat, Y.; Wu, L.; Xiao, Z.; Li, Q.; Khan, M.A.; Khan, M.U.; Lin, S.; Lin, W. Shifts in soil microbial community, soil enzymes and crop yield under peanut/maize intercropping with reduced nitrogen levels. Appl. Soil Ecol. 2018, 124, 327–334. [Google Scholar] [CrossRef]
- Dong, Q.; Zhao, X.; Zhou, D.; Liu, Z.; Shi, X.; Yuan, Y.; Jia, P.; Liu, Y.; Song, P.; Wang, X.; et al. Maize and peanut intercropping improves the nitrogen accumulation and yield per plant of maize by promoting the secretion of flavonoids and abundance of bradyrhizobium in rhizosphere. Front. Plant Sci. 2022, 13, 957336. [Google Scholar] [CrossRef]
- Zhao, X.; Dong, Q.; Han, Y.; Zhang, K.; Shi, X.; Yang, X.; Yuan, Y.; Zhou, D.; Wang, K.; Wang, X.; et al. Maize/peanut intercropping improves nutrient uptake of side-row maize and system microbial community diversity. BMC Microbiol. 2022, 22, 14. [Google Scholar] [CrossRef] [PubMed]
- Lian, T.; Mu, Y.; Jin, J.; Ma, Q.; Cheng, Y.; Cai, Z.; Nian, H. Impact of intercropping on the coupling between soil microbial community structure, activity, and nutrient-use efficiencies. PeerJ 2019, 7, e6412. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Wang, L.C.; Xing, Y.; Ma, S.M.; Zhang, X.D.; Chen, J.; Shi, C. Effects of intercropping chinese milk vetch on functional characteristics of soil microbial community in rape rhizosphere. Ying Yong Sheng Tai Xue Bao 2018, 29, 909–914. [Google Scholar] [CrossRef]
- Yang, Y.D.; Feng, X.M.; Hu, Y.G.; Ren, C.Z.; Zeng, Z.H. Effects of legume-oat intercropping on abundance and community structure of soil n2-fixing bacteria. Ying Yong Sheng Tai Xue Bao 2017, 28, 957–965. [Google Scholar] [CrossRef]
- Pang, Z.; Fallah, N.; Weng, P.; Zhou, Y.; Tang, X.; Tayyab, M.; Liu, Y.; Liu, Q.; Xiao, Y.; Hu, C.; et al. Sugarcane-peanut intercropping system enhances bacteria abundance, diversity, and sugarcane parameters in rhizospheric and bulk soils. Front. Microbiol. 2022, 12, 815129. [Google Scholar] [CrossRef]
- Tang, X.; Jiang, J.; Huang, Z.; Wu, H.; Wang, J.; He, L.; Xiong, F.; Zhong, R.; Liu, J.; Han, Z.; et al. Sugarcane/peanut intercropping system improves the soil quality and increases the abundance of beneficial microbes. J. Basic Microbiol. 2021, 61, 165–176. [Google Scholar] [CrossRef]
- Schad, P. Word reference base for soil resources. In Reference Module in Earth Systems and Environmental Sciences; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar] [CrossRef]
- Abrams, D.; Metcalf, D.; Hojjatie, M. Determination of kjeldahl nitrogen in fertilizers by AOAC official methods 978.02: Effect of copper sulfate as a catalyst. J. Aoac. Int. 2014, 97, 764–767. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.W. Methods of soil analysis, part 2. Chemical and microbiological properties. Exchangeable cations. Soil Sci. Soc. Am. J. 1982, 1159, 159–166. [Google Scholar]
- Tang, X.; Zhang, Y.; Jiang, J.; Meng, X.; Huang, Z.; Wu, H.; He, L.; Xiong, F.; Liu, J.; Zhong, R.; et al. Sugarcane/peanut intercropping system improves physicochemical properties by changing n and p cycling and organic matter turnover in root zone soil. PeerJ 2021, 9, e10880. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, F.S.; Olsen, S.R. Test of an ascorbic acid method for determining phosphorus in water and nahco3 extracts from soil. Soil Sci. Soc. Am. Proc. 1965, 291, 677–678. [Google Scholar] [CrossRef]
- Pansu, M.; Gautheyrou, J. Handbook of Soil Analysis: Mineralogical, Organic and Inorganic Methods; Springer: Berlin/Heidelberg, Germany, 2006; p. 993. [Google Scholar]
- Fu, M.M.; Huang, B.; Jia, M.M.; Hu, W.Y.; Sun, W.X.; Weindorf, D.C.; Chang, Q. Effect of intensive greenhouse vegetable cultivation on selenium availability in soil. Pedosphere 2015, 25, 343–350. [Google Scholar] [CrossRef]
- Bao, S.D. Soil and Agricultural Chemistry Analysis; China Agriculture Press: Beijing, China, 2000; pp. 100–109. [Google Scholar]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. Megahit: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. Cd-hit: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, C.; Hoffland, E.; Kuyper, T.W.; Yu, Y.; Zhang, C.; Li, H.; Zhang, F.; van der Werf, W. Syndromes of production in intercropping impact yield gains. Nat. Plants 2020, 6, 653–660. [Google Scholar] [CrossRef]
- Chang, X.; Yan, L.; Naeem, M.; Khaskheli, M.I.; Zhang, H.; Gong, G.; Zhang, M.; Song, C.; Yang, W.; Liu, T.; et al. Maize/soybean relay strip intercropping reduces the occurrence of fusarium root rot and changes the diversity of the pathogenic fusarium species. Pathogens 2020, 9, 211. [Google Scholar] [CrossRef]
- Solanki, M.K.; Wang, F.; Wang, Z.; Li, C.; Lan, T.; Singh, R.K.; Singh, P.; Yang, L.T.; Li, Y. Rhizospheric and endospheric diazotrophs mediated soil fertility intensification in sugarcane-legume intercropping systems. J. Soils Sediments 2019, 19, 1911–1927. [Google Scholar] [CrossRef]
- Suman, A.; Lal, M.; Singh, A.K.; Gaur, A. Microbial biomass turnover in indian subtropical soils under different sugarcane intercropping systems. Agron. J. 2006, 98, 698–704. [Google Scholar] [CrossRef]
- Ablimit, R.; Li, W.; Zhang, J.; Gao, H.; Zhao, Y.; Cheng, M.; Meng, X.; An, L.; Chen, Y. Altering microbial community for improving soil properties and agricultural sustainability during a 10-year maize-green manure intercropping in northwest China. J. Environ. Manag. 2022, 321, 115859. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Zhao, J.; Yuan, J.; Hale, L.; Wen, T.; Huang, Q.; Vivanco, J.M.; Zhou, J.; Kowalchuk, G.A.; Shen, Q. Root exudates drive soil-microbe-nutrient feedbacks in response to plant growth. Plant Cell Environ. 2021, 44, 613–628. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Feng, F.; Medova, H.; Dean, J.; Koblizek, M. Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum gemmatimonadetes. Proc. Natl. Acad. Sci. USA 2014, 111, 7795–7800. [Google Scholar] [CrossRef] [PubMed]
- Koblížek, M.; Dachev, M.; Bína, D.; Nupur Piwosz, K.; Kaftan, D. Utilization of light energy in phototrophic gemmatimonadetes. J. Photochem. Photobiol. B Biol. 2020, 213, 112085. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Townsend, A.R.; Sattin, S.R.; Freeman, K.R.; Fierer, N.; Neff, J.C.; Bowman, W.D.; Schadt, C.W.; Weintraub, M.N.; Schmidt, S.K. The effects of chronic nitrogen fertilization on alpine tundra soil microbial communities: Implications for carbon and nitrogen cycling. Environ. Microbiol. 2008, 10, 3093–3105. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.P.N.; Luo, Z.; Dong, Z.; Li, Q.; Liu, B.; Guo, S.; Nie, G.; Li, W. Metagenomic analysis further extends the role of chloroflexi in fundamental biogeochemical cycles. Environ. Res. 2022, 209, 112888. [Google Scholar] [CrossRef]
- Wu, C.; Yan, B.; Wei, F.; Wang, H.; Gao, L.; Ma, H.; Liu, Q.; Liu, Y.; Liu, G.; Wang, G. Long-term application of nitrogen and phosphorus fertilizers changes the process of community construction by affecting keystone species of crop rhizosphere microorganisms. Sci. Total Environ. 2023, 897, 165239. [Google Scholar] [CrossRef]
- Cao, J.; Zhao, P.; Wang, D.; Zhao, Y.; Wang, Z.; Zhong, N. Effects of a nanonetwork-structured soil conditioner on microbial community structure. Biology 2023, 12, 668. [Google Scholar] [CrossRef]
- Schneijderberg, M.; Schmitz, L.; Cheng, X.; Polman, S.; Franken, C.; Geurts, R.; Bisseling, T. A genetically and functionally diverse group of non-diazotrophic Bradyrhizobium spp. Colonizes the root endophytic compartment of arabidopsis thaliana. BMC Plant Biol. 2018, 18, 61. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Lei, H.; Lian, Y.; Zhang, Z.; Pan, H.; Yin, C.; Dong, Y. Impact of aerated drip irrigation and nitrogen application on soil properties, soil bacterial communities and agronomic traits of cucumber in a greenhouse system. Plants 2023, 12, 3834. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ma, W.; He, H.; Wang, Z.; Cao, Y. Effects of sugarcane and soybean intercropping on the nitrogen-fixing bacterial community in the rhizosphere. Front. Microbiol. 2021, 12, 713349. [Google Scholar] [CrossRef]
- Sun, R.; Wang, F.; Hu, C.; Liu, B. Metagenomics reveals taxon-specific responses of the nitrogen-cycling microbial community to long-term nitrogen fertilization. Soil Biol. Biochem. 2021, 156, 108214. [Google Scholar] [CrossRef]
- Wu, Z.; Hao, Z.; Sun, Y.; Guo, L.; Huang, L.; Zeng, Y.; Wang, Y.; Yang, L.; Chen, B. Comparison on the structure and function of the rhizosphere microbial community between healthy and root-rot panax notoginseng. Appl. Soil Ecol. 2016, 107, 99–107. [Google Scholar] [CrossRef]
- Zeng, J.; Liu, J.; Lu, C.; Ou, X.; Luo, K.; Li, C.; He, M.; Zhang, H.; Yan, H. Intercropping with turmeric or ginger reduce the continuous cropping obstacles that affect pogostemon cablin (patchouli). Front. Microbiol. 2020, 11, 579719. [Google Scholar] [CrossRef]
- Pang, Z.; Dong, F.; Liu, Q.; Lin, W.; Hu, C.; Yuan, Z. Soil metagenomics reveals effects of continuous sugarcane cropping on the structure and functional pathway of rhizospheric microbial community. Front. Microbiol. 2021, 12, 627569. [Google Scholar] [CrossRef]
- Wang, C.; Liang, Q.; Liu, J.; Zhou, R.; Lang, X.; Xu, S.; Li, X.; Gong, A.; Mu, Y.; Fang, H.; et al. Impact of intercropping grass on the soil rhizosphere microbial community and soil ecosystem function in a walnut orchard. Front. Microbiol. 2023, 14, 1137590. [Google Scholar] [CrossRef]
- Dong, H.; Fan, S.; Sun, H.; Chen, C.; Wang, A.; Jiang, L.; Ma, D. Rhizosphere-associated microbiomes of rice (Oryza sativa L.) Under the effect of increased nitrogen fertilization. Front. Microbiol. 2021, 12, 730506. [Google Scholar] [CrossRef]
- Li, H.; Luo, L.; Tang, B.; Guo, H.; Cao, Z.; Zeng, Q.; Chen, S.; Chen, Z. Dynamic changes of rhizosphere soil bacterial community and nutrients in cadmium polluted soils with soybean-corn intercropping. BMC Microbiol. 2022, 22, 57. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.; Barth, V.; Coffey, T.; McFarland, C.; Huggins, D.; Sullivan, T. Altered bacterial communities in long-term no-till soils associated with stratification of soluble aluminum and soil ph. Soil Syst. 2018, 2, 7. [Google Scholar] [CrossRef]
- Tang, X.; He, Y.; Zhang, Z.; Wu, H.; He, L.; Jiang, J.; Meng, W.; Huang, Z.; Xiong, F.; Liu, J.; et al. Beneficial shift of rhizosphere soil nutrients and metabolites under a sugarcane/peanut intercropping system. Front. Plant Sci. 2022, 13, 1018727. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Chen, S.; Huang, Z.; Huang, T.; Tang, X.; He, L.; Li, Z.; Xiong, J.; Zhong, R.; Jiang, J.; et al. Effects of Intercropping and Nitrogen Application on Soil Fertility and Microbial Communities in Peanut Rhizosphere Soil. Agronomy 2024, 14, 635. https://doi.org/10.3390/agronomy14030635
Wu H, Chen S, Huang Z, Huang T, Tang X, He L, Li Z, Xiong J, Zhong R, Jiang J, et al. Effects of Intercropping and Nitrogen Application on Soil Fertility and Microbial Communities in Peanut Rhizosphere Soil. Agronomy. 2024; 14(3):635. https://doi.org/10.3390/agronomy14030635
Chicago/Turabian StyleWu, Haining, Shufang Chen, Zhipeng Huang, Tangwei Huang, Xiumei Tang, Liangqiong He, Zhong Li, Jun Xiong, Ruichun Zhong, Jing Jiang, and et al. 2024. "Effects of Intercropping and Nitrogen Application on Soil Fertility and Microbial Communities in Peanut Rhizosphere Soil" Agronomy 14, no. 3: 635. https://doi.org/10.3390/agronomy14030635




