An Overview of Machine Learning Applications on Plant Phenotyping, with a Focus on Sunflower
Abstract
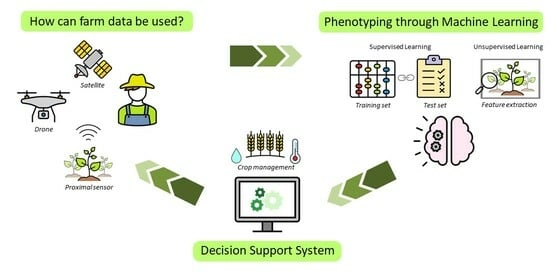
:1. Introduction
2. Machine Learning and Agriculture
3. Machine Learning Techniques for Phenotyping
3.1. Artificial Neural Networks
3.2. Support Vector Machines
3.3. Decision Trees
3.4. k-Nearest Neighbors
4. Digital Plant Phenotyping
4.1. Crop Management
4.2. Plant Health
5. Overview of Digital Sunflower Phenotyping
6. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Prosekov, A.Y.; Ivanova, S.A. Food Security: The Challenge of the Present. Geoforum 2018, 91, 73–77. [Google Scholar] [CrossRef]
- Cook, D.C.; Fraser, R.W.; Paini, D.R.; Warden, A.C.; Lonsdale, W.M.; De Barro, P.J. Biosecurity and Yield Improvement Technologies Are Strategic Complements in the Fight against Food Insecurity. PLoS ONE 2011, 6, e26084. [Google Scholar] [CrossRef]
- Fedoroff, N.V.; Battisti, D.S.; Beachy, R.N.; Cooper, P.J.M.; Fischhoff, D.A.; Hodges, C.N.; Knauf, V.C.; Lobell, D.; Mazur, B.J.; Molden, D.; et al. Radically Rethinking Agriculture for the 21st Century. Science 2010, 327, 833–834. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. The Impact of Disasters and Crises on Agriculture and Food Security; Food and Agriculture Organization of the United Nations: Rome, Italy, 2021; ISBN 978-92-5-134071-4. [Google Scholar]
- Stafford, J.V. Implementing Precision Agriculture in the 21st Century. J. Agric. Eng. Res. 2000, 76, 267–275. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, M.; Wang, N. Precision Agriculture—A Worldwide Overview. Comput. Electron. Agric. 2002, 36, 113–132. [Google Scholar] [CrossRef]
- Ilari, A.; Piancatelli, S.; Centorame, L.; Moumni, M.; Romanazzi, G.; Foppa Pedretti, E. Distribution Quality of Agrochemicals for the Revamping of a Sprayer System Based on Lidar Technology and Grapevine Disease Management. Appl. Sci. 2023, 13, 2222. [Google Scholar] [CrossRef]
- Calvitti, M.; Colonna, N.; Iannetta, M. La Relazione Cambiamenti Climatici e Sistema Agricolo: Tra Adattamento e Mitigazione. Energ. Ambiente E Innov. 2016, 1, 74–81. [Google Scholar] [CrossRef]
- FAO. FAOSTAT. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 8 March 2023).
- Giannini, V.; Maucieri, C.; Vamerali, T.; Zanin, G.; Schiavon, S.; Pettenella, D.M.; Bona, S.; Borin, M. Sunflower: From Cortuso’s Description (1585) to Current Agronomy, Uses and Perspectives. Agriculture 2022, 12, 1978. [Google Scholar] [CrossRef]
- Baldoni, R.; Giardini, L. Coltivazioni Erbacee: Piante Oleifere, Da Zucchero, Da Fibra, Orticole e Aromatiche, 3rd ed.; Patron: Bologna, Italy, 2001; Volume 2, ISBN 978-88-555-2622-7. [Google Scholar]
- Jagtap, S.; Trollman, H.; Trollman, F.; Garcia-Garcia, G.; Parra-López, C.; Duong, L.; Martindale, W.; Munekata, P.E.S.; Lorenzo, J.M.; Hdaifeh, A.; et al. The Russia-Ukraine Conflict: Its Implications for the Global Food Supply Chains. Foods 2022, 11, 2098. [Google Scholar] [CrossRef]
- Ben Hassen, T.; El Bilali, H. Impacts of the Russia-Ukraine War on Global Food Security: Towards More Sustainable and Resilient Food Systems? Foods 2022, 11, 2301. [Google Scholar] [CrossRef]
- Sun, M. The Impact of the Russia-Ukraine Conflict on Global Grain Market and Food Security: Short- and Long-Term Effects. Seed Biol. 2022, 1, 3. [Google Scholar] [CrossRef]
- Leal Filho, W.; Fedoruk, M.; Paulino Pires Eustachio, J.H.; Barbir, J.; Lisovska, T.; Lingos, A.; Baars, C. How the War in Ukraine Affects Food Security. Foods 2023, 12, 3996. [Google Scholar] [CrossRef]
- Anushree, S.; André, M.; Guillaume, D.; Frédéric, F. Stearic Sunflower Oil as a Sustainable and Healthy Alternative to Palm Oil. A Review. Agron. Sustain. Dev. 2017, 37, 18. [Google Scholar] [CrossRef]
- Pal, U.S.; Patra, R.K.; Sahoo, N.R.; Bakhara, C.K.; Panda, M.K. Effect of Refining on Quality and Composition of Sunflower Oil. J. Food Sci. Technol. 2015, 52, 4613–4618. [Google Scholar] [CrossRef]
- Gupta, M.K. Sunflower Oil and Its Applications. Lipid Technol. 2014, 26, 260–263. [Google Scholar] [CrossRef]
- Pedretti, E.F.; Del Gatto, A.; Pieri, S.; Mangoni, L.; Ilari, A.; Mancini, M.; Feliciangeli, G.; Leoni, E.; Toscano, G.; Duca, D. Experimental Study to Support Local Sunflower Oil Chains: Production of Cold Pressed Oil in Central Italy. Agriculture 2019, 9, 231. [Google Scholar] [CrossRef]
- Meijaard, E.; Brooks, T.M.; Carlson, K.M.; Slade, E.M.; Garcia-Ulloa, J.; Gaveau, D.L.A.; Lee, J.S.H.; Santika, T.; Juffe-Bignoli, D.; Struebig, M.J.; et al. The Environmental Impacts of Palm Oil in Context. Nat. Plants 2020, 6, 1418–1426. [Google Scholar] [CrossRef]
- Monzon, J.P.; Calviño, P.A.; Sadras, V.O.; Zubiaurre, J.B.; Andrade, F.H. Precision Agriculture Based on Crop Physiological Principles Improves Whole-Farm Yield and Profit: A Case Study. Eur. J. Agron. 2018, 99, 62–71. [Google Scholar] [CrossRef]
- Legrand, N. War in Ukraine: The Rational “wait and See” Mode of Global Food Markets. Appl. Econ. Perspect. Policy 2022, 45, 626–644. [Google Scholar] [CrossRef]
- Cvejić, S.; Hrnjaković, O.; Jocković, M.; Kupusinac, A.; Doroslovački, K.; Gvozdenac, S.; Jocić, S.; Miladinović, D. Oil Yield Prediction for Sunflower Hybrid Selection Using Different Machine Learning Algorithms. Sci. Rep. 2023, 13, 17611. [Google Scholar] [CrossRef]
- Pierce, F.J.; Nowak, P. Aspects of Precision Agriculture. Adv. Agron. 1999, 67, 1–85. [Google Scholar] [CrossRef]
- Roy, R.N. Plant Nutrition for Food Security: A Guide for Integrated Nutrient Management; FAO Fertilizer and Plant Nutrition Bulletin; Food and Agriculture Organization of the United Nations: Rome, Italy, 2006; ISBN 978-92-5-105490-1. [Google Scholar]
- Ministero delle Politiche Agricole Alimentari e Forestali. Linee Guida per lo Sviluppo Dell’Agricoltura di Precisione in Italia; Ministero Delle Politiche Agricole Alimentari e Forestali: Rome, Italy, 2015. [Google Scholar]
- Kumar, S.; Tiwari, P.; Zymbler, M. Internet of Things Is a Revolutionary Approach for Future Technology Enhancement: A Review. J. Big Data 2019, 6, 111. [Google Scholar] [CrossRef]
- Monteleone, S.; Moraes, E.A.D.; Tondato De Faria, B.; Aquino Junior, P.T.; Maia, R.F.; Neto, A.T.; Toscano, A. Exploring the Adoption of Precision Agriculture for Irrigation in the Context of Agriculture 4.0: The Key Role of Internet of Things. Sensors 2020, 20, 7091. [Google Scholar] [CrossRef] [PubMed]
- Araújo, S.O.; Peres, R.S.; Ramalho, J.C.; Lidon, F.; Barata, J. Machine Learning Applications in Agriculture: Current Trends, Challenges, and Future Perspectives. Agronomy 2023, 13, 2976. [Google Scholar] [CrossRef]
- Costa, C.; Schurr, U.; Loreto, F.; Menesatti, P.; Carpentier, S. Plant Phenotyping Research Trends, a Science Mapping Approach. Front. Plant Sci. 2019, 9, 1933. [Google Scholar] [CrossRef]
- Walter, A.; Studer, B.; Kölliker, R. Advanced Phenotyping Offers Opportunities for Improved Breeding of Forage and Turf Species. Ann. Bot. 2012, 110, 1271–1279. [Google Scholar] [CrossRef]
- Minervini, M.; Abdelsamea, M.M.; Tsaftaris, S.A. Image-Based Plant Phenotyping with Incremental Learning and Active Contours. Ecol. Inform. 2014, 23, 35–48. [Google Scholar] [CrossRef]
- Chawade, A.; van Ham, J.; Blomquist, H.; Bagge, O.; Alexandersson, E.; Ortiz, R. High-Throughput Field-Phenotyping Tools for Plant Breeding and Precision Agriculture. Agronomy 2019, 9, 258. [Google Scholar] [CrossRef]
- Mahlein, A.-K. Plant Disease Detection by Imaging Sensors—Parallels and Specific Demands for Precision Agriculture and Plant Phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Eron, F.; Noman, M.; De Oliveira, R.R.; Chalfun-Junior, A. Computer Vision-Aided Intelligent Monitoring of Coffee: Towards Sustainable Coffee Production. Sci. Hortic. 2024, 327, 112847. [Google Scholar] [CrossRef]
- Chlingaryan, A.; Sukkarieh, S.; Whelan, B. Machine Learning Approaches for Crop Yield Prediction and Nitrogen Status Estimation in Precision Agriculture: A Review. Comput. Electron. Agric. 2018, 151, 61–69. [Google Scholar] [CrossRef]
- McQueen, R.J.; Garner, S.R.; Nevill-Manning, C.G.; Witten, I.H. Applying Machine Learning to Agricultural Data. Comput. Electron. Agric. 1995, 12, 275–293. [Google Scholar] [CrossRef]
- Bishop, C.M. Pattern Recognition and Machine Learning; Information Science and Statistics; Corrected at 8th Printing 2009; Springer: New York, NY, USA, 2009; ISBN 978-0-387-31073-2. [Google Scholar]
- Liakos, K.; Busato, P.; Moshou, D.; Pearson, S.; Bochtis, D. Machine Learning in Agriculture: A Review. Sensors 2018, 18, 2674. [Google Scholar] [CrossRef]
- Jha, K.; Doshi, A.; Patel, P.; Shah, M. A Comprehensive Review on Automation in Agriculture Using Artificial Intelligence. Artif. Intell. Agric. 2019, 2, 1–12. [Google Scholar] [CrossRef]
- Benos, L.; Tagarakis, A.C.; Dolias, G.; Berruto, R.; Kateris, D.; Bochtis, D. Machine Learning in Agriculture: A Comprehensive Updated Review. Sensors 2021, 21, 3758. [Google Scholar] [CrossRef]
- Pokhariyal, S.; Patel, N.R.; Govind, A. Machine Learning-Driven Remote Sensing Applications for Agriculture in India—A Systematic Review. Agronomy 2023, 13, 2302. [Google Scholar] [CrossRef]
- Dash, S.S.; Nayak, S.K.; Mishra, D. A Review on Machine Learning Algorithms. In Intelligent and Cloud Computing; Mishra, D., Buyya, R., Mohapatra, P., Patnaik, S., Eds.; Smart Innovation, Systems and Technologies; Springer: Singapore, 2021; Volume 153, pp. 495–507. ISBN 9789811562013. [Google Scholar]
- Chhaya, K.; Khanzode, A.; Sarode, R.D. Advantages and Disadvantages of Artificial Intelligence and Machine Learning: A Literature Review. J. Rank. Libr. Inf. Sci. 2020, 9, 30–36. [Google Scholar]
- Araújo, S.O.; Peres, R.S.; Barata, J.; Lidon, F.; Ramalho, J.C. Characterising the Agriculture 4.0 Landscape—Emerging Trends, Challenges and Opportunities. Agronomy 2021, 11, 667. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. The Future of Food and Agriculture: Trends and Challenges; Food and Agriculture Organization of the United Nations: Rome, Italy, 2017; ISBN 978-92-5-109551-5. [Google Scholar]
- Mitchell, R.S.; Sherlock, R.A.; Smith, L.A. An Investigation into the Use of Machine Learning for Determining Oestrus in Cows. Comput. Electron. Agric. 1996, 15, 195–213. [Google Scholar] [CrossRef]
- Pietersma, D.; Lacroix, R.; Lefebvre, D.; Wade, K.M. Decision-Tree Introduction to Interpret Lactation Curves. Can. Biosyst. Eng. 2002, 44, 7.1–7.4. [Google Scholar]
- Holmes, G.; Cunningham, S.J.; Dela Rue, B.T.; Bollen, A.F. Predicting Apple Bruising Using Machine Learning. Acta Hortic. 1998, 476, 289–298. [Google Scholar] [CrossRef]
- Gualtieri, J.A.; Cromp, R.F. Support Vector Machines for Hyperspectral Remote Sensing Classification. Proc. SPIE-Int. Soc. Opt. Eng. 1999, 3584, 221–232. [Google Scholar]
- Sharma, A.; Jain, A.; Gupta, P.; Chowdary, V. Machine Learning Applications for Precision Agriculture: A Comprehensive Review. IEEE Access 2021, 9, 4843–4873. [Google Scholar] [CrossRef]
- Coopersmith, E.J.; Minsker, B.S.; Wenzel, C.E.; Gilmore, B.J. Machine Learning Assessments of Soil Drying for Agricultural Planning. Comput. Electron. Agric. 2014, 104, 93–104. [Google Scholar] [CrossRef]
- Raza, S.-A.; Smith, H.K.; Clarkson, G.J.J.; Taylor, G.; Thompson, A.J.; Clarkson, J.; Rajpoot, N.M. Automatic Detection of Regions in Spinach Canopies Responding to Soil Moisture Deficit Using Combined Visible and Thermal Imagery. PLoS ONE 2014, 9, e97612. [Google Scholar] [CrossRef]
- Lavanya, K.; Jaya Subalakshmi, R.; Tamizharasi, T.; Jane, L.; Victor, A. Unsupervised Unmixing and Segmentation of Hyper Spectral Images Accounting for Soil Fertility. SCPE 2022, 23, 291–301. [Google Scholar] [CrossRef]
- Papageorgiou, E.I.; Markinos, A.T.; Gemtos, T.A. Fuzzy Cognitive Map Based Approach for Predicting Yield in Cotton Crop Production as a Basis for Decision Support System in Precision Agriculture Application. Appl. Soft Comput. 2011, 11, 3643–3657. [Google Scholar] [CrossRef]
- Kumar, R.; Singh, M.P.; Kumar, P.; Singh, J.P. Crop Selection Method to Maximize Crop Yield Rate Using Machine Learning Technique. In Proceedings of the 2015 International Conference on Smart Technologies and Management for Computing, Communication, Controls, Energy and Materials (ICSTM), Chennai, India, 6–8 May 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 138–145. [Google Scholar]
- Kim, N.; Lee, Y.-W. Machine Learning Approaches to Corn Yield Estimation Using Satellite Images and Climate Data: A Case of Iowa State. J. Korean Soc. Surv. Geod. Photogramm. Cartogr. 2016, 34, 383–390. [Google Scholar] [CrossRef]
- Gandhi, N.; Armstrong, L.J.; Petkar, O.; Tripathy, A.K. Rice Crop Yield Prediction in India Using Support Vector Machines. In Proceedings of the 2016 13th International Joint Conference on Computer Science and Software Engineering (JCSSE), Khon Kaen, Thailand, 13–15 July 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 1–5. [Google Scholar]
- Elbasi, E.; Zaki, C.; Topcu, A.E.; Abdelbaki, W.; Zreikat, A.I.; Cina, E.; Shdefat, A.; Saker, L. Crop Prediction Model Using Machine Learning Algorithms. Appl. Sci. 2023, 13, 9288. [Google Scholar] [CrossRef]
- Römer, C.; Bürling, K.; Hunsche, M.; Rumpf, T.; Noga, G.; Plümer, L. Robust Fitting of Fluorescence Spectra for Pre-Symptomatic Wheat Leaf Rust Detection with Support Vector Machines. Comput. Electron. Agric. 2011, 79, 180–188. [Google Scholar] [CrossRef]
- Yeh, Y.-H.F.; Chung, W.-C.; Liao, J.-Y.; Chung, C.-L.; Kuo, Y.-F.; Lin, T.-T. A Comparison of Machine Learning Methods on Hyperspectral Plant Disease Assessments. IFAC Proc. Vol. 2013, 46, 361–365. [Google Scholar] [CrossRef]
- Akhtar, A.; Khanum, A.; Khan, S.A.; Shaukat, A. Automated Plant Disease Analysis (APDA): Performance Comparison of Machine Learning Techniques. In Proceedings of the 2013 11th International Conference on Frontiers of Information Technology, Islamabad, Pakistan, 16–18 December 2013; IEEE: Piscataway, NJ, USA, 2013; pp. 60–65. [Google Scholar]
- Latif, G.; Alghazo, J.; Maheswar, R.; Vijayakumar, V.; Butt, M. Deep Learning Based Intelligence Cognitive Vision Drone for Automatic Plant Diseases Identification and Spraying. IFS 2020, 39, 8103–8114. [Google Scholar] [CrossRef]
- Ahmed, F.; Bari, A.S.M.H.; Shihavuddin, A.; Al-Mamun, H.A.; Kwan, P. A Study on Local Binary Pattern for Automated Weed Classification Using Template Matching and Support Vector Machine. In Proceedings of the 2011 IEEE 12th International Symposium on Computational Intelligence and Informatics (CINTI), Budapest, Hungary, 21–22 November 2011; IEEE: Piscataway, NJ, USA, 2011; pp. 329–334. [Google Scholar]
- Wang, A.; Xu, Y.; Wei, X.; Cui, B. Semantic Segmentation of Crop and Weed Using an Encoder-Decoder Network and Image Enhancement Method under Uncontrolled Outdoor Illumination. IEEE Access 2020, 8, 81724–81734. [Google Scholar] [CrossRef]
- Pathak, H.; Igathinathane, C.; Howatt, K.; Zhang, Z. Machine Learning and Handcrafted Image Processing Methods for Classifying Common Weeds in Corn Field. Smart Agric. Technol. 2023, 5, 100249. [Google Scholar] [CrossRef]
- Goumopoulos, C.; O’Flynn, B.; Kameas, A. Automated Zone-Specific Irrigation with Wireless Sensor/Actuator Network and Adaptable Decision Support. Comput. Electron. Agric. 2014, 105, 20–33. [Google Scholar] [CrossRef]
- Vij, A.; Vijendra, S.; Jain, A.; Bajaj, S.; Bassi, A.; Sharma, A. IoT and Machine Learning Approaches for Automation of Farm Irrigation System. Procedia Comput. Sci. 2020, 167, 1250–1257. [Google Scholar] [CrossRef]
- Elhussiny, K.T.; Hassan, A.M.; Habssa, A.A.; Mokhtar, A. Prediction of Water Distribution Uniformity of Sprinkler Irrigation System Based on Machine Learning Algorithms. Sci. Rep. 2023, 13, 20885. [Google Scholar] [CrossRef]
- Ayadi, S.; Ben Said, A.; Jabbar, R.; Aloulou, C.; Chabbouh, A.; Achballah, A.B. Dairy Cow Rumination Detection: A Deep Learning Approach. In Proceedings of the Distributed Computing for Emerging Smart Networks; Jemili, I., Mosbah, M., Eds.; Springer International Publishing: Cham, Switzerland, 2020; Volume 1348, pp. 123–139. [Google Scholar]
- Haldar, A.; Mandal, S.N.; Deb, S.; Roy, R.; Laishram, M. Application of Information and Electronic Technology for Best Practice Management in Livestock Production System. In Agriculture, Livestock Production and Aquaculture; Kumar, A., Kumar, P., Singh, S.S., Trisasongko, B.H., Rani, M., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 173–218. ISBN 978-3-030-93261-9. [Google Scholar]
- Dineva, K.; Atanasova, T. Health Status Classification for Cows Using Machine Learning and Data Management on AWS Cloud. Animals 2023, 13, 3254. [Google Scholar] [CrossRef]
- Behmann, J.; Mahlein, A.-K.; Rumpf, T.; Römer, C.; Plümer, L. A Review of Advanced Machine Learning Methods for the Detection of Biotic Stress in Precision Crop Protection. Precis. Agric 2015, 16, 239–260. [Google Scholar] [CrossRef]
- Rumpf, T.; Mahlein, A.-K.; Steiner, U.; Oerke, E.-C.; Dehne, H.-W.; Plümer, L. Early Detection and Classification of Plant Diseases with Support Vector Machines Based on Hyperspectral Reflectance. Comput. Electron. Agric. 2010, 74, 91–99. [Google Scholar] [CrossRef]
- Jiao, L.; Luo, X.; Zha, L.; Bao, H.; Zhang, J.; Gu, X. Machine Learning Assisted Water Management Strategy on a Self-Sustaining Seawater Desalination and Vegetable Cultivation Platform. Comput. Electron. Agric. 2024, 217, 108569. [Google Scholar] [CrossRef]
- Jhajharia, K.; Mathur, P. Prediction of Crop Yield Using Satellite Vegetation Indices Combined with Machine Learning Approaches. Adv. Space Res. 2023, 72, 3998–4007. [Google Scholar] [CrossRef]
- Asgari, S.; Hasanlou, M. A Comparative Study of Machine Learning Classifiers for Crop Type Mapping Using Vegetation Indices. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2023, X-4/W1-2022, 79–85. [Google Scholar] [CrossRef]
- Amankulova, K.; Farmonov, N.; Mukhtorov, U.; Mucsi, L. Sunflower Crop Yield Prediction by Advanced Statistical Modeling Using Satellite-Derived Vegetation Indices and Crop Phenology. Geocarto Int. 2023, 38, 2197509. [Google Scholar] [CrossRef]
- Moscovini, L.; Ortenzi, L.; Pallottino, F.; Figorilli, S.; Violino, S.; Pane, C.; Capparella, V.; Vasta, S.; Costa, C. An Open-Source Machine-Learning Application for Predicting Pixel-to-Pixel NDVI Regression from RGB Calibrated Images. Comput. Electron. Agric. 2024, 216, 11. [Google Scholar] [CrossRef]
- Berenstein, R.; Shahar, O.B.; Shapiro, A.; Edan, Y. Grape Clusters and Foliage Detection Algorithms for Autonomous Selective Vineyard Sprayer. Intell. Serv. Robot. 2010, 3, 233–243. [Google Scholar] [CrossRef]
- Kim, S.; Kim, S. Performance Estimation Modeling via Machine Learning of an Agrophotovoltaic System in South Korea. Energies 2021, 14, 6724. [Google Scholar] [CrossRef]
- Zohdi, T.I. A Digital-Twin and Machine-Learning Framework for the Design of Multiobjective Agrophotovoltaic Solar Farms. Comput. Mech. 2021, 68, 357–370. [Google Scholar] [CrossRef]
- Ladisa, C.; Capolupo, A.; Ripa, M.N.; Tarantino, E. Combining OBIA Approach and Machine Learning Algorithm to Extract Photovoltaic Panels from Sentinel-2 Images Automatically. In Proceedings of the Remote Sensing for Agriculture, Ecosystems, and Hydrology XXIV, Berlin, Germany, 5–7 September 2022; Neale, C.M., Maltese, A., Eds.; SPIE: Berlin, Germany, 2022; p. 18. [Google Scholar]
- Haglin, J.M.; Jimenez, G.; Eltorai, A.E.M. Artificial Neural Networks in Medicine. Health Technol. 2019, 9, 1–6. [Google Scholar] [CrossRef]
- El-Shishiny, H.; Deraz, S.; Bahy, O. Mining Software Aging Patterns by Artificial Neural Networks. In Artificial Neural Networks in Pattern Recognition; Prevost, L., Marinai, S., Schwenker, F., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2008; Volume 5064, pp. 252–262. ISBN 978-3-540-69938-5. [Google Scholar]
- Mcculloch, W.S.; Pitts, W.A. A Logical Calculus of the Ideas Immanent in Nervous Activity. Bull. Math. Biophys. 1943, 5, 115–133. [Google Scholar] [CrossRef]
- Hornik, K.; Stinchcombe, M.; White, H. Multilayer Feedforward Networks Are Universal Approximators. Neural Netw. 1989, 2, 359–366. [Google Scholar] [CrossRef]
- Meng, Z.; Hu, Y.; Ancey, C. Using a Data Driven Approach to Predict Waves Generated by Gravity Driven Mass Flows. Water 2020, 12, 600. [Google Scholar] [CrossRef]
- Dastres, R.; Soori, M. Artificial Neural Network Systems. Int. J. Imaging Robot. 2021, 21, 14. [Google Scholar]
- Rahman, M.A.; Muniyandi, R.C.; Islam, K.T.; Rahman, M.M. Ovarian Cancer Classification Accuracy Analysis Using 15-Neuron Artificial Neural Networks Model. In Proceedings of the 2019 IEEE Student Conference on Research and Development (SCOReD), Bandar Seri Iskandar, Malaysia, 15–17 October 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 33–38. [Google Scholar]
- Zupan, J.; Novič, M.; Ruisánchez, I. Kohonen and Counterpropagation Artificial Neural Networks in Analytical Chemistry. Chemom. Intell. Lab. Syst. 1997, 38, 1–23. [Google Scholar] [CrossRef]
- Goldberg, Y. A Primer on Neural Network Models for Natural Language Processing. J. Artif. Intell. Res. 2016, 57, 345–420. [Google Scholar] [CrossRef]
- Li, S.; Choi, K.; Lee, Y. Artificial Neural Network Implementation in FPGA: A Case Study. In Proceedings of the 2016 International SoC Design Conference (ISOCC), Jeju, Republic of Korea, 23–26 October 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 297–298. [Google Scholar]
- Arun Balaji, S.; Baskaran, K. Design and Development of Artificial Neural Networking (ANN) System Using Sigmoid Activation Function to Predict Annual Rice Production in Tamilnadu. IJCSEIT 2013, 3, 13–31. [Google Scholar] [CrossRef]
- Giussani, B.; Roncoroni, S.; Recchia, S.; Pozzi, A. Bidimensional and Multidimensional Principal Component Analysis in Long Term Atmospheric Monitoring. Atmosphere 2016, 7, 155. [Google Scholar] [CrossRef]
- Agatonovic-Kustrin, S.; Beresford, R. Basic Concepts of Artificial Neural Network (ANN) Modeling and Its Application in Pharmaceutical Research. J. Pharm. Biomed. Anal. 2000, 22, 717–727. [Google Scholar] [CrossRef]
- Graupe, D. Principles of Artificial Neural Networks, 2nd ed.; Advanced Series on Circuits and Systems; World Scientific: Singapore; Hackensack, NJ, USA, 2007; Volume 6, ISBN 978-981-270-624-9. [Google Scholar]
- Suliman, A.; Zhang, Y. A Review on Back-Propagation Neural Networks in the Application of Remote Sensing Image Classification. J. Earth Sci. Eng. 2015, 5, 52–65. [Google Scholar] [CrossRef]
- Teuwen, J.; Moriakov, N. Convolutional Neural Networks. In Handbook of Medical Image Computing and Computer Assisted Intervention; Elsevier: Amsterdam, The Netherlands, 2020; pp. 481–501. ISBN 978-0-12-816176-0. [Google Scholar]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J.; et al. Recent Advances in Convolutional Neural Networks. Pattern Recognit. 2018, 77, 354–377. [Google Scholar] [CrossRef]
- Frazão, X.; Alexandre, L.A. Weighted Convolutional Neural Network Ensemble. In Progress in Pattern Recognition, Image Analysis, Computer Vision, and Applications; Bayro-Corrochano, E., Hancock, E., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2014; Volume 8827, pp. 674–681. ISBN 978-3-319-12567-1. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Wang, H.; Xiong, J.; Yao, Z.; Lin, M.; Ren, J. Research Survey on Support Vector Machine. In Proceedings of the 10th EAI International Conference on Mobile Multimedia Communications, Chongqing, China, 13 July 2017; EAI: Chongqing, China, 2017. [Google Scholar]
- Srivastava, D.K.; Bhambhu, L. Data Classification Using Support Vector Machine. J. Theor. Appl. Inf. Technol. 2010, 12, 7. [Google Scholar]
- Bhavsar, H.; Panchal, M.H. A Review on Support Vector Machine for Data Classification. IJARCET 2012, 1, 185–189. [Google Scholar]
- Noble, W.S. What Is a Support Vector Machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef]
- Cervantes, J.; Garcia-Lamont, F.; Rodríguez-Mazahua, L.; Lopez, A. A Comprehensive Survey on Support Vector Machine Classification: Applications, Challenges and Trends. Neurocomputing 2020, 408, 189–215. [Google Scholar] [CrossRef]
- Sheykhmousa, M.; Mahdianpari, M.; Ghanbari, H.; Mohammadimanesh, F.; Ghamisi, P.; Homayouni, S. Support Vector Machine Versus Random Forest for Remote Sensing Image Classification: A Meta-Analysis and Systematic Review. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 6308–6325. [Google Scholar] [CrossRef]
- Wu, X.; Kumar, V. The Top Ten Algorithms in Data Mining, 1st ed.; Chapman & Hall/CRC data mining and knowledge discovery series; Chapman & Hall/CRC Press: Boca Raton, FL, USA, 2009; ISBN 978-1-4200-8964-6. [Google Scholar]
- Li, J.; Castagna, J. Support Vector Machine (SVM) Pattern Recognition to AVO Classification. Geophys. Res. Lett. 2004, 31, 2003GL018299. [Google Scholar] [CrossRef]
- Fielding, A.; O’Muircheartaigh, C.A. Binary Segmentation in Survey Analysis with Particular Reference to AID. J. R. Stat. Soc. 1977, 26, 17–28. [Google Scholar] [CrossRef]
- Loh, W. Classification and Regression Trees. WIREs Data Min. Knowl. 2011, 1, 14–23. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer Series in Statistics; Springer: Berlin/Heidelberg, Germany, 2016; ISBN 978-0-387-84857-0. [Google Scholar]
- Laber, E.; Murtinho, L. Minimization of Gini Impurity: NP-Completeness and Approximation Algorithm via Connections with the k-Means Problem. Electron. Notes Theor. Comput. Sci. 2019, 346, 567–576. [Google Scholar] [CrossRef]
- Singh Kushwah, J.; Kumar, A.; Patel, S.; Soni, R.; Gawande, A.; Gupta, S. Comparative Study of Regressor and Classifier with Decision Tree Using Modern Tools. Mater. Today Proc. 2022, 56, 3571–3576. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Biau, G.; Scornet, E. A Random Forest Guided Tour. TEST 2016, 25, 197–227. [Google Scholar] [CrossRef]
- Tyralis, H.; Papacharalampous, G.; Langousis, A. A Brief Review of Random Forests for Water Scientists and Practitioners and Their Recent History in Water Resources. Water 2019, 11, 910. [Google Scholar] [CrossRef]
- Prasad, A.M.; Iverson, L.R.; Liaw, A. Newer Classification and Regression Tree Techniques: Bagging and Random Forests for Ecological Prediction. Ecosystems 2006, 9, 181–199. [Google Scholar] [CrossRef]
- Jiang, S.; Pang, G.; Wu, M.; Kuang, L. An Improved K-Nearest-Neighbor Algorithm for Text Categorization. Expert Syst. Appl. 2012, 39, 1503–1509. [Google Scholar] [CrossRef]
- Zhang, S. Challenges in KNN Classification. IEEE Trans. Knowl. Data Eng. 2022, 34, 4663–4675. [Google Scholar] [CrossRef]
- Zhang, S.; Li, X.; Zong, M.; Zhu, X.; Cheng, D. Learning k for kNN Classification. ACM Trans. Intell. Syst. Technol. 2017, 8, 1–19. [Google Scholar] [CrossRef]
- Varmuza, K.; Filzmoser, P.; Hilchenbach, M.; Krüger, H.; Silén, J. KNN Classification—Evaluated by Repeated Double Cross Validation: Recognition of Minerals Relevant for Comet Dust. Chemom. Intell. Lab. Syst. 2014, 138, 64–71. [Google Scholar] [CrossRef]
- Johansen, K.; Morton, M.J.L.; Malbeteau, Y.; Aragon, B.; Al-Mashharawi, S.; Ziliani, M.G.; Angel, Y.; Fiene, G.; Negrão, S.; Mousa, M.A.A.; et al. Predicting Biomass and Yield in a Tomato Phenotyping Experiment Using UAV Imagery and Random Forest. Front. Artif. Intell. 2020, 3, 28. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Ghulam, A.; Sidike, P.; Hartling, S.; Maimaitiyiming, M.; Peterson, K.; Shavers, E.; Fishman, J.; Peterson, J.; Kadam, S.; et al. Unmanned Aerial System (UAS)-Based Phenotyping of Soybean Using Multi-Sensor Data Fusion and Extreme Learning Machine. ISPRS J. Photogramm. Remote Sens. 2017, 134, 43–58. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Sagan, V.; Sidike, P.; Hartling, S.; Esposito, F.; Fritschi, F.B. Soybean Yield Prediction from UAV Using Multimodal Data Fusion and Deep Learning. Remote Sens. Environ. 2020, 237, 111599. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Sagan, V.; Sidike, P.; Daloye, A.M.; Erkbol, H.; Fritschi, F.B. Crop Monitoring Using Satellite/UAV Data Fusion and Machine Learning. Remote Sens. 2020, 12, 1357. [Google Scholar] [CrossRef]
- Impollonia, G.; Croci, M.; Blandinières, H.; Marcone, A.; Amaducci, S. Comparison of PROSAIL Model Inversion Methods for Estimating Leaf Chlorophyll Content and LAI Using UAV Imagery for Hemp Phenotyping. Remote Sens. 2022, 14, 5801. [Google Scholar] [CrossRef]
- Fei, S.; Xiao, S.; Li, Q.; Shu, M.; Zhai, W.; Xiao, Y.; Chen, Z.; Yu, H.; Ma, Y. Enhancing Leaf Area Index and Biomass Estimation in Maize with Feature Augmentation from Unmanned Aerial Vehicle-Based Nadir and Cross-Circling Oblique Photography. Comput. Electron. Agric. 2023, 215, 108462. [Google Scholar] [CrossRef]
- Feng, D.; Xu, W.; He, Z.; Zhao, W.; Yang, M. Advances in Plant Nutrition Diagnosis Based on Remote Sensing and Computer Application. Neural Comput. Appl. 2020, 32, 16833–16842. [Google Scholar] [CrossRef]
- Moghimi, A.; Yang, C.; Anderson, J.A. Aerial Hyperspectral Imagery and Deep Neural Networks for High-Throughput Yield Phenotyping in Wheat. Comput. Electron. Agric. 2020, 172, 105299. [Google Scholar] [CrossRef]
- Li, K.-Y.; Burnside, N.G.; Sampaio De Lima, R.; Villoslada Peciña, M.; Sepp, K.; Yang, M.-D.; Raet, J.; Vain, A.; Selge, A.; Sepp, K. The Application of an Unmanned Aerial System and Machine Learning Techniques for Red Clover-Grass Mixture Yield Estimation under Variety Performance Trials. Remote Sens. 2021, 13, 1994. [Google Scholar] [CrossRef]
- Dilmurat, K.; Sagan, V.; Moose, S. AI-Driven Maize Yield Forecasting Using Unmanned Aerial Vehicle-Based Hyperspectral and LiDAR Data Fusion. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2022, V-3-2022, 193–199. [Google Scholar] [CrossRef]
- Alabi, T.R.; Abebe, A.T.; Chigeza, G.; Fowobaje, K.R. Estimation of Soybean Grain Yield from Multispectral High-Resolution UAV Data with Machine Learning Models in West Africa. Remote Sens. Appl. Soc. Environ. 2022, 27, 100782. [Google Scholar] [CrossRef]
- Camenzind, M.P.; Yu, K. Multi Temporal Multispectral UAV Remote Sensing Allows for Yield Assessment across European Wheat Varieties Already before Flowering. Front. Plant Sci. 2024, 14, 1214931. [Google Scholar] [CrossRef] [PubMed]
- Medic, T.; Manser, N.; Kirchgessner, N.; Roth, L. Towards Wheat Yield Estimation in Plant Breeding from Inhomogeneous LiDAR Point Clouds Using Stochastic Features. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2023, XLVIII-1/W2-2023, 741–747. [Google Scholar] [CrossRef]
- Masjedi, A.; Crawford, M.M.; Carpenter, N.R.; Tuinstra, M.R. Multi-Temporal Predictive Modelling of Sorghum Biomass Using UAV-Based Hyperspectral and LiDAR Data. Remote Sens. 2020, 12, 3587. [Google Scholar] [CrossRef]
- Masjedi, A.; Crawford, M.M. Prediction of Sorghum Biomass Using Time Series UAV-Based Hyperspectral and LiDAR Data. In Proceedings of the IGARSS 2020—2020 IEEE International Geoscience and Remote Sensing Symposium, Waikoloa, HI, USA, 26 September–2 October 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 3912–3915. [Google Scholar]
- Freitas Moreira, F.; Rojas De Oliveira, H.; Lopez, M.A.; Abughali, B.J.; Gomes, G.; Cherkauer, K.A.; Brito, L.F.; Rainey, K.M. High-Throughput Phenotyping and Random Regression Models Reveal Temporal Genetic Control of Soybean Biomass Production. Front. Plant Sci. 2021, 12, 715983. [Google Scholar] [CrossRef]
- Revenga, J.C.; Trepekli, K.; Oehmcke, S.; Jensen, R.; Li, L.; Igel, C.; Gieseke, F.C.; Friborg, T. Above-Ground Biomass Prediction for Croplands at a Sub-Meter Resolution Using UAV–LiDAR and Machine Learning Methods. Remote Sens. 2022, 14, 3912. [Google Scholar] [CrossRef]
- Bhadra, S.; Sagan, V.; Skobalski, J.; Grignola, F.; Sarkar, S.; Vilbig, J. End-to-End 3D CNN for Plot-Scale Soybean Yield Prediction Using Multitemporal UAV-Based RGB Images. Precis. Agric. 2023, 25, 834–864. [Google Scholar] [CrossRef]
- Ji, Y.; Liu, R.; Xiao, Y.; Cui, Y.; Chen, Z.; Zong, X.; Yang, T. Faba Bean Above-Ground Biomass and Bean Yield Estimation Based on Consumer-Grade Unmanned Aerial Vehicle RGB Images and Ensemble Learning. Precis. Agric. 2023, 24, 1439–1460. [Google Scholar] [CrossRef]
- Fu, P.; Meacham-Hensold, K.; Guan, K.; Bernacchi, C.J. Hyperspectral Leaf Reflectance as Proxy for Photosynthetic Capacities: An Ensemble Approach Based on Multiple Machine Learning Algorithms. Front. Plant Sci. 2019, 10, 730. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Tardaguila, J.; Fernández-Novales, J.; Diago, M. Data Mining and NIR Spectroscopy in Viticulture: Applications for Plant Phenotyping under Field Conditions. Sensors 2016, 16, 236. [Google Scholar] [CrossRef]
- Garcia, L.C.; Concepcion, R.; Dadios, E.; Dulay, A.E. Spectro-Morphological Feature-Based Machine Learning Approach for Grape Leaf Variety Classification. In Proceedings of the 2022 IEEE 14th International Conference on Humanoid, Nanotechnology, Information Technology, Communication and Control, Environment, and Management (HNICEM), Boracay Island, Philippines, 1–4 December 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–6. [Google Scholar]
- Scharr, H.; Minervini, M.; French, A.P.; Klukas, C.; Kramer, D.M.; Liu, X.; Luengo, I.; Pape, J.-M.; Polder, G.; Vukadinovic, D.; et al. Leaf Segmentation in Plant Phenotyping: A Collation Study. Mach. Vis. Appl. 2016, 27, 585–606. [Google Scholar] [CrossRef]
- Ampatzidis, Y.; Partel, V. UAV-Based High Throughput Phenotyping in Citrus Utilizing Multispectral Imaging and Artificial Intelligence. Remote Sens. 2019, 11, 410. [Google Scholar] [CrossRef]
- Ampatzidis, Y.; Partel, V.; Meyering, B.; Albrecht, U. Citrus Rootstock Evaluation Utilizing UAV-Based Remote Sensing and Artificial Intelligence. Comput. Electron. Agric. 2019, 164, 104900. [Google Scholar] [CrossRef]
- Ampatzidis, Y.; Partel, V.; Costa, L. Agroview: Cloud-Based Application to Process, Analyze and Visualize UAV-Collected Data for Precision Agriculture Applications Utilizing Artificial Intelligence. Comput. Electron. Agric. 2020, 174, 105457. [Google Scholar] [CrossRef]
- Hati, A.J.; Ranjan Singh, R. Towards Smart Agriculture: A Deep Learning Based Phenotyping Scheme for Leaf Counting. In Proceedings of the 2020 International Conference on Smart Technologies in Computing, Electrical and Electronics (ICSTCEE), Bengaluru, India, 9–10 October 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 510–514. [Google Scholar]
- Uryasheva, A.; Kalashnikova, A.; Shadrin, D.; Evteeva, K.; Moskovtsev, E.; Rodichenko, N. Computer Vision-Based Platform for Apple Leaves Segmentation in Field Conditions to Support Digital Phenotyping. Comput. Electron. Agric. 2022, 201, 107269. [Google Scholar] [CrossRef]
- Yahata, S.; Onishi, T.; Yamaguchi, K.; Ozawa, S.; Kitazono, J.; Ohkawa, T.; Yoshida, T.; Murakami, N.; Tsuji, H. A Hybrid Machine Learning Approach to Automatic Plant Phenotyping for Smart Agriculture. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1787–1793. [Google Scholar]
- Hu, P.; Chapman, S.C.; Zheng, B. Coupling of Machine Learning Methods to Improve Estimation of Ground Coverage from Unmanned Aerial Vehicle (UAV) Imagery for High-Throughput Phenotyping of Crops. Funct. Plant Biol. 2021, 48, 766–779. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi-Tehran, P.; Virlet, N.; Ampe, E.M.; Reyns, P.; Hawkesford, M.J. DeepCount: In-Field Automatic Quantification of Wheat Spikes Using Simple Linear Iterative Clustering and Deep Convolutional Neural Networks. Front. Plant Sci. 2019, 10, 1176. [Google Scholar] [CrossRef] [PubMed]
- Misra, T.; Arora, A.; Marwaha, S.; Chinnusamy, V.; Rao, A.R.; Jain, R.; Sahoo, R.N.; Ray, M.; Kumar, S.; Raju, D.; et al. SpikeSegNet-a Deep Learning Approach Utilizing Encoder-Decoder Network with Hourglass for Spike Segmentation and Counting in Wheat Plant from Visual Imaging. Plant Methods 2020, 16, 40. [Google Scholar] [CrossRef] [PubMed]
- Alkhudaydi, T.; De La Lglesia, B. Counting Spikelets from Infield Wheat Crop Images Using Fully Convolutional Networks. Neural Comput. Appl. 2022, 34, 17539–17560. [Google Scholar] [CrossRef]
- Bauer, A.; Bostrom, A.G.; Ball, J.; Applegate, C.; Cheng, T.; Laycock, S.; Rojas, S.M.; Kirwan, J.; Zhou, J. Combining Computer Vision and Deep Learning to Enable Ultra-Scale Aerial Phenotyping and Precision Agriculture: A Case Study of Lettuce Production. Hortic. Res. 2019, 6, 70. [Google Scholar] [CrossRef]
- Haque, S.; Lobaton, E.; Nelson, N.; Yencho, G.C.; Pecota, K.V.; Mierop, R.; Kudenov, M.W.; Boyette, M.; Williams, C.M. Computer Vision Approach to Characterize Size and Shape Phenotypes of Horticultural Crops Using High-Throughput Imagery. Comput. Electron. Agric. 2021, 182, 106011. [Google Scholar] [CrossRef]
- Mbaye, M.; Audebert, A. Identification and Counting of Sorghum Panicles Using Artificial Intelligence Based Drone Field Phenotyping. Adv. Artif. Intell. Mach. Learn. 2021, 01, 234–240. [Google Scholar] [CrossRef]
- Karami, A.; Crawford, M.; Delp, E.J. Automatic Plant Counting and Location Based on a Few-Shot Learning Technique. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 5872–5886. [Google Scholar] [CrossRef]
- Zaji, A.; Liu, Z.; Xiao, G.; Bhowmik, P.; Sangha, J.S.; Ruan, Y. AutoOLA: Automatic Object Level Augmentation for Wheat Spikes Counting. Comput. Electron. Agric. 2023, 205, 107623. [Google Scholar] [CrossRef]
- Yalcin, H. Phenology Recognition Using Deep Learning. In Proceedings of the 2018 Electric Electronics, Computer Science, Biomedical Engineerings’ Meeting (EBBT), Istanbul, Turkey, 18–19 April 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–5. [Google Scholar]
- Davis Ii, R.L.; Greene, J.K.; Dou, F.; Jo, Y.-K.; Chappell, T.M. A Practical Application of Unsupervised Machine Learning for Analyzing Plant Image Data Collected Using Unmanned Aircraft Systems. Agronomy 2020, 10, 633. [Google Scholar] [CrossRef]
- Ashapure, A.; Jung, J.; Oh, S.; Chang, A.; Dube, N.; Landivar, J. Combining UAS and Sentinel-2 Data to Estimate Canopy Parameters of a Cotton Crop Using Machine Learning. In Proceedings of the IGARSS 2020—2020 IEEE International Geoscience and Remote Sensing Symposium, Waikoloa, HI, USA, 26 September–2 October 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 5199–5202. [Google Scholar]
- Koh, J.C.O.; Spangenberg, G.; Kant, S. Automated Machine Learning for High-Throughput Image-Based Plant Phenotyping. Remote Sens. 2021, 13, 858. [Google Scholar] [CrossRef]
- Li, K.-Y.; Burnside, N.G.; De Lima, R.S.; Peciña, M.V.; Sepp, K.; Cabral Pinheiro, V.H.; De Lima, B.R.C.A.; Yang, M.-D.; Vain, A.; Sepp, K. An Automated Machine Learning Framework in Unmanned Aircraft Systems: New Insights into Agricultural Management Practices Recognition Approaches. Remote Sens. 2021, 13, 3190. [Google Scholar] [CrossRef]
- Tan, S.; Liu, J.; Lu, H.; Lan, M.; Yu, J.; Liao, G.; Wang, Y.; Li, Z.; Qi, L.; Ma, X. Machine Learning Approaches for Rice Seedling Growth Stages Detection. Front. Plant Sci. 2022, 13, 914771. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, S.; Blystone, D.; Singh, A.K.; Ganapathysubramanian, B.; Singh, A.; Sarkar, S. An Explainable Deep Machine Vision Framework for Plant Stress Phenotyping. Proc. Natl. Acad. Sci. USA 2018, 115, 4613–4618. [Google Scholar] [CrossRef] [PubMed]
- Backhaus, A.; Bollenbeck, F.; Seiffert, U. Robust Classification of the Nutrition State in Crop Plants by Hyperspectral Imaging and Artificial Neural Networks. In Proceedings of the 2011 3rd Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Lisbon, Portugal, 6–9 June 2011; IEEE: Piscataway, NJ, USA, 2011; pp. 1–4. [Google Scholar]
- Aquino, H.; Sybingco, E.; Mendigoria, C.H.; Concepcion, R.; Bandala, A.; Alajas, O.J.; Dadios, E.; Vicerra, R.R. On-Demand Healthy and Chlorotic Lactuca Sativa Leaf Classification Using Support Vector Machine in a Rotating Hydroponic System. In Proceedings of the 2022 IEEE 14th International Conference on Humanoid, Nanotechnology, Information Technology, Communication and Control, Environment, and Management (HNICEM), Boracay Island, Philippines, 1–4 December 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–5. [Google Scholar]
- Abdalla, A.; Cen, H.; Wan, L.; Mehmood, K.; He, Y. Nutrient Status Diagnosis of Infield Oilseed Rape via Deep Learning-Enabled Dynamic Model. IEEE Trans. Ind. Inf. 2021, 17, 4379–4389. [Google Scholar] [CrossRef]
- Okyere, F.G.; Cudjoe, D.; Sadeghi-Tehran, P.; Virlet, N.; Riche, A.B.; Castle, M.; Greche, L.; Simms, D.; Mhada, M.; Mohareb, F.; et al. Modeling the Spatial-Spectral Characteristics of Plants for Nutrient Status Identification Using Hyperspectral Data and Deep Learning Methods. Front. Plant Sci. 2023, 14, 1209500. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, X.; Cheng, Q.; Fei, S.; Chen, Z. A Machine-Learning Model Based on the Fusion of Spectral and Textural Features from UAV Multi-Sensors to Analyse the Total Nitrogen Content in Winter Wheat. Remote Sens. 2023, 15, 2152. [Google Scholar] [CrossRef]
- Yao, J.; Sun, D.; Cen, H.; Xu, H.; Weng, H.; Yuan, F.; He, Y. Phenotyping of Arabidopsis Drought Stress Response Using Kinetic Chlorophyll Fluorescence and Multicolor Fluorescence Imaging. Front. Plant Sci. 2018, 9, 603. [Google Scholar] [CrossRef] [PubMed]
- Sankararao, A.U.G.; Rajalakshmi, P.; Kaliamoorthy, S.; Choudhary, S. Water Stress Detection in Pearl Millet Canopy with Selected Wavebands Using UAV Based Hyperspectral Imaging and Machine Learning. In Proceedings of the 2022 IEEE Sensors Applications Symposium (SAS), Sundsvall, Sweden, 1–3 August 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–6. [Google Scholar]
- Ruszczak, B. Reducing High-Dimensional Feature Set of Hyperspectral Measurements for Plant Phenotype Classification. In Proceedings of the Companion Conference on Genetic and Evolutionary Computation, Lisbon, Portugal, 15–19 July 2023; ACM: New York, NY, USA, 2023; pp. 77–78. [Google Scholar]
- Chandel, N.S.; Chakraborty, S.K.; Rajwade, Y.A.; Dubey, K.; Tiwari, M.K.; Jat, D. Identifying Crop Water Stress Using Deep Learning Models. Neural Comput. Appl. 2021, 33, 5353–5367. [Google Scholar] [CrossRef]
- Gupta, E.; Azimi, S.; Gandhi, T.K. Characterizing Water Deficiency Induced Stress in Plants Using Gabor Filter Based CNN. In Proceedings of the 2022 IEEE IAS Global Conference on Emerging Technologies (GlobConET), Arad, Romania, 20–22 May 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 91–95. [Google Scholar]
- Brewer, K.; Clulow, A.; Sibanda, M.; Gokool, S.; Odindi, J.; Mutanga, O.; Naiken, V.; Chimonyo, V.G.P.; Mabhaudhi, T. Estimation of Maize Foliar Temperature and Stomatal Conductance as Indicators of Water Stress Based on Optical and Thermal Imagery Acquired Using an Unmanned Aerial Vehicle (UAV) Platform. Drones 2022, 6, 169. [Google Scholar] [CrossRef]
- Zahid, A.; Dashtipour, K.; Abbas, H.T.; Mabrouk, I.B.; Al-Hasan, M.; Ren, A.; Imran, M.A.; Alomainy, A.; Abbasi, Q.H. Machine Learning Enabled Identification and Real-Time Prediction of Living Plants’ Stress Using Terahertz Waves. Def. Technol. 2022, 18, 1330–1339. [Google Scholar] [CrossRef]
- Wahabzada, M.; Mahlein, A.-K.; Bauckhage, C.; Steiner, U.; Oerke, E.-C.; Kersting, K. Metro Maps of Plant Disease Dynamics—Automated Mining of Differences Using Hyperspectral Images. PLoS ONE 2015, 10, e0116902. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Bueno, M.L.; Pineda, M.; Cabeza, F.M.; Barón, M. Multicolor Fluorescence Imaging as a Candidate for Disease Detection in Plant Phenotyping. Front. Plant Sci. 2016, 7, 1790. [Google Scholar] [CrossRef] [PubMed]
- Pineda, M.; Pérez-Bueno, M.L.; Paredes, V.; Barón, M. Use of Multicolour Fluorescence Imaging for Diagnosis of Bacterial and Fungal Infection on Zucchini by Implementing Machine Learning. Funct. Plant Biol. 2017, 44, 563. [Google Scholar] [CrossRef] [PubMed]
- Pineda, M.; Pérez-Bueno, M.L.; Barón, M. Detection of Bacterial Infection in Melon Plants by Classification Methods Based on Imaging Data. Front. Plant Sci. 2018, 9, 164. [Google Scholar] [CrossRef]
- Islam, M.; Dinh, A.; Wahid, K.; Bhowmik, P. Detection of Potato Diseases Using Image Segmentation and Multiclass Support Vector Machine. In Proceedings of the 2017 IEEE 30th Canadian Conference on Electrical and Computer Engineering (CCECE), Windsor, ON, USA, 30 April–3 May 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–4. [Google Scholar]
- Żelazny, W.R.; Chrpová, J.; Hamouz, P. Fusarium Head Blight Detection from Spectral Measurements in a Field Phenotyping Setting—A Pre-Registered Study. Biosyst. Eng. 2021, 211, 97–113. [Google Scholar] [CrossRef]
- Simões, I.O.P.S.; Rios Do Amaral, L. UAV-Based Multispectral Data for Sugarcane Resistance Phenotyping of Orange and Brown Rust. Smart Agric. Technol. 2023, 4, 100144. [Google Scholar] [CrossRef]
- Mendigoria, C.H.; Concepcion, R.; Bandala, A.; Alajas, O.J.; Aquino, H.; Dadios, E. OryzaNet: Leaf Quality Assessment of Oryza Sativa Using Hybrid Machine Learning and Deep Neural Network. In Proceedings of the 2021 IEEE 13th International Conference on Humanoid, Nanotechnology, Information Technology, Communication and Control, Environment, and Management (HNICEM), Manila, Philippines, 28–30 November 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1–6. [Google Scholar]
- Noola, D.A.; Basavaraju, D.R. Corn Leaf Disease Detection with Pertinent Feature Selection Model Using Machine Learning Technique with Efficient Spot Tagging Model. RIA 2021, 35, 477–482. [Google Scholar] [CrossRef]
- El Abidine, M.Z.; Merdinoglu-Wiedemann, S.; Rasti, P.; Dutagaci, H.; Rousseau, D. Machine Learning-Based Classification of Powdery Mildew Severity on Melon Leaves. In Image and Signal Processing; El Moataz, A., Mammass, D., Mansouri, A., Nouboud, F., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2020; Volume 12119, pp. 74–81. ISBN 978-3-030-51934-6. [Google Scholar]
- Dong, X.; Zhao, K.; Wang, Q.; Wu, X.; Huang, Y.; Wu, X.; Zhang, T.; Dong, Y.; Gao, Y.; Chen, P.; et al. PlantPAD: A Platform for Large-Scale Image Phenomics Analysis of Disease in Plant Science. Nucleic Acids Res. 2024, 52, D1556–D1568. [Google Scholar] [CrossRef]
- Arribas, J.I.; Sánchez-Ferrero, G.V.; Ruiz-Ruiz, G.; Gómez-Gil, J. Leaf Classification in Sunflower Crops by Computer Vision and Neural Networks. Comput. Electron. Agric. 2011, 78, 9–18. [Google Scholar] [CrossRef]
- Pérez-Ortiz, M.; Peña, J.M.; Gutiérrez, P.A.; Torres-Sánchez, J.; Hervás-Martínez, C.; López-Granados, F. A Semi-Supervised System for Weed Mapping in Sunflower Crops Using Unmanned Aerial Vehicles and a Crop Row Detection Method. Appl. Soft Comput. 2015, 37, 533–544. [Google Scholar] [CrossRef]
- Tun, E.E.M. Comparison Analysis of Oil Crop Yield Prediction in Magway Region Using Machine Learning Method. In Proceedings of the 2023 IEEE Conference on Computer Applications (ICCA), Yangon, Myanmar, 27–28 February 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 86–90. [Google Scholar]
- Amankulova, K.; Farmonov, N.; Mucsi, L. Time-Series Analysis of Sentinel-2 Satellite Images for Sunflower Yield Estimation. Smart Agric. Technol. 2023, 3, 100098. [Google Scholar] [CrossRef]
- Song, Z.; Zhang, Z.; Yang, S.; Ding, D.; Ning, J. Identifying Sunflower Lodging Based on Image Fusion and Deep Semantic Segmentation with UAV Remote Sensing Imaging. Comput. Electron. Agric. 2020, 179, 105812. [Google Scholar] [CrossRef]
- Atsmon, G.; Nehurai, O.; Kizel, F.; Eizenberg, H.; Nisim Lati, R. Hyperspectral Imaging Facilitates Early Detection of Orobanche Cumana Below-Ground Parasitism on Sunflower under Field Conditions. Comput. Electron. Agric. 2022, 196, 106881. [Google Scholar] [CrossRef]
- Pinto, L.S.; Ray, A.; Reddy, M.U.; Perumal, P.; Aishwarya, P. Crop Disease Classification Using Texture Analysis. In Proceedings of the 2016 IEEE International Conference on Recent Trends in Electronics, Information & Communication Technology (RTEICT), Bangalore, India, 20–21 May 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 825–828. [Google Scholar]
- Liu, J.; Lv, F.; Di, P. Identification of Sunflower Leaf Diseases Based on Random Forest Algorithm. In Proceedings of the 2019 International Conference on Intelligent Computing, Automation and Systems (ICICAS), Chongqing, China, 6–8 December 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 459–463. [Google Scholar]
- Dawod, R.G.; Dobre, C. Classification of Sunflower Foliar Diseases Using Convolutional Neural Network. In Proceedings of the 2021 23rd International Conference on Control Systems and Computer Science (CSCS), Bucharest, Romania, 26–28 May 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 476–481. [Google Scholar]
- Ghosh, P.; Mondal, A.K.; Chatterjee, S.; Masud, M.; Meshref, H.; Bairagi, A.K. Recogcnition of Sunflower Diseases Using Hybrid Deep Learning and Its Explainability with AI. Mathematics 2023, 11, 2241. [Google Scholar] [CrossRef]









| Learning Type | Used for | Algorithms |
|---|---|---|
| Supervised Learning | Classification Regression Prediction | Bayesian networks Support Vector Machine Random Forest Neural networks Decision tree Hidden Markov model Naïve Bayes |
| Unsupervised Learning | Clustering Dimensionality reduction | k-means x-means Principal Component Analysis Independent Component Analysis Gaussian mixture models |
| Semi-supervised Learning | Hybrid | Self-training Transductive Support Vector Machine Generative models |
| Reinforcement Learning | Real-time decision making | Q-learning Markov decision process |
| Advantages | Disadvantages |
|---|---|
| Ability to identify trends and patterns that are invisible to humans. | Algorithms require huge unbiased datasets for training. |
| Automation due to the ability of algorithms to self-learn and improve. | Requires a lot of time and resources. |
| Continuous improvement in accuracy and efficiency. | Specific skills are required to achieve good interpretation of results. |
| Possibility to handle multi-dimensional and multi-sources data. | High error-susceptibility, especially when the algorithm is trained on insufficient datasets that generate bias. |
| Wide applications (engineering, medicine, agriculture, etc.). |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Centorame, L.; Gasperini, T.; Ilari, A.; Del Gatto, A.; Foppa Pedretti, E. An Overview of Machine Learning Applications on Plant Phenotyping, with a Focus on Sunflower. Agronomy 2024, 14, 719. https://doi.org/10.3390/agronomy14040719
Centorame L, Gasperini T, Ilari A, Del Gatto A, Foppa Pedretti E. An Overview of Machine Learning Applications on Plant Phenotyping, with a Focus on Sunflower. Agronomy. 2024; 14(4):719. https://doi.org/10.3390/agronomy14040719
Chicago/Turabian StyleCentorame, Luana, Thomas Gasperini, Alessio Ilari, Andrea Del Gatto, and Ester Foppa Pedretti. 2024. "An Overview of Machine Learning Applications on Plant Phenotyping, with a Focus on Sunflower" Agronomy 14, no. 4: 719. https://doi.org/10.3390/agronomy14040719
APA StyleCentorame, L., Gasperini, T., Ilari, A., Del Gatto, A., & Foppa Pedretti, E. (2024). An Overview of Machine Learning Applications on Plant Phenotyping, with a Focus on Sunflower. Agronomy, 14(4), 719. https://doi.org/10.3390/agronomy14040719