Pan-Genome Analysis and Secondary Metabolic Pathway Mining of Biocontrol Bacterium Brevibacillus brevis
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Average Nucleotide Identity (ANI) and Digital DNA–DNA Hybridization (dDDH) Analysis
2.3. Phylogenetic Analyses Based on Genomic Sequences
2.4. Pan-Genome Analysis
2.5. Analysis of Secondary Metabolite Biosynthetic Gene Clusters
3. Results
3.1. General Genomic Characteristics of B. brevis
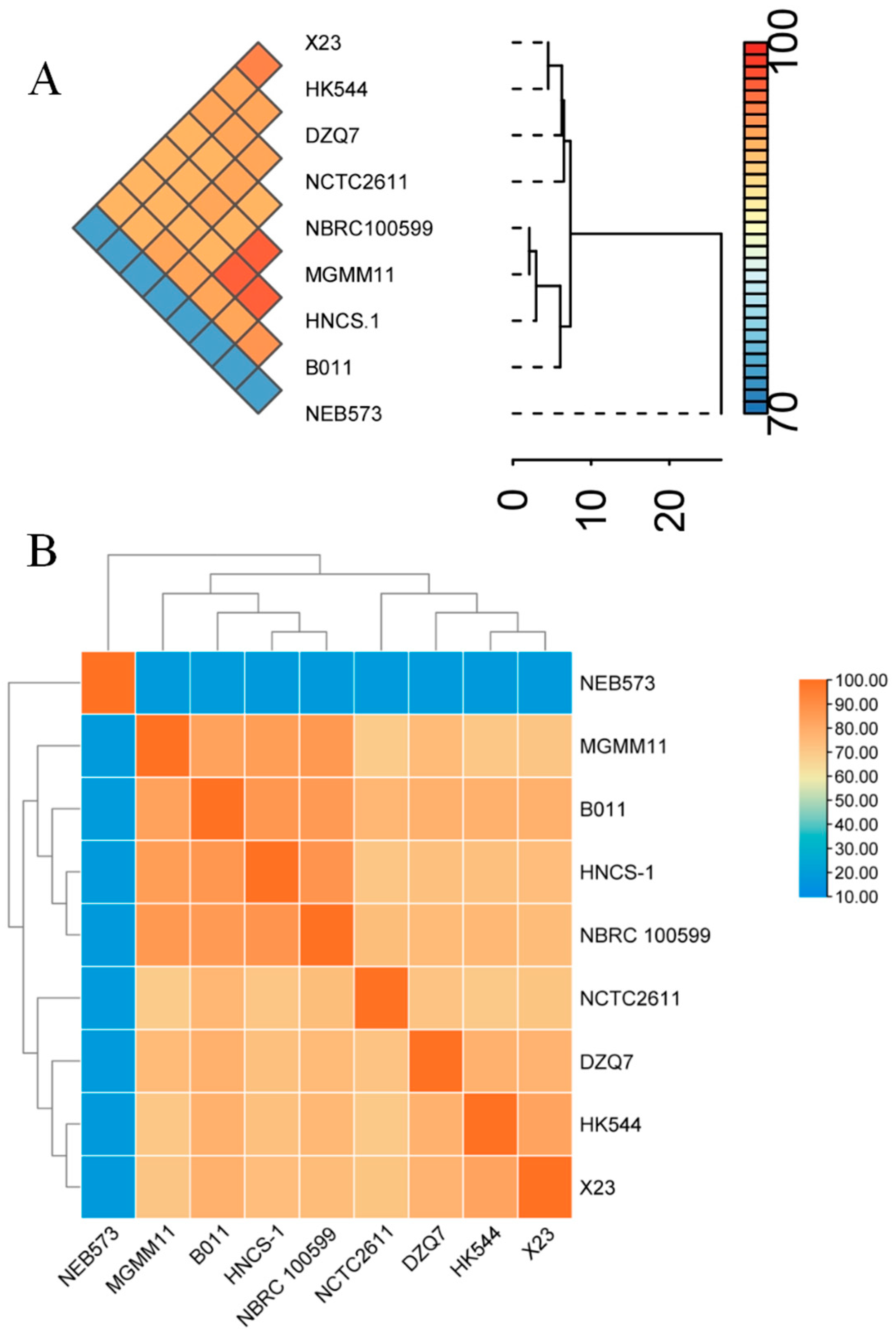
3.2. Genetic Diversity of the Nine B. brevis Strains Based on ANI and dDDH Analyses
3.3. Phylogenetic Analysis of the Nine B. brevis Strains
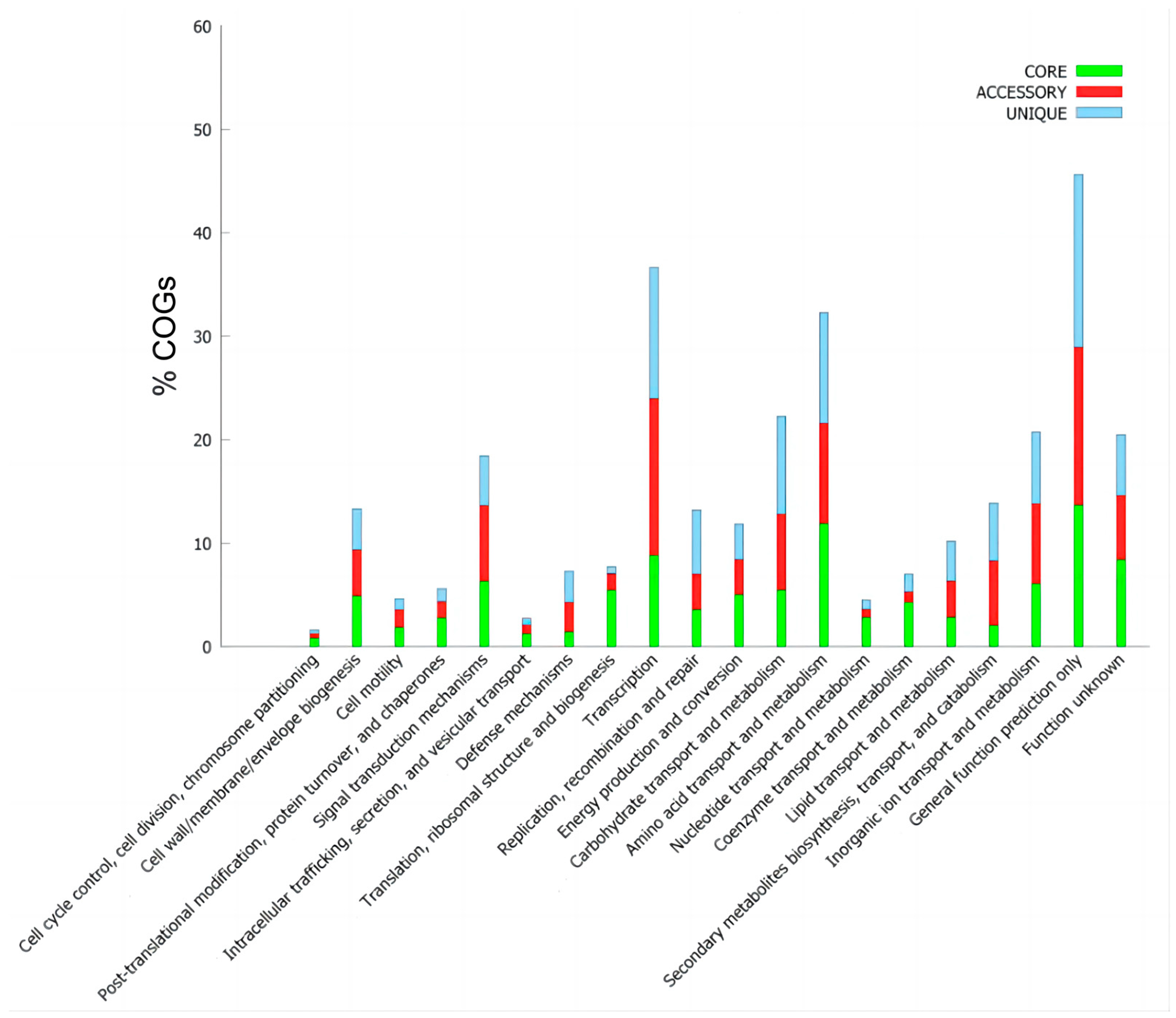
3.4. Pan-Genome Characteristics of B. brevis
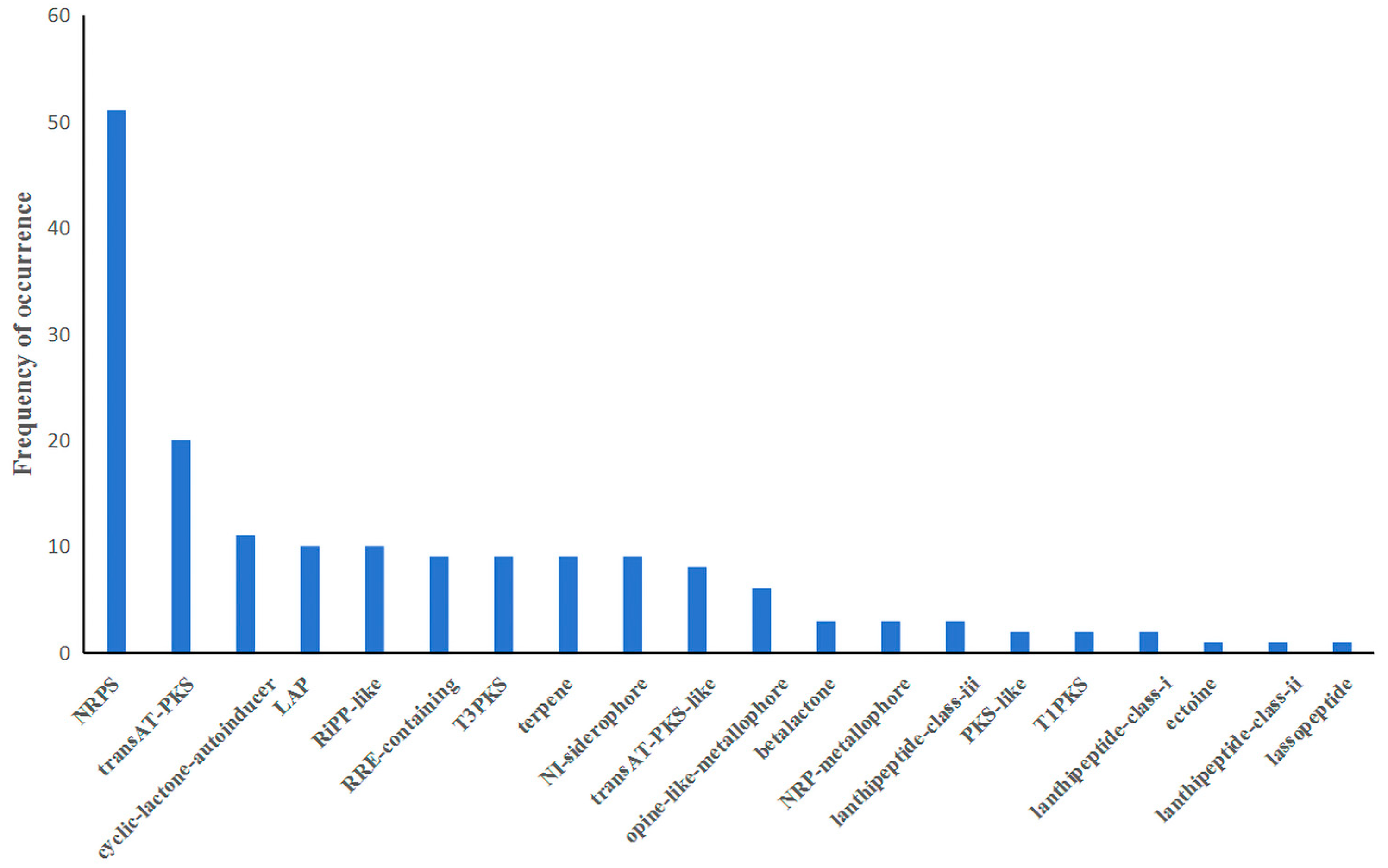
3.5. Secondary Metabolite Biosynthetic Gene Cluster of B. brevis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Panda, A.K.; Bisht, S.S.; DeMondal, S.; Senthil Kumar, N.; Gurusubramanian, G.; Panigrahi, A.K. Brevibacillus as a biological tool: A short review. Antonie Van Leeuwenhoek 2014, 105, 623–639. [Google Scholar] [CrossRef] [PubMed]
- Che, J.; Ye, S.; Liu, B.; Deng, Y.; Chen, Q.; Ge, C.; Liu, G.; Wang, J. Effects of Brevibacillus brevis FJAT-1501-BPA on growth performance, faecal microflora, faecal enzyme activities and blood parameters of weaned piglets. Antonie Van Leeuwenhoek 2016, 109, 1545–1553. [Google Scholar] [CrossRef] [PubMed]
- Samrot, A.V.; Prasad, R.J.; Rio, A.J.; Sneha, S.J. Bioprospecting of Brevibacillus brevis isolated from soil. Recent Pat. Biotechnol. 2015, 9, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, L.; Wang, R.; Wang, X.; Ye, J.; Long, Y. Biosorption and degradation of decabromodiphenyl ether by Brevibacillus brevis and the influence of decabromodiphenyl ether on cellular metabolic responses. Environ. Sci. Pollut. Res. Int. 2016, 23, 5166–5178. [Google Scholar] [CrossRef] [PubMed]
- Che, J.; Liu, B.; Lin, Y.; Tang, W.; Tang, J. Draft genome sequence of biocontrol bacterium Brevibacillus brevis strain FJAT-0809-GLX. Genome Announc. 2013, 1, e0016013. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Wang, C.; Hou, X.; Xia, Z.; Ye, J.; Liu, K.; Liu, H.; Wang, J.; Guo, H.; Yu, X.; et al. Draft genome sequence of Brevibacillus brevis DZQ7, a plant growth-promoting rhizobacterium with broad-spectrum antimicrobial activity. Genome Announc. 2015, 3, e00831-15. [Google Scholar] [CrossRef] [PubMed]
- Marche, M.G.; Mura, M.E.; Falchi, G.; Ruiu, L. Spore surface proteins of Brevibacillus laterosporus are involved in insect pathogenesis. Sci. Rep. 2017, 7, 43805. [Google Scholar] [CrossRef] [PubMed]
- Jianmei, C.; Bo, L.; Zheng, C.; Huai, S.; Guohong, L.; Cibin, G. Identification of ethylparaben as the antimicrobial substance produced by Brevibacillus brevis FJAT-0809-GLX. Microbiol. Res. 2015, 172, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.T.; Bowman, M.J.; Dunlap, C.A. Brevibacillus fortis NRS-1210 produces edeines that inhibit the in vitro growth of conidia and chlamydospores of the onion pathogen Fusarium oxysporum f. sp. cepae. Antonie Van Leeuwenhoek 2020, 113, 973–987. [Google Scholar] [CrossRef]
- Kierońska, D.; Rózalska, B.; Zablocki, B. Immunosuppressive properties of the antibiotic edeine. Bull. Acad. Pol. Sci. Biol. 1976, 24, 705–710. [Google Scholar]
- Li, S.; Zhao, Z.; Li, M.; Gu, Z.; Bai, C.; Huang, W. Purification and characterization of a novel chitinase from Bacillus brevis. Acta Biochim. Biophys. Sin. 2002, 34, 690–696. [Google Scholar]
- Rautenbach, M.; Vlok, N.M.; Stander, M.; Hoppe, H.C. Inhibition of malaria parasite blood stages by tyrocidines, membrane-active cyclic peptide antibiotics from Bacillus brevis. Biochim. Biophys. Acta 2007, 1768, 1488–1497. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Haddad, N.; Yang, S.; Mu, B. Structural characterization of lipopeptides from Brevibacillus brevis HOB1. Appl. Biochem. Biotechnol. 2010, 160, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Liu, Q.; Guo, H.; Ju, R.; Zhao, Y.; Li, J.; Liu, X. Tostadin, a novel antibacterial peptide from an antagonistic microorganism Brevibacillus brevis XDH. Bioresour. Technol. 2012, 111, 504–506. [Google Scholar] [CrossRef]
- Ghadbane, M.; Harzallah, D.; Laribi, A.I.; Jaouadi, B.; Belhadj, H. Purification and biochemical characterization of a highly thermostable bacteriocin isolated from Brevibacillus brevis strain GM100. Biosci. Biotechnol. Biochem. 2013, 77, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, S.A.; Ali, A.; Naz, A.; Hassan, A.; Riaz, N.; Saeed-ul-Hassan, S.; Andleeb, S.; Barh, D. A New Broad-Spectrum Peptide Antibiotic Produced by Bacillus brevis Strain MH9 Isolated from Margalla Hills of Islamabad, Pakistan. Int. J. Pept. Res. Ther. 2016, 22, 271–279. [Google Scholar] [CrossRef]
- Sheng, M.; Jia, H.; Zhang, G.; Zeng, L.; Zhang, T.; Long, Y.; Lan, J.; Hu, Z.; Zeng, Z.; Wang, B. Siderophore production by rhizosphere biological control bacteria Brevibacillus brevis GZDF3 of Pinellia ternata and its antifungal effects on Candida albicans. J. Microbiol. Biotechnol. 2020, 30, 689. [Google Scholar] [CrossRef]
- Binnewies, T.T.; Motro, Y.; Hallin, P.F.; Lund, O.; Dunn, D.; La, T.; Hampson, D.J.; Bellgard, M.; Wassenaar, T.M.; Ussery, D.W. Ten years of bacterial genome sequencing: Comparative-genomics-based discoveries. Funct. Integr. Genom. 2006, 6, 165–185. [Google Scholar] [CrossRef]
- Vernikos, G.; Medini, D.; Riley, D.R.; Tettelin, H. Ten years of pan-genome analyses. Curr. Opin. Microbiol. 2015, 23, 148–154. [Google Scholar] [CrossRef]
- Kim, Y.; Koh, I.; Young Lim, M.; Chung, W.-H.; Rho, M. Pan-genome analysis of Bacillus for microbiome profiling. Sci. Rep. 2017, 7, 10984. [Google Scholar] [CrossRef]
- Xing, J.; Li, X.; Sun, Y.; Zhao, J.; Miao, S.; Xiong, Q.; Zhang, Y.; Zhang, G. Comparative genomic and functional analysis of Akkermansia muciniphila and closely related species. Genes Genom. 2019, 41, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Belbahri, L.; Chenari Bouket, A.; Rekik, I.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Petrovova, E.; Oszako, T.; Cherrad, S.; Vacher, S. Comparative genomics of Bacillus amyloliquefaciens strains reveals a core genome with traits for habitat adaptation and a secondary metabolites rich accessory genome. Front. Microbiol. 2017, 8, 1438. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.-A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, Y.; Fan, G.; Sun, D.; Zhang, X.; Yu, Z.; Wang, J.; Wu, L.; Shi, W.; Ma, J. IPGA: A handy integrated prokaryotes genome and pan-genome analysis web service. iMeta 2022, 1, e55. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Al-Saari, N.; Gao, F.; AKM Rohul, A.; Sato, K.; Sato, K.; Mino, S.; Suda, W.; Oshima, K.; Hattori, M.; Ohkuma, M. Advanced microbial taxonomy combined with genome-based-approaches reveals that Vibrio astriarenae sp. nov., an agarolytic marine bacterium, forms a new clade in Vibrionaceae. PLoS ONE 2015, 10, e0136279. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, K.T.; Tiedje, J.M. Prokaryotic taxonomy and phylogeny in the genomic era: Advancements and challenges ahead. Curr. Opin. Microbiol. 2007, 10, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA-an ultra-fast pan-genome analysis pipeline. Sci Rep. 2016, 6, 24373. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 1, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Medema, M.H.; Kottmann, R.; Lee, S.Y.; Weber, T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2017, 45, D555–D559. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Kim, Y.S.; Han, J.W.; Choi, G.J.; Kim, H. Genome Sequence of Brevibacillus brevis HK544, an Antimicrobial Bacterium Isolated from Soil in Daejeon, South Korea. Microbiol. Resour. Announc. 2021, 10, e0041721. [Google Scholar] [CrossRef]
- Chen, W.; Wang, Y.; Li, D.; Li, L.; Xiao, Q.; Zhou, Q. Draft genome sequence of Brevibacillus brevis strain X23, a biocontrol agent against bacterial wilt. J. Bacteriol. 2012, 194, 6634–6635. [Google Scholar] [CrossRef]
- Yang, W.; Yang, H.; Hussain, M.; Qin, X. Brevibacillus brevis HNCS-1: A biocontrol bacterium against tea plant diseases. Front. Microbiol. 2023, 14, 1198747. [Google Scholar] [CrossRef]
- Yang, X.; Li, Y.; Zang, J.; Li, Y.; Bie, P.; Lu, Y.; Wu, Q. Analysis of pan-genome to identify the core genes and essential genes of Brucella spp. Mol. Genet. Genom. 2016, 291, 905–912. [Google Scholar] [CrossRef]
- Nehra, V.; Saharan, B.S.; Choudhary, M. Evaluation of Brevibacillus brevis as a potential plant growth promoting rhizobacteria for cotton (Gossypium hirsutum) crop. Springerplus 2016, 5, 948. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Fu, J.; Qin, Q. Pan-genomics analysis of 30 Escherichia coli genomes. Yi Chuan 2012, 34, 765–772. [Google Scholar] [CrossRef]
- Jeske, O.; Jogler, M.; Petersen, J.; Sikorski, J.; Jogler, C. From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules. Antonie Van Leeuwenhoek 2013, 104, 551–567. [Google Scholar] [CrossRef] [PubMed]
- Letzel, A.-C.; Pidot, S.J.; Hertweck, C. Genome mining for ribosomally synthesized and post-translationally modified peptides (RiPPs) in anaerobic bacteria. BMC Genom. 2014, 15, 983. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Kuipers, O.P. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genom. 2016, 17, 119. [Google Scholar] [CrossRef] [PubMed]


| Strain | Size Mb | GC % | Gene | CDS | Source | Country | Accession No. | Reference |
|---|---|---|---|---|---|---|---|---|
| NCTC2611 | 6.73 | 47.5 | 6592 | 6425 | — | — | LR134338.1 | — |
| HK544 | 6.49 | 47.5 | 6133 | 5766 | Soil | South Korea | CP042161.1 | [34] |
| DZQ7 | 6.44 | 47.5 | 6087 | 5752 | Tobacco rhizosphere soil | China | CP030117.1 | [6] |
| NBRC 100599 | 6.30 | 47.5 | 6123 | 5949 | — | — | AP008955.1 | — |
| X23 | 6.64 | 47.0 | 6450 | 6144 | Soil of vegetable field | China | CP023474.1 | [35] |
| B011 | 6.16 | 47.5 | 5784 | 5455 | Tobacco roots | China | CP041767.1 | — |
| NEB573 | 6.23 | 54.0 | 6106 | 5856 | Cell culture | — | CP134050.1 | — |
| HNCS-1 | 6.35 | 47.0 | 6041 | 5770 | Soil | China | CP128411.1 | [36] |
| MGMM11 | 6.32 | 47.0 | 5893 | 5776 | Rhizospheric soil | Russia | CP124547.1 | — |
| Strain | Accession No. | Gene Cluster | Start | End | Homolog of Known Cluster | Similarity |
|---|---|---|---|---|---|---|
| NCTC2611 | LR134338.1 | Cluster 1 | 1476119 | 1536630 | ulbactin F/ulbactin G | 100% |
| Cluster 2 | 3005749 | 3016132 | ectoine | 100% | ||
| Cluster 3 | 3024715 | 3133409 | gramicidin | 91% | ||
| Cluster 4 | 3177936 | 3200077 | bacillopaline | 100% | ||
| Cluster 5 | 3202856 | 3276565 | tyrocidine | 75% | ||
| Cluster 6 | 4016684 | 4048395 | petrobactin | 83% | ||
| HK544 | CP042161.1 | Cluster 7 | 361636 | 422151 | ulbactin F/ulbactin G | 100% |
| Cluster 8 | 1975342 | 2049051 | tyrocidine | 81% | ||
| Cluster 9 | 2814515 | 2846223 | petrobactin | 83% | ||
| Cluster 10 | 5799588 | 5988436 | macrobrevin | 100% | ||
| DZQ7 | CP030117.1 | Cluster 11 | 2108577 | 2140291 | petrobactin | 83% |
| Cluster 12 | 2955322 | 3051077 | tyrocidine | 81% | ||
| Cluster 13 | 3161865 | 3399925 | marthiapeptide A | 83% | ||
| Cluster 14 | 5529371 | 5713939 | macrobrevin | 100% | ||
| NBRC 100599 | AP008955.1 | Cluster 15 | 2116926 | 2148625 | petrobactin | 83% |
| Cluster 16 | 2856081 | 2929792 | tyrocidine | 100% | ||
| Cluster 17 | 2932793 | 2954934 | bacillopaline | 100% | ||
| Cluster 18 | 3002439 | 3150775 | gramicidin | 100% | ||
| X23 | CP023474.1 | Cluster 19 | 2255634 | 2287345 | petrobactin | 83% |
| Cluster 20 | 3097361 | 3167698 | tyrocidine | 81% | ||
| Cluster 21 | 4897981 | 4958493 | ulbactin F/ulbactin G | 100% | ||
| B011 | CP041767.1 | Cluster 22 | 2141737 | 2173451 | petrobactin | 83% |
| Cluster 23 | 2896872 | 2988295 | tyrocidine | 81% | ||
| Cluster 24 | 3019366 | 3143800 | gramicidin | 91% | ||
| HNCS-1 | CP128411.1 | Cluster 25 | 2178678 | 2210377 | petrobactin | 83% |
| Cluster 26 | 2946955 | 3020665 | tyrocidine | 87% | ||
| Cluster 27 | 3022626 | 3044767 | bacillopaline | 100% | ||
| Cluster 28 | 3088959 | 3197575 | gramicidin | 91% | ||
| MGMM11 | CP124547.1 | Cluster 29 | 2772174 | 2803873 | petrobactin | 83% |
| Cluster 30 | 3518961 | 3592672 | tyrocidine | 81% | ||
| Cluster 31 | 3595633 | 3617774 | bacillopaline | 100% | ||
| Cluster 32 | 3673595 | 3781752 | gramicidin | 91% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, J.; Huang, B.; Huang, J.; Long, Q.; Zhang, C.; Guo, Z.; Wang, Y.; Chen, W.; Tan, S.; Liu, Q. Pan-Genome Analysis and Secondary Metabolic Pathway Mining of Biocontrol Bacterium Brevibacillus brevis. Agronomy 2024, 14, 1024. https://doi.org/10.3390/agronomy14051024
Du J, Huang B, Huang J, Long Q, Zhang C, Guo Z, Wang Y, Chen W, Tan S, Liu Q. Pan-Genome Analysis and Secondary Metabolic Pathway Mining of Biocontrol Bacterium Brevibacillus brevis. Agronomy. 2024; 14(5):1024. https://doi.org/10.3390/agronomy14051024
Chicago/Turabian StyleDu, Jie, Binbin Huang, Jun Huang, Qingshan Long, Cuiyang Zhang, Zhaohui Guo, Yunsheng Wang, Wu Chen, Shiyong Tan, and Qingshu Liu. 2024. "Pan-Genome Analysis and Secondary Metabolic Pathway Mining of Biocontrol Bacterium Brevibacillus brevis" Agronomy 14, no. 5: 1024. https://doi.org/10.3390/agronomy14051024
APA StyleDu, J., Huang, B., Huang, J., Long, Q., Zhang, C., Guo, Z., Wang, Y., Chen, W., Tan, S., & Liu, Q. (2024). Pan-Genome Analysis and Secondary Metabolic Pathway Mining of Biocontrol Bacterium Brevibacillus brevis. Agronomy, 14(5), 1024. https://doi.org/10.3390/agronomy14051024







