PGRFA Management of Outcrossing Plants Propagated by Seed: From On-Farm to Ex Situ Conservation and Some Italian Maize Case Studies
Abstract
:1. Introduction
2. On-Farm Conservation
2.1. Guidelines for On-Farm Conservation
2.2. The Role of Custodian Farmers
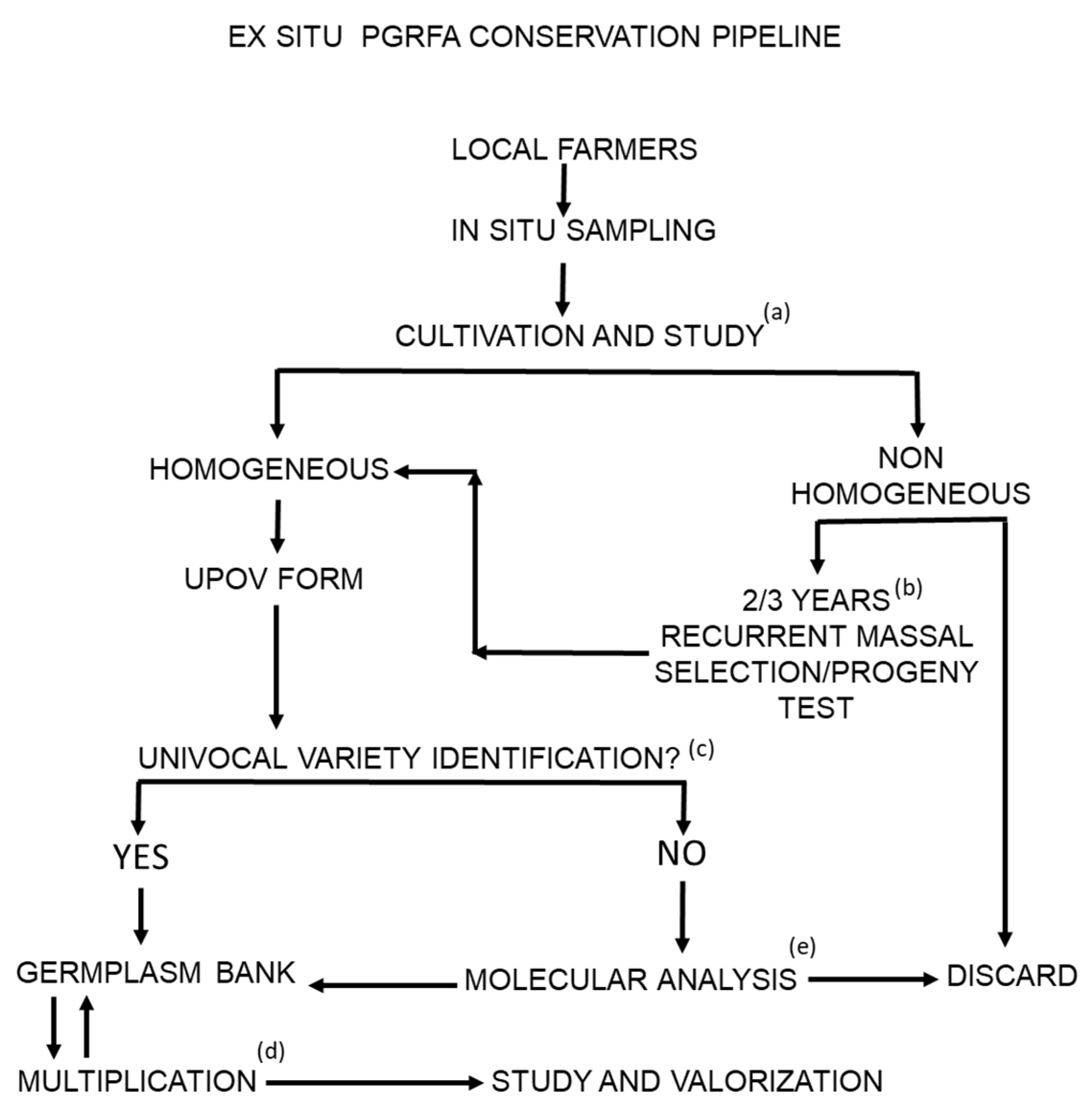
3. Ex Situ Conservation, from Sampling to Germplasm Bank Processing and Storing
3.1. Guidelines for Ex Situ Conservation
3.2. Examples of Italian Germplasm Banks
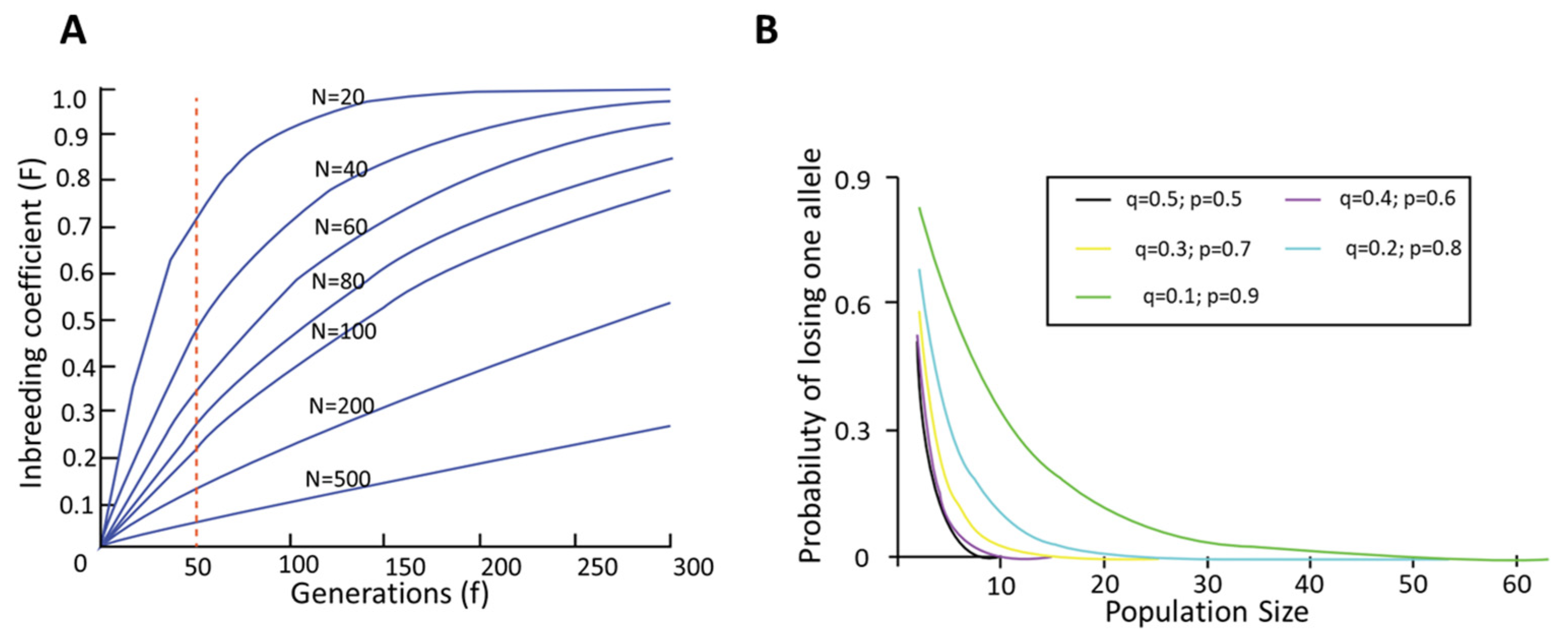
4. Evaluation of Inbreeding Depression and Genetic Diversity by Classical Methods
5. Evaluation of Genetic Diversity by Molecular Markers
- (i)
- genetic identity verification: molecular markers help ensure the accuracy of germplasm identification and prevent mislabeling or mix-ups within germplasm collections. For example, agrobiodiversity can either be conserved [78] or lost [79,80] by the pressures of selection for adaptation that has favored the emergence of new alleles introduced through crosses with modern varieties [24,80].
- (ii)
- genetic diversity management: molecular markers enable the assessment of genetic diversity within ex situ collections, allowing curators to prioritize the conservation of genetically distinct or rare accessions.
- (iii)
- (iv)
- conservation prioritization: molecular markers assist in prioritizing accessions for conservation based on their genetic uniqueness, adaptive potential, or significance for breeding programs.
- (v)
- germplasm utilization and breeding: molecular markers aid in the efficient utilization of ex situ germplasm resources for breeding purposes [81]. They facilitate the identification of desirable traits, genetic mapping of important genes, and the selection of suitable parental lines for breeding programs, thereby accelerating the development of improved cultivars with desired traits.
Choice of Molecular Markers
6. Study, Promotion and Valorization of PGRFA: Some Case Studies
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. Agricultural Biodiversity, Multifunctional Character of Agriculture and Land Conference, Background Paper 1. September 1999. Available online: https://www.fao.org/3/x2775e/x2775e03.htm (accessed on 5 March 2024).
- Cardinale, B.J.; Duffy, J.E.; Gonzalez, A.; Hooper, D.U.; Perrings, C.; Venail, P.; Narwani, A.; MacE, G.M.; Tilman, D.; Wardle, D.A.; et al. Biodiversity loss and its impact on humanity. Nature 2012, 486, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.C.; Begg, G.S.; Swanston, J.S. Deployment of diversity for enhanced crop function. Ann. Appl. Biol. 2009, 154, 309–322. [Google Scholar] [CrossRef]
- FAO. The state of the world’s biodiversity for food and agriculture. In FAO Commission on Genetic Resources for Food and Agriculture Assessments; B’elanger, J., Pilling, D., Eds.; FAO: Rome, Italy, 2019; Available online: https://www.fao.org/documents/card/en?details=ca3129en (accessed on 5 March 2024).
- Maxted, N.; Hawkes, J.G.; Ford-Lloyd, B.V.; Williams, J.T. A practical model for in situ genetic conservation. In Plant Genetic Conservation: The In Situ Approach; Maxted, N., Ford-Lloyd, B.V., Hawkes, J.G., Eds.; Chapman & Hall: London, UK, 1997; pp. 339–367. [Google Scholar]
- Wang, Y.; Wang, Y.; Sun, X.; Caiji, Z.; Yang, J.; Cui, D.; Cao, G.; Ma, X.; Han, B.; Xue, D.; et al. Influence of ethnic traditional cultures on genetic diversity of rice landraces under on-farm conservation in southwest China. J. Ethnobiol. Ethnomed. 2016, 12, 51. [Google Scholar] [CrossRef]
- Tiranti, B.; Negri, V. Selective microenvironmental effects play a role in shaping genetic diversity and structure in a Phaseolus vulgaris L. landrace: Implications for on-farm conservation. Mol. Ecol. 2007, 16, 4942–4955. [Google Scholar] [CrossRef] [PubMed]
- Vigouroux, Y.; Barnaud, A.; Scarcelli, N.; Thuillet, A.C. Biodiversity, evolution and adaptation of cultivated crops. C. R. Biol. 2011, 334, 450–457. [Google Scholar] [CrossRef]
- Curry, H.A. The history of seed banking and the hazards of backup. Soc. Stud. Sci. 2022, 52, 664–688. [Google Scholar] [CrossRef] [PubMed]
- Maxted, N.; Hunter, D.; Ortiz Ríos, R. Plant Genetic Conservation; Cambridge University Press: New York, NY, USA, 2020. [Google Scholar]
- ECPGR. ECPGR Concept for On-Farm Conservation and Management of Plant Genetic Resources for Food and Agriculture; ECPGR: Rome, Italy, 2017; Available online: www.ecpgr.cgiar.org/fileadmin/bioversity/publications/pdfs/ECPGR_Concept_for_on_farm_final__05_05_2017_bis.pdf (accessed on 5 March 2024).
- Casañas, F.; Simó, J.; Casals, J.; Prohens, J. Toward an evolved concept of landrace. Front. Plant Sci. 2017, 8, 145. [Google Scholar] [CrossRef] [PubMed]
- FAO. What Is Happening to Agrobiodiversity? Available online: https://www.fao.org/3/y5609e/y5609e02.htm (accessed on 5 March 2024).
- Dulloo, M.E.; Hunter, D.; Borelli, T. Ex situ and in situ conservation of agricultural biodiversity: Major advances and research needs. Not. Bot. Horti Agrobo 2010, 38, 123–135. [Google Scholar]
- Volis, S. Complementarities of two existing intermediate conservation approaches. Plant Divers. 2017, 39, 379–382. [Google Scholar] [CrossRef]
- Zegeye, H. In situ and ex situ conservation: Complementary approaches for maintaining biodiversity. Int. J. Res. Env. Std. 2016, 4, 1–12. [Google Scholar]
- Bellon, M.R.; Pham, J.L.; Jackson, M.T. Genetic conservation: A role for rice farmers. In Plant Conservation: The In Situ Approach; Maxted, N., Ford-Lloyd, B.V., Hawkes, J.G., Eds.; Chapman and Hall: London, UK, 1997; pp. 263–289. [Google Scholar]
- Bellon, M.R.; van Etten, J. Climate change and on-farm conservation of crop landraces in centres of diversity. In Plant Genetic Resources and Climate Change; Jackson, M., Ford-Lloyd, B., Parry, M., Eds.; CABI: Oxford, OK, 2013; pp. 137–150. [Google Scholar]
- Zeven, A.C. Landraces: A review of definitions and classifications. Euphytica 1998, 104, 127–139. [Google Scholar] [CrossRef]
- Negri, V.; Maxted, N.; Veteläinen, M. European landrace conservation: An introduction. In European Landraces: On farm Conservation, Management and Use: Biodiversity Technical Bulletin No 15; Veteläinen, M., Negri, V., Maxted, N., Eds.; European Cooperative Programme for Plant Genetic Resources: Rome, Italy, 2009. [Google Scholar]
- Villa, T.C.; Maxted, N.; Scholten, M.A.; Ford-Lloyd, B.V. Defining and identifying crop landraces. Plant Genet. Res. 2005, 3, 373–384. [Google Scholar] [CrossRef]
- Tin, H.Q.; Berg, T.; Bjørnstad, Å. Diversity and adaptation in rice varieties under static (ex situ) and dynamic (in situ) management. A case study in the Mekong Delta, Vietnam. Euphytica 2001, 122, 491–502. [Google Scholar] [CrossRef]
- Gómez, O.J.; Blair, M.W.; Frankow-Lindberg, B.E.; Gullberg, U. Comparative study of common bean (Phaseolus vulgaris L.) landraces conserved ex situ in genebanks and in situ by farmers. Genet. Resour. Crop Evol. 2005, 52, 371–380. [Google Scholar]
- McLean-Rodriguez, F.D.; Costich, D.E.; Camacho-Villa, T.C.; Pe, M.E.; Dell’Acqua, M. Genetic diversity and selection signatures in maize landraces compared across 50 years of in situ and ex situ conservation. Heredity 2021, 126, 913–928. [Google Scholar] [CrossRef] [PubMed]
- Bretting, P.K.; Duvick, D.N. Dynamic conservation of plant genetic resources. Adv. Agron. 1997, 61, 1–51. [Google Scholar]
- Brush, S.B. Farmers’ Bounty. Locating Crop Diversity in the Contemporary World; Yale University Press: New Haven, CT, USA, 2004. [Google Scholar]
- Gepts, P. Plant genetic resources conservation and utilization: The accomplishments and future of a societal insurance policy. Crop. Sci. 2006, 46, 2278–2292. [Google Scholar] [CrossRef]
- Valorizzazione e Salvaguardia delle Risorse Genetiche Vegetali D’interesse Agrario e Alimentare (RGVAA). Available online: http://www.geneticagraria.it/attachment/SocietaScuolaRicerca/Documento_SIGA_Agrobiodiversita_2023.pdf (accessed on 5 March 2024).
- Convention on Biodiversity (CBD). Available online: https://www.cbd.int/ (accessed on 8 March 2024).
- FAO. International Treaty on Plant Genetic Resources for Food and Agriculture (ITPGRFA); FAO: Rome, Italy, 2009; Available online: https://www.fao.org/3/i0510e/i0510e.pdf (accessed on 8 March 2024).
- Spataro, G.; Negri, V. The European seed legislation on conservation varieties: Focus, implementation, present and future impact on landrace on farm conservation. Genet. Resour. Crop Evol. 2013, 60, 2421–2430. [Google Scholar] [CrossRef]
- EURISCO. Available online: https://eurisco.ipk-gatersleben.de/apex/eurisco_ws/r/eurisco/home (accessed on 8 March 2024).
- Kotni, P.; Van Hintum, T.; Maggioni, L.; Oppermann, M.; Weise, S. EURISCO update 2023: The European Search Catalogue for Plant Genetic Resources, a pillar for documentation of genebank material. Nucleic Acids Res. 2023, 51, 1465–1469. [Google Scholar] [CrossRef]
- EVA Maize Network. Available online: https://www.ecpgr.cgiar.org/eva/eva-networks/maize (accessed on 8 March 2024).
- Italian Law for the Ratification and Execution of the Convention on Biodiversity, with Annexes (Rio de Janeiro, 5 June 1992). Available online: https://www.mase.gov.it/normative/l-14-febbraio-1994-n-124-ratifica-ed-esecuzione-della-convenzione-sulla-biodiversita-conannessi (accessed on 8 March 2024).
- Italian Law n° 194, Provisions for the Protection and Enhancement of Agricultural and Food Biodiversity. 2015. Available online: https://www.gazzettaufficiale.it/eli/id/2015/12/11/15G00210/sg%20 (accessed on 23 March 2024).
- FAO/ITPGRFA. Views, Experiences and Best Practices as an Example of Possible Options for the National Implementation of Article 9 of the International Treaty. Available online: www.fao.org/3/ca7834en/ca7834en.pdf (accessed on 8 March 2024).
- Byrnes, A.; Sthapit, B. A Guide to the Identification and Importance of Custodian Farmers; Bioversity International: Rome, Italy, 2016; 2p, Available online: https://cgspace.cgiar.org/server/api/core/bitstreams/707ead85-7b15-406f-b017-8479c0d7486a/content (accessed on 5 March 2024).
- Barcaccia, G.; Falcinelli, M. Miglioramento Genetico In Genetica e Genomica, 2nd ed.; Liguori ed Srl: Napoli, Italia, 2005; Volume II. [Google Scholar]
- Bannert, M.; Stamp, P. Cross-pollination of maize at long distance. Eur. J. Agron. 2007, 27, 44–51. [Google Scholar] [CrossRef]
- Bøhn, T.; Aheto, D.W.; Mwangala, F.S.; Fischer, K.; Bones, I.L.; Simoloka, C.; Mbeule, I.; Schmidt, G.; Breckling, B. Pollen-mediated gene flow and seed exchange in small-scale Zambian maize farming, implications for biosafety assessment. Sci. Rep. 2016, 6, 34483. [Google Scholar] [CrossRef] [PubMed]
- Poudel, D. On Farm Conservation of Crop Genetic Resource: Declining De Facto Diversity and Optimal Funding Strategy. Nat. Resour. J. 2015, 6, 196–207. [Google Scholar]
- Amancah, E.; Mercado, W.; Gómez-Pando, L.; Escalante, R.; Sotomayor, D.A. Integrating in situ conservation of plant genetic resources with ex situ conservation management: Involving custodian farmers, benefits and their willingness to accept compensation. Sci. Agropecu. 2023, 14, 447–464. [Google Scholar] [CrossRef]
- Wale, E. Costing on-farm conservation of crop diversity: The case of sorghum and wheat in Ethiopia and implications for policy. Afr. J. Agric. Res. 2011, 6, 401–406. [Google Scholar]
- FAO. Second Report on the World’s Plant Genetic Resources for Food and Agriculture; FAO: Rome, Italy, 2010; p. 299. Available online: https://www.fao.org/3/i1500e/i1500e00.htm (accessed on 5 March 2024).
- Hay, F.R.; Probert, R.J. Advances in seed conservation of wild plant species: A review of recent research. Conserv. Physiol. 2013, 1, cot030. [Google Scholar] [CrossRef] [PubMed]
- Wambugu, P.W.; Ndjiondjop, M.N.; Henry, R.J. Role of genomics in promoting the utilization of plant genetic resources in genebanks. Brief. Funct. Genom. 2018, 17, 198–206. [Google Scholar] [CrossRef]
- FAO/IPGRI. Genebank Standards; Food and Agriculture Organization of the United Nations: Rome, Italy; International Plant Genetic Resources Institute: Rome, Italy, 1994; Available online: https://cropgenebank.sgrp.cgiar.org/images/file/learning_space/genebank_standards.pdf (accessed on 5 March 2024).
- FAO. Genebank Standards for Plant Genetic Resources for Food and Agriculture, Rev. Ed.; FAO: Rome, Italy, 2014; Available online: www.fao.org/3/i3704e/i3704e.pdf (accessed on 8 March 2024).
- Walters, C.; Wheeler, L.M.; Grotenhuis, J.M. Longevity of seeds stored in a genebank: Species characteristics. Seed Sci. Res. 2005, 15, 1–20. [Google Scholar] [CrossRef]
- Center for Plant Conservation. Best Plant Conservation Practices. Available online: https://saveplants.org/best-practices/difference-between-orthodox-intermediate-and-recalcitrantseed/#:~:text=Recalcitrant%20seeds%20cannot%20be%20stored,between%2045%20and%2065%25%20R (accessed on 23 April 2024).
- Hoban, S. New guidance for ex situ gene conservation: Sampling realistic population systems and accounting for collection attrition. Biol. Conserv. 2019, 235, 199–208. [Google Scholar] [CrossRef]
- Kew Seed Bank Resources. Available online: https://brahmsonline.kew.org/msbp/Training/Resources (accessed on 5 March 2024).
- Ex Situ Conservation of Plant Genetic Resources. Biodiversity International. Available online: https://cgspace.cgiar.org/server/api/core/bitstreams/f66c1a4b-b390-42ab-9478-a6ba439cb35d/content (accessed on 8 March 2024).
- Hawkes, J.G.; Maxted, N.; Ford-Lloyd, B.V. The Ex Situ Conservation of Plant Genetic Resources; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2000. [Google Scholar]
- Rao, N.K.; Hanson, J.; Dulloo, M.E.; Ghosh, K.; Nowell, D.; Larinde, M. Manual of Seed Handling in Genebanks; Handbook for Genebanks No. 8.; Biodiversity International: Rome, Italy, 2006. [Google Scholar]
- Harrington, J.F. Seed storage and longevity. In Seed Biology, Insects, and Seed Collection, Storage, Testing and Certification; Kozlowski, T.T., Ed.; Academic Press: New York, NY, USA, 1972; pp. 145–245. [Google Scholar]
- Ranganathan, U.; Groot, S.P.C. Seed Longevity and Deterioration. In Seed Science and Technology; Dadlani, M., Yadava, D.K., Eds.; Springer: Singapore, 2023. [Google Scholar]
- Basey, A.C.; Fant, J.B.; Kramer, A.T. Producing native plant materials for restoration: 10 rules to collect and maintain genetic diversity. Native Plants J. 2015, 16, 37–52. [Google Scholar] [CrossRef]
- Ellis, R.H.; Roberts, E.H. Improved Equations for the Prediction of Seed Longevity. Ann. Bot. 1980, 45, 13–30. [Google Scholar] [CrossRef]
- Engels, J.M.M.; Ebert, A.W. A Critical Review of the Current Global Ex Situ Conservation System for Plant Agrobiodiversity. II. Strengths and Weaknesses of the Current System and Recommendations for Its Improvement. Plants 2021, 10, 1904. [Google Scholar] [CrossRef]
- Laboratorio di Ecologia Vegetale (University of Pavia). Available online: http://www.labecove.it/index.php?option=com_content&view=article&id=55&Itemid=225&lang=en (accessed on 5 March 2024).
- ENSCONET (European Native Seed Conservation Network). Seed Collecting Manual for Wild Species. 2009. Available online: http://ensconet.maich.gr/ (accessed on 5 March 2024).
- A European Genebank Integrated System (AEGIS). Available online: https://www.cgiar.org/aegis (accessed on 8 March 2024).
- ISTAT. 2023. Available online: https://dati.istat.it/Index.aspx?DataSetCode=DCSP_COLTIVAZIONI (accessed on 21 February 2024).
- Butrón, A.; Revilla, P.; Sandoya, G.; Ordás, A.; Malvar, R.A. Resistance to reduce corn borer damage in maize for bread, in Spain. J. Crop Prot. 2009, 28, 134–138. [Google Scholar] [CrossRef]
- Santiago, R.; Cao, A.; Malvar, R.A.; Butrón, A. Assessment of corn resistance to fumonisin accumulation in a broad collection of inbred lines. Field Crop Res. 2013, 149, 193–202. [Google Scholar] [CrossRef]
- Balconi, C.; Berardo, N.; Locatelli, S.; Lanzanova, C.; Torri, A.; Redaelli, R. Evaluation of ear rot (Fusarium verticilliodes) resistance and fumonisinaccumulation in Italian maize inbred lines. Phytopathol. Med. 2014, 53, 1426. [Google Scholar]
- Torri, A.; Lanzanova, C.; Locatelli, S.; Valoti, P.; Balconi, C. Screening of local Italian maize varieties for resistance to Fusarium verticillioides. Maydica 2015, 60, 1–8. [Google Scholar]
- Brandolini, A.; Brandolini, A. Maize introduction, evolution and diffusion in Italy. Maydica 2009, 54, 233–242. [Google Scholar]
- Wang, J.; Crossa, J.; van Ginkel, M.; Taba, S. Statistical genetics and simulation models in genetic resource conservation and regeneration. Crop Sci. 2004, 44, 2246–2253. [Google Scholar] [CrossRef]
- IBPGR. Descriptors for Maize; International Maize and Wheat Improvement Center: Mexico City, Mexico; International Board for Plant Genetic Resources: Rome, Italy, 1991; Available online: https://www.google.com/url?sa=i&url=https%3A%2F%2Fwww.genebanks.org%2Fresources%2Fpublications%2Fdescriptors-for-maize%2F&psig=AOvVaw1swVN6EOl6YBRbs2LSP5XD&ust=1714492561029000&source=images&cd=vfe&opi=89978449&ved=0CAUQn5wMahcKEwjgtfrR5OeFAxUAAAAAHQAAAAAQBA (accessed on 29 April 2024).
- ECPGR: European Accessions. Available online: www.ecpgr.cgiar.org/aegis/european-collection/european-accessions (accessed on 23 March 2024).
- Mastrangelo, A.M.; Hartings, H.; Lanzanova, C.; Balconi, C.; Locatelli, S.; Cassol, H.; Valoti, P.; Petruzzino, G.; Pecchioni, N. Genetic Diversity within a Collection of Italian Maize Inbred Lines: A Resource for Maize Genomics and Breeding. Plants 2024, 13, 336. [Google Scholar] [CrossRef]
- McDonald, B.A. Population Genetics of Plant Pathogens. Plant Health Instr. 2004, 4. [Google Scholar]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics, 4th ed.; Cambridge University Press: Cambridge, UK, 2007; pp. 52–75. [Google Scholar]
- Lamkey, K.R.; Smith, O.S. Performance and Inbreeding Depression of Populations Representing Seven Eras of Maize Breeding. Crop Sci. 1987, 27, 695–699. [Google Scholar] [CrossRef]
- Bezançon, G.; Pham, J.L.; Deu, M.; Vigouroux, Y.; Sagnard, F.; Mariac, C.; Kapran, I.; Mamadou, A.; Gérard, B.; Ndjeunga, J.; et al. Changes in the diversity and geographic distribution of cultivated millet (Pennisetum glaucum (L.) R. Br.) and sorghum (Sorghum bicolor (L.) Moench) varieties in Niger between 1976 and 2003. Genet. Resour. Crop Evol. 2009, 56, 223–236. [Google Scholar] [CrossRef]
- Hammer, K.; Knupffer, H.; Xhuveli, L.; Perrino, P. Estimating genetic erosion in landraces—two case studies. Genet. Resour. Crop Evol. 1996, 43, 329–336. [Google Scholar] [CrossRef]
- Bitocchi, E.; Bellucci, E.; Rau, D.; Albertini, E.; Rodriguez, M.; Veronesi, F.; Attene, G.; Nanni, L. European flint landraces grown in situ reveal adaptive introgression from modern maize. PLoS ONE 2015, 10, e0121381. [Google Scholar] [CrossRef] [PubMed]
- Cassani, E.; Puglisi, D.; Cantaluppi, E.; Landoni, M.; Giupponi, L.; Giorgi, A.; Pilu, R. Genetic studies regarding the control of seed pigmentation of an ancient European pointed maize (Zea mays L.) rich in phlobaphenes: The “Nero Spinoso” from the Camonica valley. Genet. Resour. Crop Evol. 2017, 64, 761–773. [Google Scholar] [CrossRef]
- Heslot, N.; Rutkoski, J.; Poland, J.; Jannink, J.L.; Sorrells, M.E. Impact of marker ascertainment bias on genomic selection accuracy and estimates of genetic diversity. PLoS ONE 2013, 8, e74612. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Sapkota, M.; Lin, M.; Beil, C.; Sheehan, M.; Greene, S.; Irish, B.M. Genetic diversity, population structure, and taxonomic confirmation in annual medic (Medicago spp.) collections from Crimea, Ukraine. Front. Plant Sci. 2024, 15, 1339298. [Google Scholar] [CrossRef]
- Dou, T.; Wang, C.; Ma, Y.; Chen, Z.; Zhang, J.; Guo, G. CoreSNP: An efficient pipeline for core marker profile selection from genome-wide SNP datasets in crops. BMC Plant Biol. 2023, 23, 580. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Šajgalík, M.; Ondreičková, K.; Hauptvogel, P.; Mihálik, D.; Glasa, M.; Kraic, J. Higher Effectiveness of New Common Bean (Phaseolus vulgaris L.) Germplasm Acquisition by Collecting Expeditions Associated with Molecular Analyses. Sustainability 2019, 11, 5270. [Google Scholar] [CrossRef]
- Nagy, S.; Poczai, P.; Cernák, I.; Gorji, A.M.; Hegedus, G.; Taller, J. PICcalc: An online program to calculate polymorphic information content for molecular genetic studies. Biochem. Genet. 2012, 50, 670–672. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.A.; Churchill, G.A.; Autrique, J.E.; Tanksley, S.D.; Sorrells, M.E. Optimizing parental selection for genetic linkage maps. Genome Dyn. 1993, 36, 181–186. [Google Scholar] [CrossRef]
- Galli, Z.; Halász, G.; Kiss, E.; Heszky, L.; Dobránszki, J. Molecular identification of commercial apple cultivars with microsatellite markers. Hortscience 2005, 40, 1974–1977. [Google Scholar] [CrossRef]
- Vietina, M.; Agrimonti, C.; Marmiroli, M.; Bonas, U.; Marmiroli, N. Applicability of SSR markers to the traceability of monovarietal olive oils. J. Sci. Food Agric. 2011, 91, 1381–1391. [Google Scholar] [CrossRef] [PubMed]
- Registro Varietà Vegetali. Available online: https://cns.sian.it/portale-sian/sottosezione.jsp?pid=6 (accessed on 8 March 2024).
- Giupponi, L.; Leoni, V.; Colombo, F.; Cassani, E.; Hejna, M.; Rossi, L.; Pilu, R. Characterization of “Mais delle Fiorine” (Zea mays L.) and nutritional, morphometric and genetic comparison with other maize landraces of Lombardy region (Northern Italy). Genet. Resour. Crop Evol. 2021, 68, 2075–2091. [Google Scholar] [CrossRef]
- Petroni, K.; Pilu, R.; Tonelli, C. Anthocyanins in corn: A wealth of genes for human health. Planta 2014, 240, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Revilla, P.; Soenga, P.; Malvar, R.A. Effects of antioxidant activity of black maize in corn borer larval survival and growth. Spanish J. Agric. Res. 2018, 16, 1004–1011. [Google Scholar] [CrossRef]
- Casas, M.I.; Duarte, S.; Doseff, A.I.; Grotewold, E. Flavone-rich maize: An opportunity to improve the nutritional value of an important commodity crop. Front. Plant Sci. 2014, 5, 440. [Google Scholar] [CrossRef] [PubMed]
- Landoni, M.; Puglisi, D.; Cassani, E.; Borlini, G.; Brunoldi, G.; Comaschi, C.; Pilu, R. Phlobaphenes modify pericarp thickness in maize and accumulation of the fumonisin mycotoxins. Sci. Rep. 2020, 10, 1417. [Google Scholar] [CrossRef] [PubMed]
- Lago, C.; Cassani, E.; Zanzi, C.; Landoni, M.; Trovato, R.; Pilu, R. Development and study of a maize cultivar rich in anthocyanins: Coloured polenta, a new functional food. Plant Breed. 2014, 133, 210–217. [Google Scholar] [CrossRef]
- Suriano, S.; Balconi, C.; Valoti, P.; Redaelli, R. Comparison of total polyphenols, profile anthocyanins, color analysis, carotenoids and tocols in pigmented maize. LWT 2021, 144, 111257. [Google Scholar] [CrossRef]
- Revilla, P.; Alves, M.L.; Andjelkovic, V.; Balconi, C.; Dinis, I.; Reis Mendes-Moreira, P.M.; Redaelli, R.; Ruiz De Galarreta, J.I.; Vaz Patto, M.C.; Vaz Patto, M.C.; et al. Traditional maize (Zea mays L.) foods in Europe. Front. Nutr. 2022, 8, 683399. [Google Scholar] [CrossRef]

| Population | Yield (Mg ha−1) | Inbreeding Depression | |||
|---|---|---|---|---|---|
| S0 | S1 | Difference (i) | Percent (ii) | Rate (iii) | |
| Reid opv | 2.91 | 2.25 | 0.66 | 22.7 | 0.013 |
| Lancaster opv | 3.14 | 2.27 | 0.87 | 27.7 | 0.017 |
| Era 1 (1930’) | 3.05 | 2.23 | 0.82 | 26.9 | 0.016 |
| Era 2 (1940’) | 3.91 | 2.92 | 0.99 | 25.3 | 0.020 |
| Era 3 (1950’) | 4.42 | 3.08 | 1.34 | 30.3 | 0.027 |
| Era 4 (1960’) | 4.77 | 3.38 | 1.39 | 29.1 | 0.028 |
| Era 5 (1970’) | 5.63 | 4.25 | 1.38 | 24.5 | 0.028 |
| Era 6 (1980’) | 6.07 | 4.41 | 1.66 | 27.3 | 0.033 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landoni, M.; Bertoncini, A.; Ghidoli, M.; Rossi, G.; Cassani, E.; Locatelli, S.; Balconi, C.; Pilu, R. PGRFA Management of Outcrossing Plants Propagated by Seed: From On-Farm to Ex Situ Conservation and Some Italian Maize Case Studies. Agronomy 2024, 14, 1030. https://doi.org/10.3390/agronomy14051030
Landoni M, Bertoncini A, Ghidoli M, Rossi G, Cassani E, Locatelli S, Balconi C, Pilu R. PGRFA Management of Outcrossing Plants Propagated by Seed: From On-Farm to Ex Situ Conservation and Some Italian Maize Case Studies. Agronomy. 2024; 14(5):1030. https://doi.org/10.3390/agronomy14051030
Chicago/Turabian StyleLandoni, Michela, Anna Bertoncini, Martina Ghidoli, Graziano Rossi, Elena Cassani, Sabrina Locatelli, Carlotta Balconi, and Roberto Pilu. 2024. "PGRFA Management of Outcrossing Plants Propagated by Seed: From On-Farm to Ex Situ Conservation and Some Italian Maize Case Studies" Agronomy 14, no. 5: 1030. https://doi.org/10.3390/agronomy14051030
APA StyleLandoni, M., Bertoncini, A., Ghidoli, M., Rossi, G., Cassani, E., Locatelli, S., Balconi, C., & Pilu, R. (2024). PGRFA Management of Outcrossing Plants Propagated by Seed: From On-Farm to Ex Situ Conservation and Some Italian Maize Case Studies. Agronomy, 14(5), 1030. https://doi.org/10.3390/agronomy14051030








