Genome-Wide Identification and Analysis of Plasma Membrane H+-ATPases Associated with Waterlogging in Prunus persica (L.) Batsch
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Waterlogging Treatment
2.2. Identification of H+-ATP Family Members in the P. persica Genome
2.3. Chromosomal Localization and the Ka/Ks Calculation
2.4. Phylogenetic Tree, Gene Structure, and Conserved Motif Analysis
2.5. Expression Profiles of the Plasma Membrane H+-ATPase Genes in P. persica Using RNA Sequencing
2.6. Total RNA Isolation and cDNA Synthesis
2.7. Quantitative Real-Time PCR (RT-qPCR) Validation
2.8. Statistical Analysis
3. Results
3.1. Identification and Characteristics of Plasma Membrane H+-ATPase Family Genes in the Peach Genome
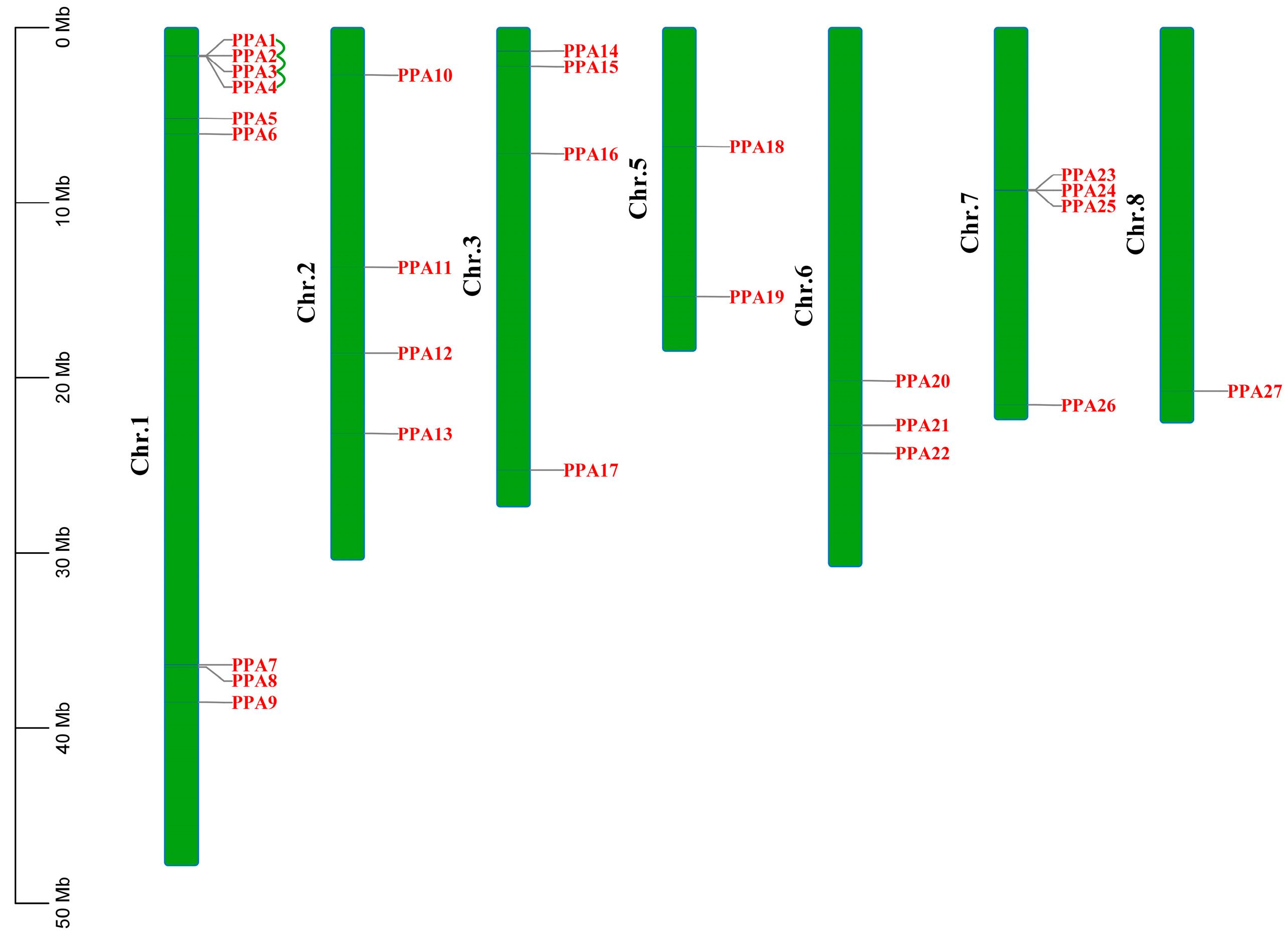
3.2. Chromosomal Distribution and Gene Duplication of Plasma Membrane H+-ATPase Genes
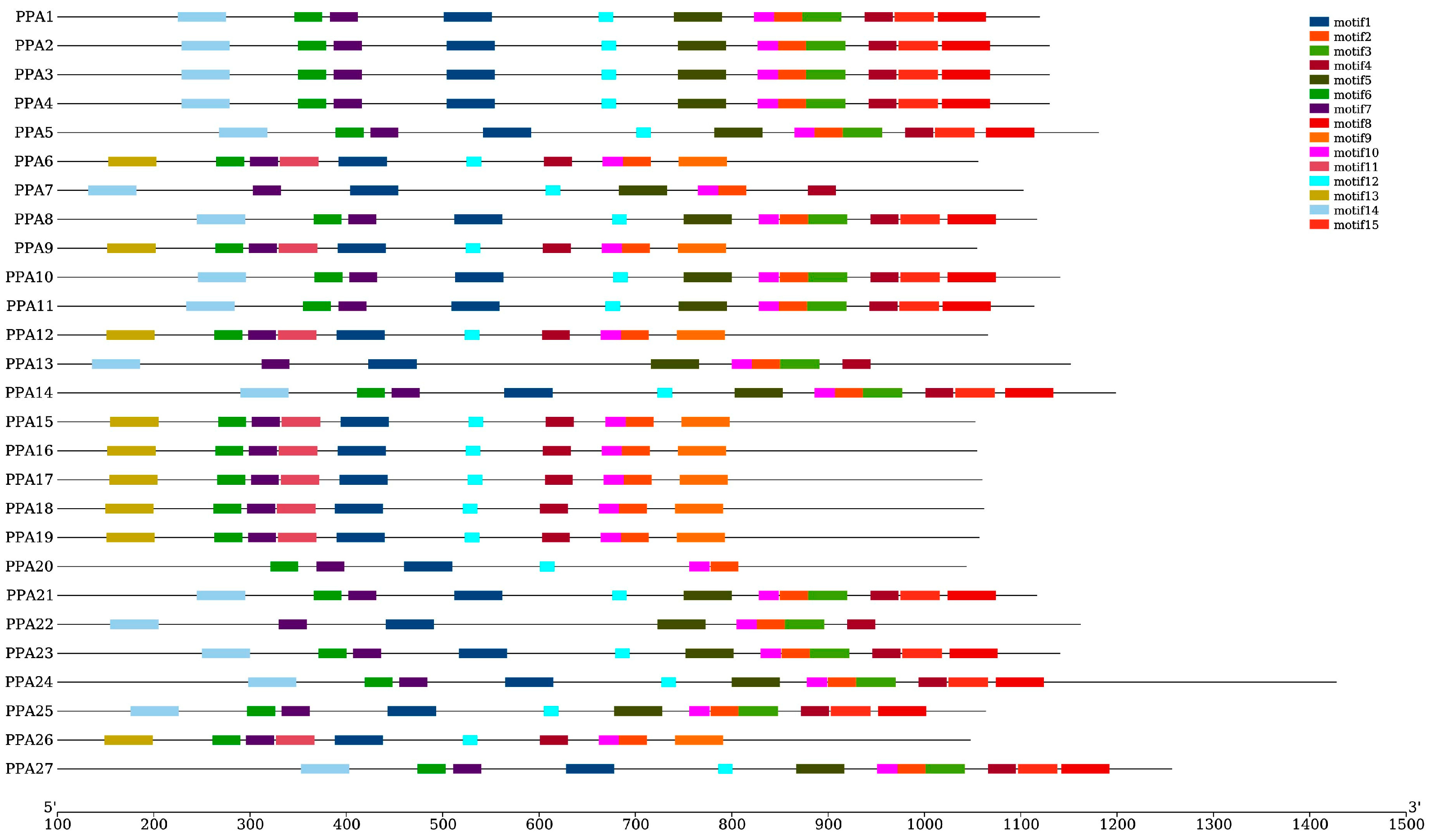
3.3. Phylogenetic Analysis, Gene Structure, and Motif Composition of Plasm Membrane H+-ATPase Genes
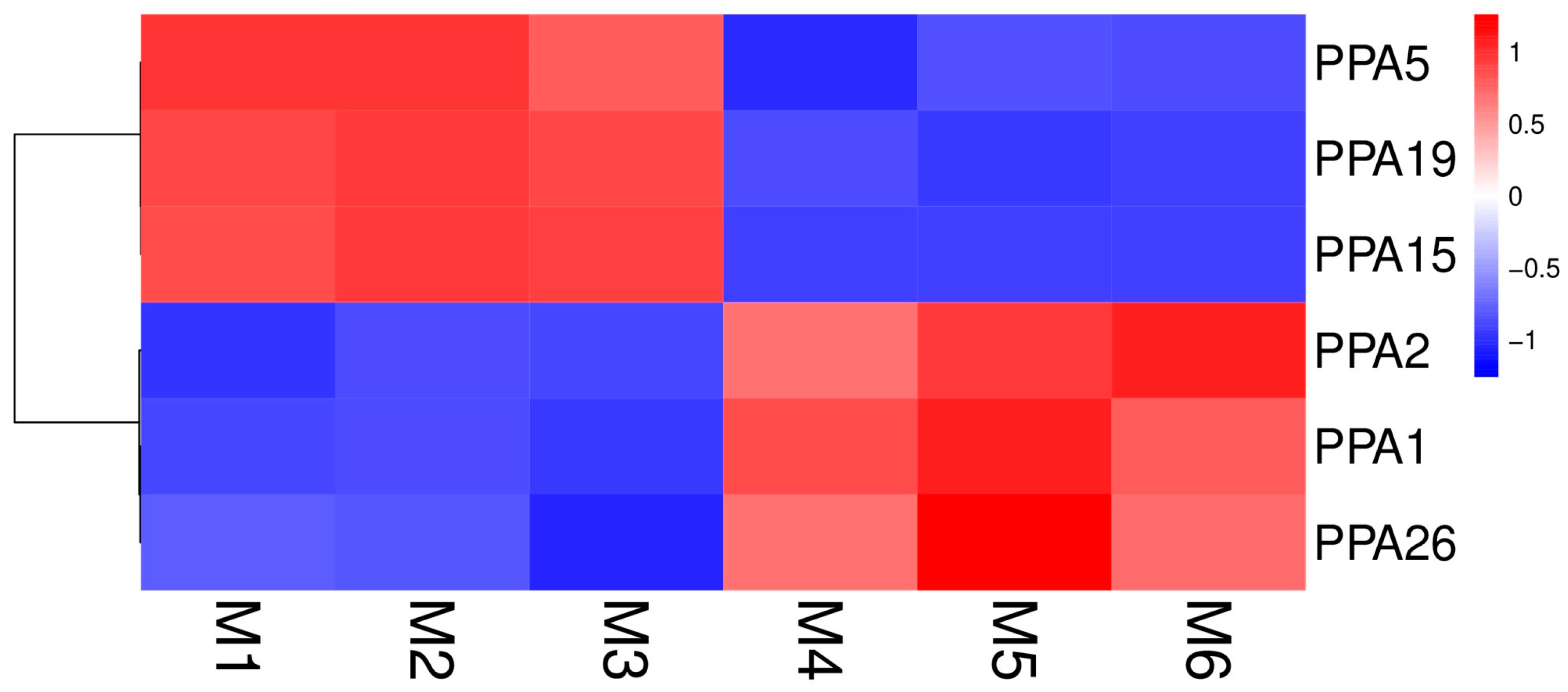
3.4. Expression Analysis of Plasma Membrane H+-ATPase Genes after Flooding Stress
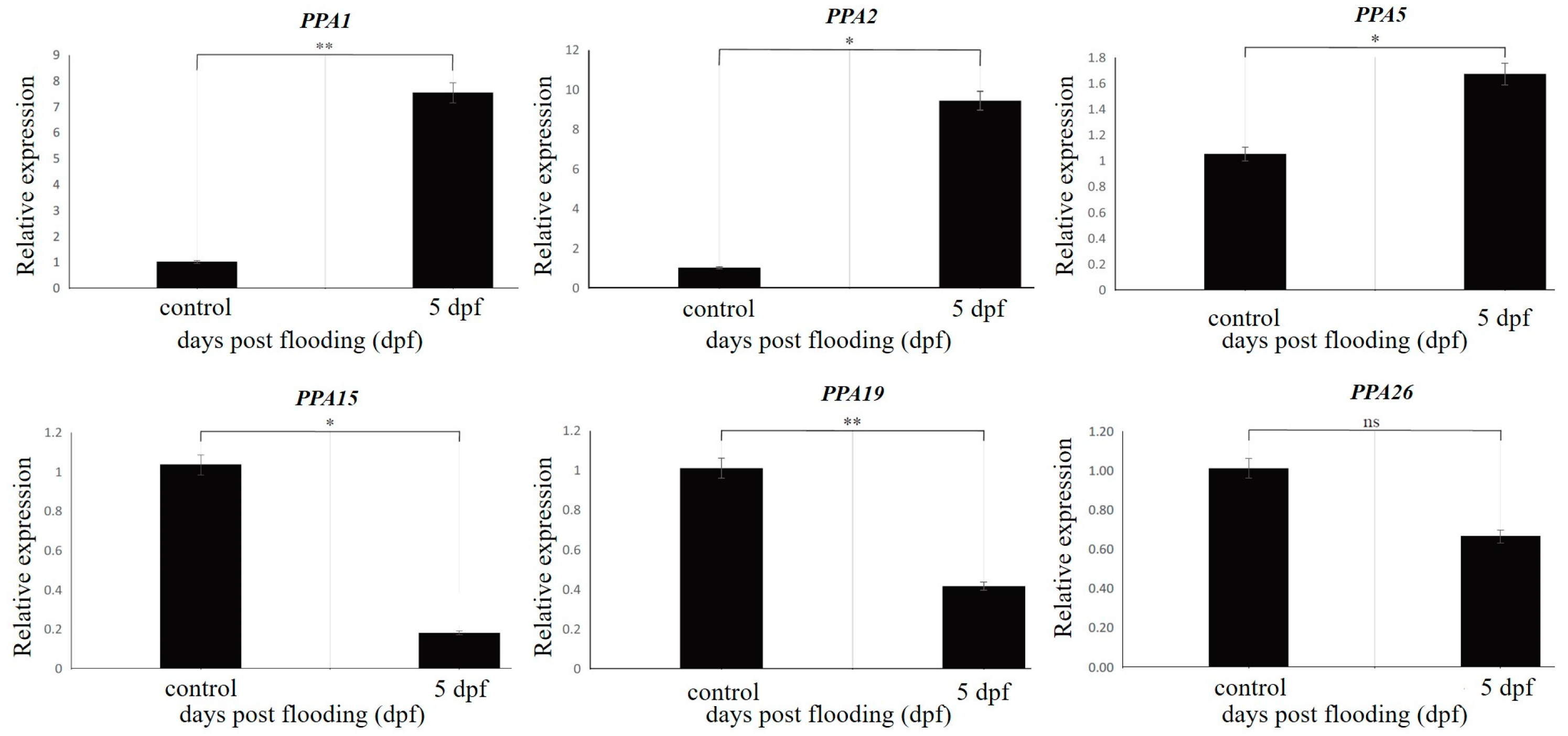
3.5. Real-Time Quantitative PCR (RT-qPCR) Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Vartapetian, B.B.; Jackson, M.B. Plant adaptations to anaerobic stress. Ann. Bot. 1997, 79 (Suppl. S1), 3–20. [Google Scholar] [CrossRef]
- Jackson, M.; Armstrong, W. Formation of aerenchyma and the processes of plant ventilation in relation to soil flooding and submergence. Plant Biol. 1999, 1, 274–287. [Google Scholar] [CrossRef]
- Xiao, Y.; Wu, X.; Sun, M.; Peng, F. Hydrogen sulfide alleviates waterlogging-induced damage in peach seedlings via enhancing antioxidative system and inhibiting ethylene synthesis. Front. Plant Sci. 2020, 11, 696. [Google Scholar] [CrossRef] [PubMed]
- Kreuzwieser, J.; Gessler, A. Global climate change and tree nutrition: Influence of water availability. Tree Physiol. 2010, 30, 1221–1234. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Hill, C.B.; Zhou, G.; Zhang, X.Q.; Jia, Y.; Li, C. Opportunities for improving waterlogging tolerance in cereal crops—Physiological traits and genetic mechanisms. Plants 2021, 10, 1560. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Sharif, R.; Xu, X.; Chen, X. Mechanisms of waterlogging tolerance in plants: Research progress and prospects. Front. Plant Sci. 2021, 11, 627331. [Google Scholar] [CrossRef]
- Lin, C.; Zhu, T.; Ogorek, L.L.P.; Wang, Y.; Sauter, M.; Pedersen, O. The pyramiding of three key root traits aid breeding of flood-tolerant rice. Plants 2022, 11, 2033. [Google Scholar] [CrossRef] [PubMed]
- Luan, H.; Li, H.; Li, Y.; Chen, C.; Li, S.; Wang, Y.; Yang, J.; Xu, M.; Shen, H.; Qiao, H. Transcriptome analysis of barley (Hordeum vulgare L.) under waterlogging stress, and overexpression of the HvADH4 gene confers waterlogging tolerance in transgenic Arabidopsis. BMC Plant Biol. 2023, 23, 62. [Google Scholar]
- Du, H.; Shen, X.; Huang, Y.; Huang, M.; Zhang, Z. Overexpression of Vitreoscilla hemoglobin increases waterlogging tolerance in Arabidopsis and maize. BMC Plant Biol. 2016, 16, 35. [Google Scholar] [CrossRef] [PubMed]
- Kumutha, D.; Sairam, R.K.; Ezhilmathi, K.; Chinnusamy, V.; Meena, R.C. Effect of waterlogging on carbohydrate metabolism in pigeon pea (Cajanus cajan L.): Upregulation of sucrose synthase and alcohol dehydrogenase. Plant Sci. 2008, 175, 706–716. [Google Scholar] [CrossRef]
- Pedersen, O.; Malik, A.I.; Colmer, T.D. Submergence tolerance in Hordeum marinum: Dissolved CO2 determines underwater photosynthesis and growth. Funct. Plant Biol. 2010, 37, 524–531. [Google Scholar] [CrossRef]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Van Dongen, J.T.; Giuntoli, B.; Novi, G.; Santaniello, A.; Geigenberger, P.; Perata, P. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 2010, 62, 302–315. [Google Scholar] [CrossRef]
- Lee, K.W.; Chen, P.W.; Lu, C.A.; Chen, S.; Ho, T.H.D.; Yu, S.M. Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci. Signal. 2009, 2, ra61. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Gao, H.; Wu, X.; Zhang, S.; Han, Z.; Chen, X.; Zhang, G.; Zeng, F. The ability to regulate transmembrane potassium transport in root is critical for drought tolerance in barley. Int. J. Mol. Sci. 2019, 20, 4111. [Google Scholar] [CrossRef] [PubMed]
- Gill, M.B.; Zeng, F.; Shabala, L.; Böhm, J.; Zhang, G.; Zhou, M.; Shabala, S. The ability to regulate voltage-gated K+-permeable channels in the mature root epidermis is essential for waterlogging tolerance in barley. J. Exp. Bot. 2018, 69, 667–680. [Google Scholar] [CrossRef]
- Gill, M.B.; Zeng, F.; Shabala, L.; Zhang, G.; Fan, Y.; Shabala, S.; Zhou, M. Cell-based phenotyping reveals QTL for membrane potential maintenance associated with hypoxia and salinity stress tolerance in barley. Front. Plant Sci. 2017, 8, 1941. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Konnerup, D.; Shabala, L.; Zhou, M.; Colmer, T.D.; Zhang, G.; Shabala, S. Linking oxygen availability with membrane potential maintenance and K+ retention of barley roots: Implications for waterlogging stress tolerance. Plant Cell Environ. 2014, 37, 2325–2338. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Sun, J.; Wang, K.; Liu, D.; Li, Z.; Zhang, J. Spermidine enhances waterlogging tolerance via regulation of antioxidant defence, heat shock protein expression and plasma membrane H+-ATPase activity in Zea mays. J. Agron. Crop Sci. 2014, 200, 199–211. [Google Scholar] [CrossRef]
- Xu, X.; Liu, M.; Hu, Q.; Yan, W.; Pan, J.; Yan, Y.; Chen, X. A CsEIL3-CsARN6. 1 module promotes waterlogging-triggered adventitious root formation in cucumber by activating the expression of CsPrx5. Plant J. 2023, 114, 824–835. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.H.; Khan, M.N.; Mukherjee, S.; Alamri, S.; Basahi, R.A.; Al-Amri, A.A.; Alsubaie, Q.D.; Al-Munqedhi, B.M.; Ali, H.M.; Almohisen, I.A. Hydrogen sulfide (H2S) and potassium (K+) synergistically induce drought stress tolerance through regulation of H+-ATPase activity, sugar metabolism, and antioxidative defense in tomato seedlings. Plant Cell Rep. 2021, 40, 1543–1564. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Li, B.; Lu, X.; Yuan, L.; Yang, Y.; Yuan, Y.; Du, J.; Guo, S. The effect of exogenous calcium on mitochondria, respiratory metabolism enzymes and ion transport in cucumber roots under hypoxia. Sci. Rep. 2015, 5, 11391. [Google Scholar] [CrossRef] [PubMed]
- Su, N.; Wu, Q.; Chen, J.; Shabala, L.; Mithöfer, A.; Wang, H.; Qu, M.; Yu, M.; Cui, J.; Shabala, S. GABA operates upstream of H+-ATPase and improves salinity tolerance in Arabidopsis by enabling cytosolic K+ retention and Na+ exclusion. J. Exp. Bot. 2019, 70, 6349–6361. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, Z.H.; Liu, X.; Colmer, T.D.; Shabala, L.; Salih, A.; Zhou, M.; Shabala, S. Revealing the roles of GORK channels and NADPH oxidase in acclimation to hypoxia in Arabidopsis. J. Exp. Bot. 2017, 68, 3191–3204. [Google Scholar] [CrossRef] [PubMed]
- Planes, M.D.; Niñoles, R.; Rubio, L.; Bissoli, G.; Bueso, E.; García-Sánchez, M.J.; Alejandro, S.; Gonzalez-Guzmán, M.; Hedrich, R.; Rodriguez, P.L. A mechanism of growth inhibition by abscisic acid in germinating seeds of Arabidopsis thaliana based on inhibition of plasma membrane H+-ATPase and decreased cytosolic pH, K+, and anions. J. Exp. Bot. 2015, 66, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Brault, M.; Amiar, Z.; Pennarun, A.M.; Monestiez, M.; Zhang, Z.; Cornel, D.; Dellis, O.; Knight, H.; Bouteau, F.; Rona, J.P. Plasma membrane depolarization induced by abscisic acid in Arabidopsis suspension cells involves reduction of proton pumping in addition to anion channel activation, which are both Ca2+ dependent. Plant Physiol. 2004, 135, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Kanczewska, J.; Marco, S.; Vandermeeren, C.; Maudoux, O.; Rigaud, J.-L.; Boutry, M. Activation of the plant plasma membrane H+-ATPase by phosphorylation and binding of 14-3-3 proteins converts a dimer into a hexamer. Proc. Natl. Acad. Sci. USA 2005, 102, 11675–11680. [Google Scholar] [CrossRef] [PubMed]
- Palmgren, M.G. Plant plasma membrane H+-ATPases: Powerhouses for nutrient uptake. Annu. Rev. Plant Biol. 2001, 52, 817–845. [Google Scholar] [CrossRef] [PubMed]
- Michelet, B.; Boutry, M. The plasma membrane H+-ATPase (A highly regulated enzyme with multiple physiological functions). Plant Physiol. 1995, 108, 1. [Google Scholar] [CrossRef] [PubMed]
- Sondergaard, T.E.; Schulz, A.; Palmgren, M.G. Energization of transport processes in plants. Roles of the plasma membrane H+-ATPase. Plant Physiol. 2004, 136, 2475–2482. [Google Scholar] [CrossRef] [PubMed]
- Serrano, R. Salt tolerance in plants and microorganisms: Toxicity targets and defense responses. Int. Rev. Cytol. 1996, 165, 1–52. [Google Scholar] [PubMed]
- Reuveni, M.; Bressan, R.A.; Hasegawa, P.M. Modification of proton transport kinetics of the plasma membrane H+-ATPase after adaptation of tobacco cells to NaCI. J. Plant Physiol. 1993, 142, 312–318. [Google Scholar] [CrossRef]
- Niu, X.; Damsz, B.; Kononowicz, A.K.; Bressan, R.A.; Hasegawa, P.M. NaCl-induced alterations in both cell structure and tissue-specific plasma membrane H+-ATPase gene expression. Plant Physiol. 1996, 111, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Seliskar, D.M. Salinity adaptation of plasma membrane H+-ATPase in the salt marsh plant Spartina patens: ATP hydrolysis and enzyme kinetics. J. Exp. Bot. 1998, 49, 1005–1013. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, A.; Li, X.; Kou, M.; Wang, W.; Chen, X.; Xu, T.; Zhu, M.; Ma, D.; Li, Z. Melatonin-stimulated triacylglycerol breakdown and energy turnover under salinity stress contributes to the maintenance of plasma membrane H+–ATPase activity and K+/Na+ homeostasis in sweet potato. Front. Plant Sci. 2018, 9, 256. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, J.; Zhang, F.; Zhao, L.; Zhang, L. NaCl induced changes of the H+-ATPase in root plasma membrane of two wheat cultivars. Plant Sci. 2004, 166, 913–918. [Google Scholar] [CrossRef]
- Jakubowska, D.; Janicka, M. The role of brassinosteroids in the regulation of the plasma membrane H+-ATPase and NADPH oxidase under cadmium stress. Plant Sci. 2017, 264, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Bobik, K.; Boutry, M.; Duby, G. Activation of the plasma membrane H+-ATPase by acid stress: Antibodies as a tool to follow the phosphorylation status of the penultimate activating Thr. Plant Signal. Behav. 2010, 5, 681–683. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Gray, W.M.; Sussman, M.R. Regulation of the plasma membrane proton pump (H+-ATPase) by phosphorylation. Curr. Opin. Plant Biol. 2015, 28, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, E.; Kerkeb, B.; Donaire, J.P.; Belver, A. Effects of salt stress on H+-ATPase activity of plasma membrane-enriched vesicles isolated from sunflower roots. Plant Sci. 1998, 134, 181–190. [Google Scholar] [CrossRef]
- Arango, M.; Gévaudant, F.; Oufattole, M.; Boutry, M. The plasma membrane proton pump ATPase: The significance of gene subfamilies. Planta 2003, 216, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Falhof, J.; Pedersen, J.T.; Fuglsang, A.T.; Palmgren, M. Plasma membrane H+-ATPase regulation in the center of plant physiology. Mol. Plant 2016, 9, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Baxter, I.; Tchieu, J.; Sussman, M.R.; Boutry, M.; Palmgren, M.G.; Gribskov, M.; Harper, J.F.; Axelsen, K.B. Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 2003, 132, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Sibole, J.V.; Cabot, C.; Michalke, W.; Poschenrieder, C.; Barceló, J. Relationship between expression of the PM H+-ATPase, growth and ion partitioning in the leaves of salt-treated Medicago species. Planta 2005, 221, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Ewing, N.N.; Bennett, A.B. Assessment of the number and expression of P-type H+-ATPase genes in tomato. Plant Physiol. 1994, 106, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Gévaudant, F.; Pétel, G.; Guilliot, A. Differential expression of four members of the H+-ATPase gene family during dormancy of vegetative buds of peach trees. Planta 2001, 212, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Sammut, S.J.; Finn, R.D.; Bateman, A. Pfam 10 years on: 10,000 families and still growing. Brief. Bioinform. 2008, 9, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud Se Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Bork, P.; Koonin, E.V. Protein sequence motifs. Curr. Opin. Struct. Biol. 1996, 6, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37 (Suppl. S2), W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, A.J. Java Treeview—Extensible visualization of microarray data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Manley, J.L. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009, 10, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Zhang, M.; Zeng, H.; Hayashi, Y.; Zhu, Y.; Kinoshita, T. Molecular basis of plasma membrane H+-ATPase function and potential application in the agricultural production. Plant Physiol. Biochem. 2021, 168, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Bramley, H.; Turner, D.; Tyerman, S.; Turner, N. Water flow in the roots of crop species: The influence of root structure, aquaporin activity, and waterlogging. Adv. Agron. 2007, 96, 133–196. [Google Scholar]
- Yamauchi, A.; Pardales, J.R., Jr.; Kono, Y. Root system structure and its relation to stress tolerance. In Proceedings of the Roots and Nitrogen in Cropping Systems of the Semi-Arid Tropics, Patancheru, India, 21–25 November 1994; pp. 211–233. [Google Scholar]
- Haruta, M.; Burch, H.L.; Nelson, R.B.; Barrett-Wilt, G.; Kline, K.G.; Mohsin, S.B.; Young, J.C.; Otegui, M.S.; Sussman, M.R. Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 2010, 285, 17918–17929. [Google Scholar] [CrossRef] [PubMed]
- Robertson, W.R.; Clark, K.; Young, J.C.; Sussman, M.R. An Arabidopsis thaliana plasma membrane proton pump is essential for pollen development. Genetics 2004, 168, 1677–1687. [Google Scholar] [CrossRef]
- Lan, P.; Li, W.; Lin, W.-D.; Santi, S.; Schmidt, W. Mapping gene activity of Arabidopsis root hairs. Genome Biol. 2013, 14, R67. [Google Scholar] [CrossRef] [PubMed]
- Baxter, I.R.; Young, J.C.; Armstrong, G.; Foster, N.; Bogenschutz, N.; Cordova, T.; Peer, W.A.; Hazen, S.P.; Murphy, A.S.; Harper, J.F. A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 2649–2654. [Google Scholar] [CrossRef] [PubMed]
- Schaller, A.; Oecking, C. Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell 1999, 11, 263–272. [Google Scholar] [PubMed]
- Fan, Y.; Wan, S.; Jiang, Y.; Xia, Y.; Chen, X.; Gao, M.; Cao, Y.; Luo, Y.; Zhou, Y.; Jiang, X. Over-expression of a plasma membrane H+-ATPase SpAHA1 conferred salt tolerance to transgenic Arabidopsis. Protoplasma 2018, 255, 1827–1837. [Google Scholar] [CrossRef] [PubMed]
- Młodzińska, E.; Kłobus, G.; Christensen, M.D.; Fuglsang, A.T. The plasma membrane H+-ATPase AHA2 contributes to the root architecture in response to different nitrogen supply. Physiol. Plant. 2015, 154, 270–282. [Google Scholar] [CrossRef]
- Hsu, J.L.; Wang, L.Y.; Wang, S.Y.; Lin, C.H.; Ho, K.C.; Shi, F.K.; Chang, I.F. Functional phosphoproteomic profiling of phosphorylation sites in membrane fractions of salt-stressed Arabidopsis thaliana. Proteome Sci. 2009, 7, 42. [Google Scholar] [CrossRef]
- Yamauchi, S.; Takemiya, A.; Sakamoto, T.; Kurata, T.; Tsutsumi, T.; Kinoshita, T.; Shimazaki, K.-I. The plasma membrane H+-ATPase AHA1 plays a major role in stomatal opening in response to blue light. Plant Physiol. 2016, 171, 2731–2743. [Google Scholar] [CrossRef]
- Merlot, S.; Leonhardt, N.; Fenzi, F.; Valon, C.; Costa, M.; Piette, L.; Vavasseur, A.; Genty, B.; Boivin, K.; Müller, A. Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 2007, 26, 3216–3226. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Chételat, A.; Nguyen, C.T.; Farmer, E.E. Arabidopsis H+-ATPase AHA1 controls slow wave potential duration and wound-response jasmonate pathway activation. Proc. Natl. Acad. Sci. USA 2019, 116, 20226–20231. [Google Scholar] [CrossRef]
- Young, J.C.; DeWitt, N.D.; Sussman, M.R. A transgene encoding a plasma membrane H+-ATPase that confers acid resistance in Arabidopsis thaliana seedlings. Genetics 1998, 149, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Fan, J.; Gao, Y.G.; Wang, Z.; Yang, P.; Liang, Y.; Opiyo, S.; Xia, Y. Arabidopsis plasma membrane ATPase AHA5 is negatively involved in PAMP-triggered immunity. Int. J. Mol. Sci. 2022, 23, 3857. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Hao, W.; Wang, Y.; Chen, Z.; Cao, S.; Jiang, L. Loss-of-function mutations in the ERF96 gene enhance iron-deficient tolerance in Arabidopsis. Plant Physiol. Biochem. 2022, 175, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Han, X.; Ma, L.; Wu, Y.; Liu, X.; Fu, H.; Liu, G.; Lei, X.; Guo, Y. Dynamic changes of phosphatidylinositol and phosphatidylinositol 4-phosphate levels modulate H+-ATPase and Na+/H+ antiporter activities to maintain ion homeostasis in Arabidopsis under salt stress. Mol. Plant 2021, 14, 2000–2014. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.S.; Cai, J.L.; Lee, J.T.; Li, Y.M.; Balladona, F.K.; Sukma, D.; Chan, M.T. Arabidopsis AtMSRB5 functions as a salt-stress protector for both Arabidopsis and rice. Front. Plant Sci. 2023, 14, 1072173. [Google Scholar] [CrossRef] [PubMed]
- Knight, C.A.; Vogel, H.; Kroymann, J.; Shumate, A.; Witsenboer, H.; Mitchell-Olds, T. Expression profiling and local adaptation of Boechera holboellii populations for water use efficiency across a naturally occurring water stress gradient. Mol. Ecol. 2006, 15, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Zhang, D.; Song, T.; Xu, F.; Lin, S.; Xu, W.; Li, Q.; Zhu, Y.; Liang, J.; Zhang, J. Arabidopsis plasma membrane H+-ATPase genes AHA2 and AHA7 have distinct and overlapping roles in the modulation of root tip H+ efflux in response to low-phosphorus stress. J. Exp. Bot. 2017, 68, 1731–1741. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Peng, Y.; Liu, E.; Ma, H.; Qiao, K.; Zhou, A.; Liu, S.; Bu, Y. Arabidopsis V-ATPase d2 subunit plays a role in plant responses to oxidative stress. Genes 2020, 11, 701. [Google Scholar] [CrossRef]
- Yuan, W.; Li, Y.; Li, L.; Siao, W.; Zhang, Q.; Zhang, Y.; Liu, J.; Xu, W.; Miao, R. BR-INSENSITIVE1 regulates hydrotropic response by interacting with plasma membrane H+-ATPases in Arabidopsis. Plant Signal. Behav. 2018, 13, e1486147. [Google Scholar] [CrossRef] [PubMed]
- Santi, S.; Schmidt, W. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 2009, 183, 1072–1084. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Primer Name | Primer Sequence |
|---|---|---|---|
| PPA1 | Prupe.1G023100 | PPA1-F | CAGACGCCTTTACAAGAACGG |
| PPA1-R | AGAGAGCCTCCTCACCAACG | ||
| PPA2 | Prupe.1G023200 | PPA2-F | CCAAAATACCTCGCTCTCCC |
| PPA2-R | TGCTGGTGGCTTCATGTATGT | ||
| PPA5 | Prupe.1G072600 | PPA5-F | TGCTGGGTATAAAGACAGAGGG |
| PPA5-R | TCTGCCGCCTCTAATGACC | ||
| PPA15 | Prupe.3G029800 | PPA15-F | TGTACAGTTTGATTTTCTACATTCC |
| PPA15-R | TTGGCTGCCCTATCTTCCTT | ||
| PPA19 | Prupe.5G185000 | PPA19-F | TGAAGTATTGGACGCTGTGTTG |
| PPA19-R | CCACGAGAGAGGGTTCCACAT | ||
| PPA26 | Prupe.7G258000 | PPA26-F | CGCACACATTTCACAGAACTCA |
| PPA26-R | CACTTTCCATCCCTCAAGACCTTAG | ||
| PPactin | PPactin | PPactin-F | GTTATTCTTCATCGGCGTCTTCG |
| PPactin-R | CTTCACCATTCCAGTTCCATTGTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Mao, Q.; Guo, X.; Ma, R.; Yu, M.; Xu, J.; Guo, S. Genome-Wide Identification and Analysis of Plasma Membrane H+-ATPases Associated with Waterlogging in Prunus persica (L.) Batsch. Agronomy 2024, 14, 908. https://doi.org/10.3390/agronomy14050908
Zhang Y, Mao Q, Guo X, Ma R, Yu M, Xu J, Guo S. Genome-Wide Identification and Analysis of Plasma Membrane H+-ATPases Associated with Waterlogging in Prunus persica (L.) Batsch. Agronomy. 2024; 14(5):908. https://doi.org/10.3390/agronomy14050908
Chicago/Turabian StyleZhang, Yuyan, Qinsi Mao, Xin Guo, Ruijuan Ma, Mingliang Yu, Jianlan Xu, and Shaolei Guo. 2024. "Genome-Wide Identification and Analysis of Plasma Membrane H+-ATPases Associated with Waterlogging in Prunus persica (L.) Batsch" Agronomy 14, no. 5: 908. https://doi.org/10.3390/agronomy14050908
APA StyleZhang, Y., Mao, Q., Guo, X., Ma, R., Yu, M., Xu, J., & Guo, S. (2024). Genome-Wide Identification and Analysis of Plasma Membrane H+-ATPases Associated with Waterlogging in Prunus persica (L.) Batsch. Agronomy, 14(5), 908. https://doi.org/10.3390/agronomy14050908






