Molecular Identification and Phylogenetic Analysis of Cymbidium Species (Orchidaceae) Based on the Potential DNA Barcodes matK, rbcL, psbA-trnH, and Internal Transcribed Spacer
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Extraction, Amplification, and Sequencing
2.3. Data Analysis
3. Results
3.1. PCR Amplification Success Rate and Sequence Characteristics
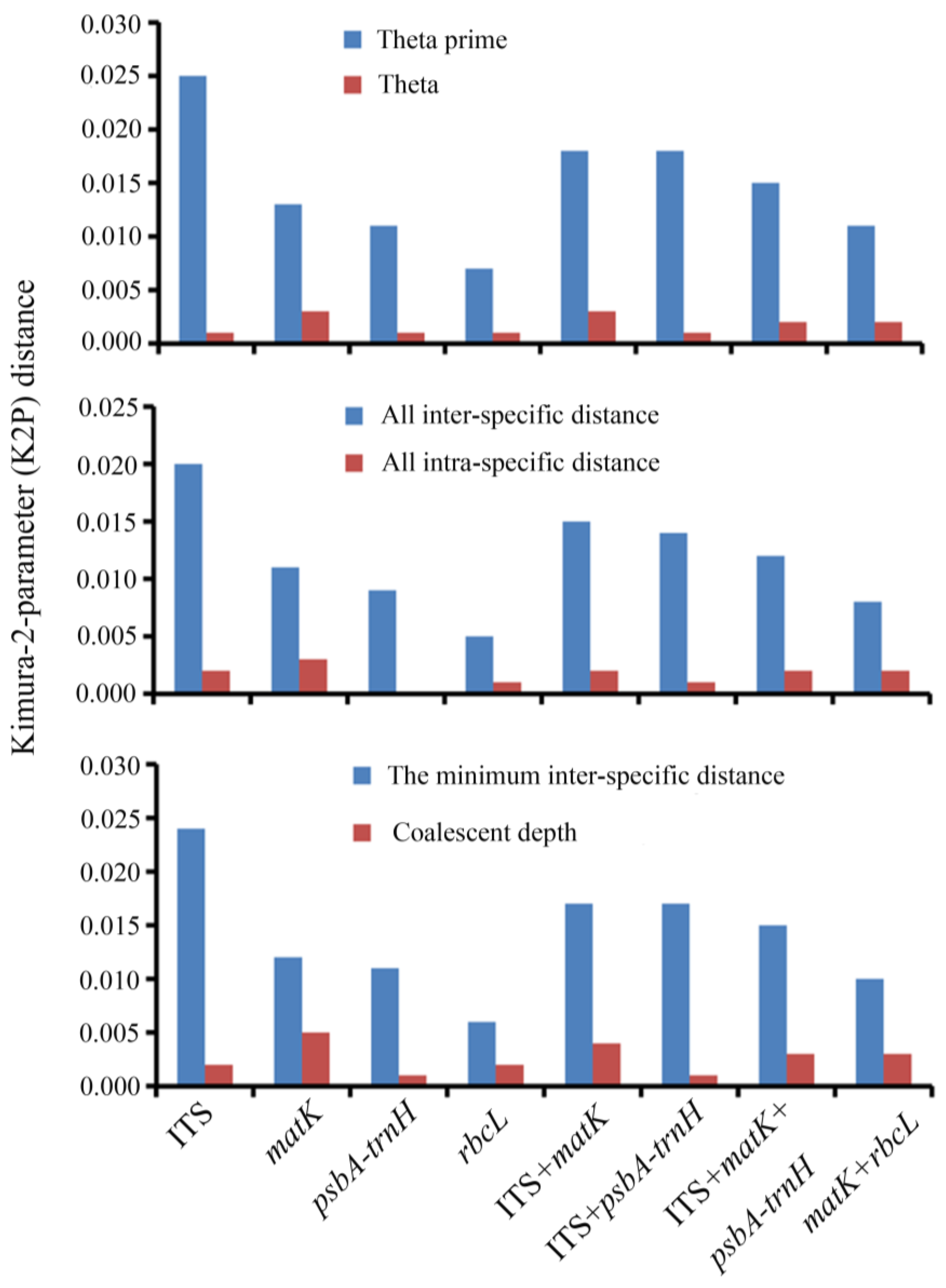
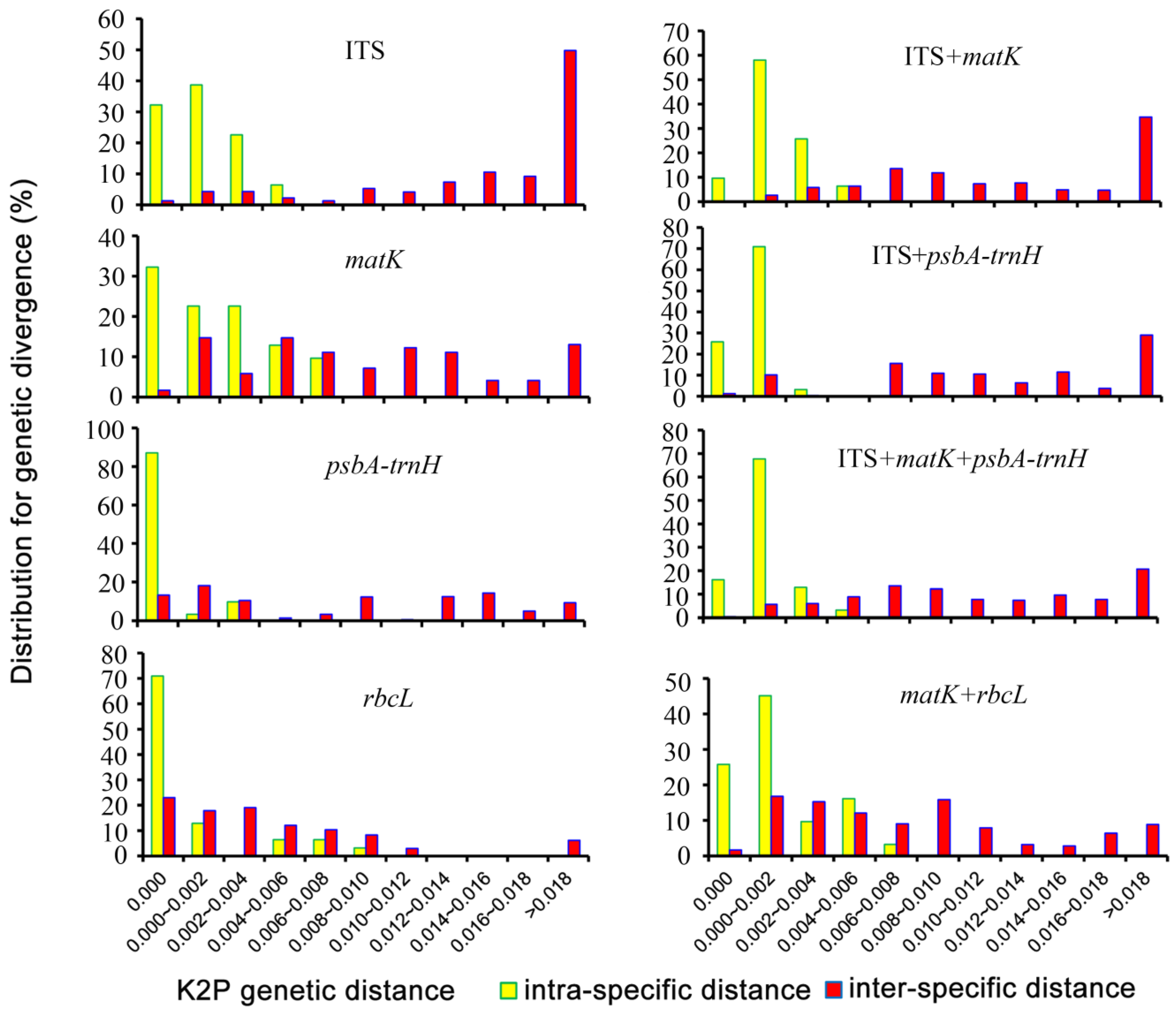
3.2. Genetic Divergence within and between Species
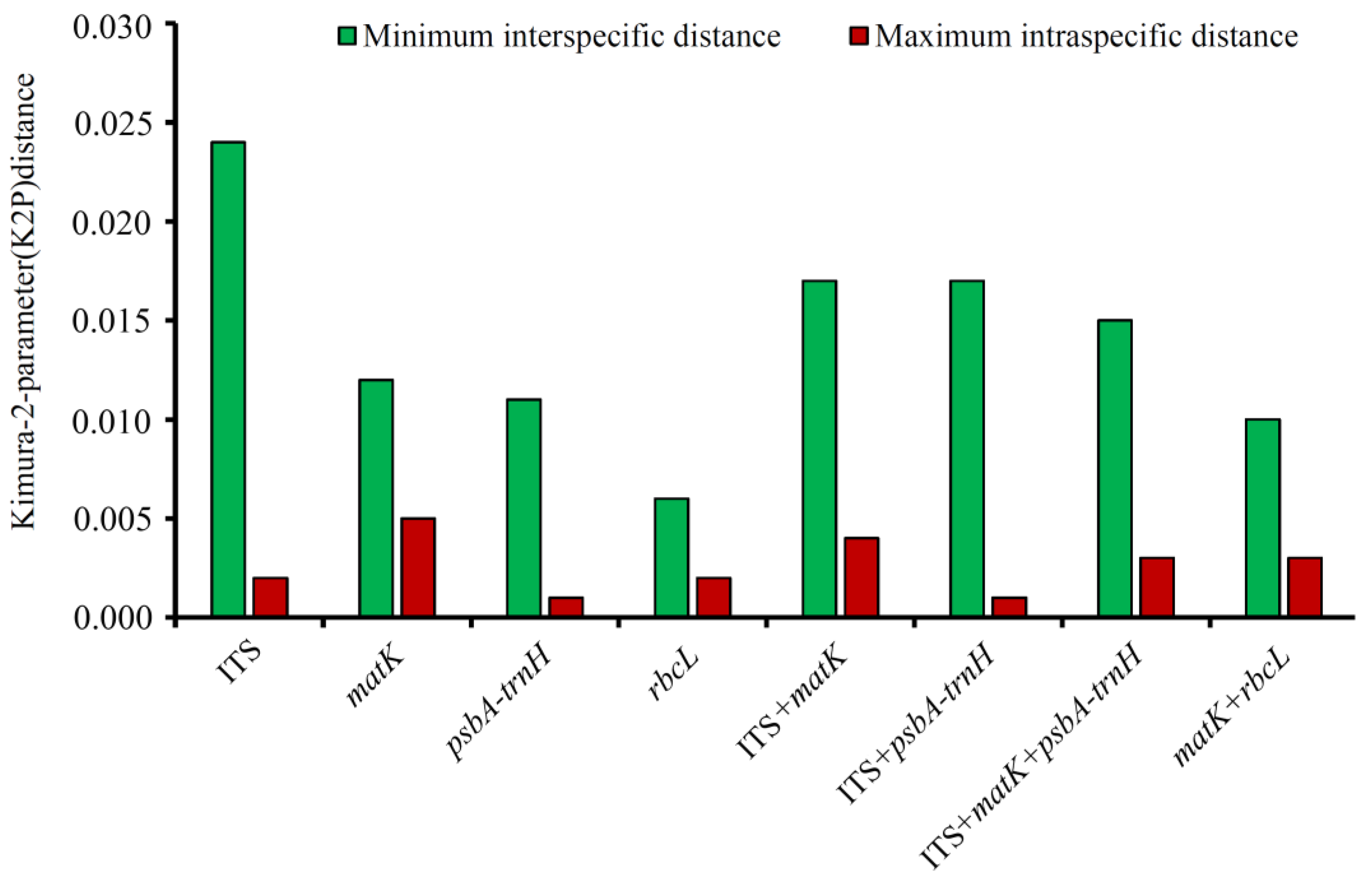
3.3. Assessment of the Barcoding Gap
3.4. Applicability for Species Authentication
3.5. Evaluation of the Discriminatory Power of Candidate Barcoding
3.6. Analysis of ITS2 Secondary Structure
3.7. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, J.B.; Tang, M.; Li, H.T.; Zhang, Z.R.; Li, D.Z. Complete chloroplast genome of the genus Cymbidium: Lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 2013, 13, 84. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.X.; Gao, J.; Wei, Y.L.; Ren, R.; Zhang, G.Q.; Lu, C.Q.; Jin, J.P.; Ai, Y.; Wang, Y.Q.; Chen, L.J.; et al. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 2021, 19, 2501–2516. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Chase, M.W. Independent degradation in genes of the plastid ndh gene family in species of the orchid genus Cymbidium (Orchidaceae; Epidendroideae). PLoS ONE 2017, 12, e0187318. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.K.; Rajkumari, K.; Kumaria, S.; Tandon, P.; Rao, S.R. Karyo-morphological characterization of natural genetic variation in some threatened Cymbidium species of Northeast India. Caryologia 2010, 63, 99–105. [Google Scholar] [CrossRef]
- Yukawa, T.; Stern, W.L. Comparative vegetative anatomy and systematics of Cymbidium (Cymbidieae: Orchidaceae). Bot. J. Linn. Soc. 2002, 138, 383–419. [Google Scholar] [CrossRef]
- Schlechter, R. Die Gattungen Cymbidium Sw. und Cyperorchis Bl. Repert. Sp. Nov. Regni. 1924, 20, e96–e110. [Google Scholar] [CrossRef]
- Hunt, P.F. Notes on Asiatic orchis 5. Kew Bull. 1970, 24, e93–e94. [Google Scholar] [CrossRef]
- Seth, C.J.; Cribb, P.J. A reassessment of the sectional limits in the genus Cymbidium Swartz. In Orchid Biology, Reviews and Prospectives 3; Cornell University Press: Ithaca, NY, USA; London, UK, 1984. [Google Scholar]
- Puy, D.D.; Cribb, P. The Genus Cymbidium; Helm, C., Ed.; The University of Chicago Press: Chicago, IL, USA, 1988. [Google Scholar]
- Liu, Z.J.; Chen, X.Q.; Ru, Z.Z. The Genus Cymbidium in China; Science Press: Beijing, China, 2006. [Google Scholar]
- Wang, H.Z.; Wu, Z.X.; Lu, J.J.; Shi, N.N.; Zhao, Y.; Zhang, Z.T.; Liu, J.J. Molecular diversity and relationships among Cymbidium goeringii cultivars based on inter-simple sequence repeat (ISSR) markers. Genetica 2009, 136, 391–399. [Google Scholar] [CrossRef]
- Chen, J.; Chen, M.K.; Zheng, Q.D.; Ma, S.H.; Liu, Z.J.; Ai, Y. Chloroplast characterization and phylogenetic relationship of Cymbidium aloifolium (Orchidaceae). Mitochondrial. DNA B Resour. 2020, 5, 478–479. [Google Scholar] [CrossRef]
- Hollingsworth, P.M. Refining the DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19451–19452. [Google Scholar] [CrossRef]
- Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; Yang, J.B.; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar]
- Schindel, D.E.; Miller, S.E. DNA barcoding a useful tool for taxonomists. Nature 2005, 435, 17. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Jin, Y.Y.; Gao, Y.D.; Zhang, Y.; Ying, Q.C.; Shen, C.J.; Lu, J.J.; Zhan, X.R.; Wang, H.Z.; Feng, S.G. The Complete chloroplast genomes of two Physalis species, Physalis macrophysa and P. ixocarpa: Comparative genomics, evolutionary dynamics and phylogenetic relationships. Agronomy 2023, 13, 135. [Google Scholar] [CrossRef]
- Bergmann, T.; Rach, J.; Damm, S.; Desalle, R.; Schierwater, B.; Hadrys, H. The potential of distance-based thresholds and character-based DNA barcoding for defining problematic taxonomic entities by CO1 and ND1. Mol. Ecol. Resour. 2013, 13, 1069–1081. [Google Scholar] [CrossRef]
- Karamat, S.; Ashraf, N.; Akhtar, T.; Rahim, F.; Shafi, N.; Khalid, S.; Shahid, B.; Khawaja, S.; Rahim, J.; Majeed, Z.; et al. CO1-Based DNA barcoding for assessing diversity of Pteropus giganteus from the state of Azad Jammu Kashmir, Pakistan. Braz. J. Biol. 2021, 81, 584–591. [Google Scholar] [CrossRef]
- Larranaga, N.; Hormaza, J.I. DNA barcoding of perennial fruit tree species of agronomic interest in the genus Annona (Annonaceae). Front. Plant Sci. 2015, 6, 589. [Google Scholar] [CrossRef] [PubMed]
- Cbol Plant Working Group. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef]
- Chen, S.L.; Yao, H.; Han, J.P.; Liu, C.; Song, J.Y.; Shi, L.C.; Zhu, Y.J.; Ma, X.Y.; Gao, T.; Pang, X.H.; et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef] [PubMed]
- Li, X.W.; Yang, Y.; Henry, R.J.; Rossetto, M.; Wang, Y.T.; Chen, S.L. Plant DNA barcoding: From gene to genome. Biol. Rev. Camb. Philos. Soc. 2015, 90, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Sun, W.; Wu, L.; Xu, R.; Zhang, Y.; Zhu, W.; Panjwani; Liu, Z.; Zhao, B. Evaluation of four commonly used DNA barcoding loci for Ardisia species identification. Front. Plant Sci. 2022, 13, 860778. [Google Scholar] [CrossRef]
- Siripiyasing, P.; Kaenratana, K.; Mokkamul, P.; Tanee, T.; Sudmoon, R.; Chaveerach, A. DNA barcoding of the Cymbidium species (Orchidaceae) in Thailand. Afr. J. Agric. Res. 2012, 7, 393–404. [Google Scholar]
- Obara-Okeyo, P.; Kako, S. Genetic diversity and identification of Cymbidium cultivars as measured by random amplified polymorphic DNA (RAPD) markers. Euphytica 1998, 99, 95–101. [Google Scholar] [CrossRef]
- Wang, H.Z.; Wang, Y.D.; Zhou, X.Y.; Ying, Q.C.; Zheng, K.L. Analysis of genetic diversity of 14 species of Cymbidium based on RAPDs and AFLPs. Shi Yan Sheng Wu Xue Bao 2004, 37, 482–486. [Google Scholar] [PubMed]
- Wu, Z.X.; Wang, H.Z.; Shi, N.N.; Zhao, Y. The genetic diversity of Cymbidium by ISSR. Yi Chuan 2008, 30, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.K.; Kumaria, S.; Tandon, P.; Rao, S.R. Assessment of genetic variation and identification of species-specific ISSR markers in five species of Cymbidium (Orchidaceae). J. Plant Biochem. Biot. 2013, 22, 250–255. [Google Scholar] [CrossRef]
- Moe, K.T.; Zhao, W.; Song, H.S.; Kim, Y.H.; Chung, J.W.; Cho, Y.I.; Park, P.H.; Park, H.S.; Chae, S.C.; Park, Y.J. Development of SSR markers to study diversity in the genus Cymbidium. Biochem. Syst. Ecol. 2010, 38, 585–594. [Google Scholar] [CrossRef]
- Van den Berg, C.; Ryan, A.; Cribb, P.; Chase, M. Molecular phylogenetics of Cymbidium (Orchidaceae: Maxillarieae) sequence data from internal transcribed spacers (ITS) of nuclear ribosomal DNA and plastid matK. Lindleyana 2002, 17, 102–111. [Google Scholar]
- Sharma, S.K.; Dkhar, J.; Kumaria, S.; Tandon, P.; Rao, S.R. Assessment of phylogenetic inter-relationships in the genus Cymbidium (Orchidaceae) based on internal transcribed spacer region of rDNA. Gene 2012, 495, 10–15. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, Y.W.; Huang, J.L.; Ya, J.D.; Zhe, M.Q.; Zeng, C.X.; Zhang, Z.R.; Zhang, S.B.; Li, D.Z.; Li, H.T.; et al. DNA barcoding of Cymbidium by genome skimming: Call for next-generation nuclear barcodes. Mol. Ecol. Resour. 2022, 23, 424–439. [Google Scholar] [CrossRef]
- Wang, X.M.; Cheng, L.J.; Li, Z.L.; Zhang, Z.C.; Wang, Y.Y. Characterization of the complete chloroplast genome of a Chinese endangered species Cymbidium wenshanense Y. S. Wu et F. Y. Liu. Mitochondrial. DNA B Resour. 2023, 8, 815–818. [Google Scholar] [CrossRef]
- Choi, S.; Kim, M.; Lee, J.; Ryu, K. Genetic diversity and phylogenetic relationships among and within species of oriental Cymbidiums based on RAPD analysis. Sci. Hortic. 2006, 108, 79–85. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Chen, G.Z.; Chen, L.J.; Zhai, J.W.; Huang, J.; Wu, X.Y.; Li, M.H.; Peng, D.H.; Rao, W.H.; Liu, Z.J.; et al. Phylogenetic incongruence in Cymbidium orchids. Plant Divers. 2021, 43, 452–461. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 1, 2–3. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Feng, S.G.; Jiang, M.Y.; Shi, Y.J.; Jiao, K.L.; Shen, C.J.; Lu, J.J.; Ying, Q.C.; Wang, H.Z. Application of the ribosomal DNA ITS2 region of Physalis (Solanaceae): DNA barcoding and phylogenetic study. Front. Plant Sci. 2016, 7, 1047. [Google Scholar] [CrossRef]
- Slabbinck, B.; Dawyndt, P.; Martens, M.; De Vos, P.; De Baets, B. TaxonGap: A visualization tool for intra- and inter-species variation among individual biomarkers. Bioinformatics 2008, 24, 866–867. [Google Scholar] [CrossRef]
- Chinese Academy of Sciences. Flora of China; Science Press: Beijing, China, 1999; Volume 18, p. 191. [Google Scholar]
- Lahaye, R.; Van der Bank, M.; Bogarin, D.; Warner, J.; Pupulin, F.; Gigot, G.; Maurin, O.; Duthoit, S.; Barraclough, T.G.; Savolainen, V. DNA barcoding the floras of biodiversity hotspots. Proc. Natl. Acad. Sci. USA 2008, 105, 2923–2928. [Google Scholar] [CrossRef]
- Xiang, X.G.; Hu, H.; Wang, W.; Jin, X.H. DNA barcoding of the recently evolved genus Holcoglossum (Orchidaceae: Aeridinae): A test of DNA barcode candidates. Mol. Ecol. Resour. 2011, 11, 1012–1021. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Cheng, L.J.; Zhao, Y.R.; Li, Y.F.; He, F.M.; Li, Z.L. Characterization of the complete chloroplast genome of a Chinese endangered species Cymbidium mannii. Mitochondrial. DNA B Resour. 2020, 5, 3199–3200. [Google Scholar] [CrossRef]
- Du, Z.H.; Yang, X.Y.; Tan, G.F.; Chen, Z.L. The complete chloroplast genome of Cymbidium dayanum (Orchidaceae). Mitochondrial. DNA B Resour. 2021, 6, 1897–1898. [Google Scholar] [CrossRef]
- Newmaster, S.G.; Fazekas, A.J.; Steeves, R.A.; Janovec, J. Testing candidate plant barcode regions in the Myristicaceae. Mol. Ecol. Resour. 2008, 8, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.G.; Jiao, K.L.; Zhu, Y.J.; Wang, H.H.; Jiang, M.Y.; Wang, H.Z. Molecular identification of species of Physalis (Solanaceae) using a candidate DNA barcode: The chloroplast psbA-trnH intergenic region. Genome 2018, 61, 15–20. [Google Scholar] [CrossRef]
- Fujii, T.; Mori, T.; Tatsuo, Y.; Takao, Y.; Fujino, H.; Tsuchida, T.; Minami, M. Identification of Valeriana fauriei and other Eurasian medicinal valerian by DNA polymorphisms in psbA-trnH intergenic spacer sequences in chloroplast DNA. J. Nat. Med. 2021, 75, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Erickson, D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef] [PubMed]
- Sass, C.; Little, D.P.; Stevenson, D.W.; Specht, C.D. DNA barcoding in the cycadales: Testing the potential of proposed barcoding markers for species identification of cycads. PLoS ONE 2007, 2, e1154. [Google Scholar] [CrossRef]
- Newmaster, S.; Fazekas, A.; Ragupathy, S. DNA barcoding in land plants: Evaluation of rbcL in a multigene tiered approach. Botany 2006, 84, 335–341. [Google Scholar] [CrossRef]
- Starr, J.R.; Naczi, R.F.; Chouinard, B.N. Plant DNA barcodes and species resolution in sedges (Carex, Cyperaceae). Mol. Ecol. Resour. 2009, 9 (Suppl. S1), 151–163. [Google Scholar] [CrossRef]
- Gao, T.; Yao, H.; Song, J.Y.; Zhu, Y.J.; Liu, C.; Chen, S.L. Evaluating the feasibility of using candidate DNA barcodes in discriminating species of the large Asteraceae family. BMC Evol. Biol. 2010, 10, 324. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C. Plant Bar Code Soon to Become Reality; Science: New York, NY, USA, 2009; Volume 325, p. 526. [Google Scholar]
- Samanta, B.; Ehrman, J.M.; Kaczmarska, I. A consensus secondary structure of ITS2 for the diatom Order Cymatosirales (Mediophyceae, Bacillariophyta) and reappraisal of the order based on DNA, morphology, and reproduction. Mol. Phylogenet. Evol. 2018, 129, 117–129. [Google Scholar] [CrossRef]




| DNA Region | Primer Name | Sequence (5′-3′) | Amplication Protocol |
|---|---|---|---|
| ITS | ITS4 | TCCTCCGCTTATTGATATGC | 94 °C 5 min; 94 °C 1 min, 55 °C 1 min; |
| ITS5 | GGAAGGAGAAGTCGTAACAAGG | 72 °C 1.5 min, 35 cycles; 72 °C 10 min | |
| matK | matK-1F | CGTACAGTACTTTTGTGTTTACGAG | 94 °C 5 min; 94 °C 1 min, 60 °C 1 min; |
| matK-1R | ACCCAGTCCATCTGGAAATCTTGGTTC | 72 °C 1.5 min, 30 cycles; 72 °C 10 min | |
| rbcL | rbcL-IF | ATGTCACCACAAACAGAGACTAAAGC | 95 °C 2 min; 94 °C 1 min, 55 °C 1 min; |
| rbcL-1R | GTAAAATCAAGTCCACCRCG | 72 °C 1.5 min, 35 cycles; 72 °C 10 min | |
| psbA-trnH | psbA-3F | GTTATGCATGAACGTAATGCTC | 94 °C 5 min; 94 °C 1 min, 55 °C 1 min; |
| trnHF | CGCGCATGGTGGATTCACAATCC | 72 °C 1.5 min, 32 cycles; 72 °C 10 min | |
| atpF-atpH | atpF-H/f | ACTCGCACACACTCCCTTTCC | 94 °C 5 min; 94 °C 1 min, 50 °C 1 min; |
| atpF-H/R | GCTTTTATGGAAGCTTTAACAAT | 72 °C 1.5 min, 35 cycles; 72 °C 10 min |
| No. of Samples/Species | PCR Success (%) | Sequencing Success (%) | Aligned Length (bp) | N Variable Characters (%Variable Characters) | G+C Ratio (%) | Ability to Discriminate | |
|---|---|---|---|---|---|---|---|
| ITS | 34/18 | 100 | 100 | 661 | 74 (11.2) | 68.4 | 93.20% |
| matK | 34/18 | 100 | 100 | 848 | 59 (7.0) | 32.1 | 75.80% |
| psbA-trnH | 34/18 | 100 | 100 | 845 | 87 (10.3) | 34 | 87.10% |
| rbcL | 34/18 | 100 | 100 | 575 | 28 (5.0) | 42.9 | 54.20% |
| atpF-atpH | 34/18 | low | low | - | - | - | - |
| ITS + matK | 34/18 | - | - | 1509 | 131 (8.7) | 47.9 | 88.50% |
| ITS + psbA-trnH | 34/18 | - | - | 1510 | 159 (10.5) | 50 | 90.90% |
| ITS + matK + psbA-trnH | 34/18 | - | - | 2362 | 213 (9.0) | 42.1 | 93.20% |
| matK + rbcL | 34/18 | - | - | 1686 | 162 (9.6) | 33 | 67.00% |
| W+ | W− | Relative Ranks, n, p Value | Result |
|---|---|---|---|
| ITS | matK | W+ = 122,851.5, W− = 12,088.5, n = 519, p ≤ 5.144 × 10−59 | ITS > matK |
| ITS | psbA-trnH | W+ = 119,137, W− = 8123, n = 504, p ≤ 1.492 × 10−64 | ITS > psbA-trnH |
| ITS | rbcL | W+ = 126,189, W− = 2082, n = 506, p ≤ 2.735 × 10−79 | ITS > rbcL |
| ITS | ITS + matK | W+ = 124,066.5, W− = 7774.5, n = 513, p ≤ 4.208 × 10−67 | ITS > ITS + matK |
| ITS | ITS + psbA-trnH | W+ = 127,702.5, W− = 3113.5, n = 511, p ≤ 1.259 × 10−77 | ITS >TS + psbA-trnH |
| ITS | ITS + matK + psbA-trnH | W+ = 122,080, W− = 4173, n = 502, p ≤ 1.982 × 10−73 | ITS > ITS + matK + psbA-trnH |
| ITS | matK + rbcL | W+ = 129,347, W− = 4039, n = 516, p ≤ 2.675 × 10−76 | ITS > matK + rbcL |
| matK | psbA-trnH | W+ = 71,961, W− = 55,804, n = 505, p ≤ 0.01381 | matK > psbA-trnH |
| matK | rbcL | W+ = 107,518, W− = 18,233, n = 501, p ≤ 4.034 × 10−43 | matK > rbcL |
| matK | ITS + matK | W+ = 20,587.5, W− = 103,663.5, n = 498, p ≤ 3.24 × 10−38 | matK< ITS + matK |
| matK | ITS + psbA-trnH | W+ = 33,000.5, W− = 93,755.5, n = 503, p ≤ 1.251 × 10−20 | matK< ITS + psbA-trnH |
| matK | ITS + matK + psbA-trnH | W+ = 37,713, W− = 87,037, n = 499, p ≤ 1.988 × 10−14 | matK < ITS + matK + psbA-trnH |
| matK | matK + rbcL | W+ = 76,946, W− = 30,007, n = 462, p ≤ 3.024 × 10−16 | matK > matK + rbcL |
| psbA-trnH | rbcL | W+ = 80,886, W− = 15,255, n = 438, p ≤ 3.479 × 10−35 | psbA-trnH > rbcL |
| psbA-trnH | ITS + matK | W+ = 25,876.5, W− = 106,993.5, n = 515, p ≤ 3.492 × 10−33 | psbA-trnH < ITS + matK |
| psbA-trnH | ITS + psbA-trnH | W+ = 22,874, W− = 100,879, n = 497, p ≤ 4.331 × 10−34 | psbA-trnH < ITS + psbA-trnH |
| psbA-trnH | ITS + matK + psbA-trnH | W+ = 35,329.5, W− = 93,448.5, n = 507, p ≤ 1.329 × 10−18 | psbA-trnH < ITS + matK + psbA-trnH |
| psbA-trnH | matK + rbcL | W+ = 70,183.5, W− = 56,069.5, n = 502, p ≤ 0.03 | psbA-trnH > matK + rbcL |
| rbcL | ITS + matK | W+ = 7126, W− = 129,900, n = 523, p ≤ 1.721 × 10−70 | rbcL< ITS + matK |
| rbcL | ITS + psbA-trnH | W+ = 4827.5, W− = 120,422.5, n = 500, p ≤ 1.772 × 10−71 | rbcL< ITS + psbA-trnH |
| rbcL | ITS + matK + psbA-trnH | W+ = 11,060, W− = 121,810, n = 515, p ≤ 2.417 × 10−60 | rbcL< ITS + matK + psbA-trnH |
| rbcL | matK + rbcL | W+ = 16,738.5, W− = 102,089.5, n = 487, p ≤ 6.601 × 10−43 | rbcL< matK + rbcL |
| ITS + matK | ITS + psbA-trnH | W+ = 69,846, W− = 40,369, n = 469, p ≤ 5.215 × 10 −7 | ITS + matK > ITS + psbA-trnH |
| ITS + matK | ITS+ matK + psbA-trnH | W+ = 112,461.5, W− = 5879.5, n = 486, p ≤ 2.658 × 10−66 | ITS + matK > ITS + matK + psbA-trnH |
| ITS + matK | matK + rbcL | W+ = 117,120.5, W− = 2195.5, n = 488, p ≤ 7.187 × 10−76 | ITS + matK> matK + rbcL |
| ITS + psbA-trnH | matK + psbA-trnH | W+ = 118,283, W− = 9988, n = 506, p ≤ 8.182 × 10−61 | ITS + psbA-trnH > matK+psbA-trnH |
| ITS + psbA-trnH | matK + rbcL | W+ = 118,735.5, W− = 8020.5, n = 503, p ≤ 1.376 × 10−64 | ITS + psbA-trnH > matK + rbcL |
| ITS+ matK + psbA-trnH | matK +rbcL | W+ = 115,863, W− = 6402, n = 494, p ≤ 1.386 × 10−66 | ITS + matK + psbA-trnH > matK + rbcL |
| W+ | W− | Relative Ranks, n, p Value | Result |
|---|---|---|---|
| ITS | matK | W+ = 161, W− = 217, n = 27, p ≤ 0.5088 | ITS = matK |
| ITS | psbA-trnH | W+ = 225, W− = 51, n = 23, p ≤ 0.008516 | ITS > psbA-trnH |
| ITS | rbcL | W+ = 182, W− = 94, n = 23, p ≤ 0.1858 | ITS = rbcL |
| ITS | ITS + matK | W+ = 161, W− = 217, n = 27, p ≤ 0.5088 | ITS = ITS + matK |
| ITS | ITS + psbA-trnH | W+ = 225, W− = 51, n = 23, p ≤ 0.008516 | ITS > ITS + psbA-trnH |
| ITS | ITS + matK + psbA-trnH | W+ = 151, W− = 102, n = 22, p ≤ 0.4359 | ITS = ITS + matK + psbA-trnH |
| ITS | matK + rbcL | W+ = 169, W− = 209, n = 27, p ≤ 0.6394 | ITS = matK + rbcL |
| matK | psbA-trnH | W+ = 231, W− = 0, n = 21, p ≤ 6.414× 10−5 | matK > psbA-trnH |
| matK | rbcL | W+ = 191, W− = 62, n = 22, p ≤ 0.03773 | matK > rbcL |
| matK | ITS + matK | W+ = 186, W− = 67, n = 22, p ≤ 0.05543 | matK = ITS + matK |
| matK | ITS + psbA-trnH | W+ = 228.5, W− = 47.5, n = 23, p ≤ 0.006194 | matK > ITS + psbA-trnH |
| matK | ITS + matK + psbA-trnH | W+ = 192.5, W− = 17.5, n = 20, p ≤ 0 | matK > ITS + matK + psbA-trnH |
| matK | matK + rbcL | W+ = 111.5, W− = 24.5, n = 16, p ≤ 0 | matK > matK + rbcL |
| psbA-trnH | rbcL | W+ = 3.5, W− = 51.5, n = 10, p ≤ 0 | psbA-trnH < rbcL |
| psbA-trnH | ITS + matK | W+ = 13, W− = 393, n = 28, p ≤ 1.596× 10−5 | psbA-trnH < ITS + matK |
| psbA-trnH | ITS + psbA-trnH | W+ = 36, W− = 264, n = 24, p ≤ 0.001183 | psbA-trnH < ITS + psbA-trnH |
| psbA-trnH | ITS + matK + psbA-trnH | W+ = 18, W− = 333, n = 26, p ≤ 6.681× 10−5 | psbA-trnH < ITS + matK + psbA-trnH |
| psbA-trnH | matK + rbcL | W+ = 0, W− = 276, n = 23, p ≤ 2.891× 10−5 | psbA-trnH < matK + rbcL |
| rbcL | ITS + matK | W+ = 134.5, W− = 330.5, n = 30, p ≤ 0.04492 | rbcL < ITS + matK |
| rbcL | ITS + psbA-trnH | W+ = 150, W− = 175, n = 25, p ≤ 0.7468 | rbcL = ITS + psbA-trnH |
| rbcL | ITS + matK + psbA-trnH | W+ = 148.5, W− = 229.5, n = 27, p ≤ 0.3366 | rbcL = ITS + matK + psbA-trnH |
| rbcL | matK + rbcL | W+ = 59.5, W− = 193.5, n = 22, p ≤ 0.03085 | rbcL < matK +rbcL |
| ITS + matK | ITS + psbA-trnH | W+ = 128.5, W− = 7.5, n = 16, p ≤ 0 | ITS + matK > ITS + psbA-trnH |
| ITS + matK | ITS + matK + psbA-trnH | W+ = 171, W− = 0, n = 18, p ≤ 0 | ITS + matK > ITS + matK + psbA-trnH |
| ITS + matK | matK + rbcL | W+ = 114, W− = 96, n = 20, p ≤ 0 | ITS + matK > matK + rbcL |
| ITS + psbA-trnH | ITS + matK + psbA-trnH | W+ = 72, W− = 138, n = 20, p ≤ 0 | ITS + psbA-trnH < ITS + matK + psbA-trnH |
| ITS + psbA-trnH | matK + rbcL | W+ = 76, W− = 224, n = 24, p ≤ 0.03573 | ITS + psbA-trnH < matK + rbcL |
| ITS + matK + psbA-trnH | matK + rbcL | W+ = 30, W− = 141, n = 18, p ≤ 0 | ITS + matK + psbA-trnH < matK + rbcL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Gao, L.; Wang, H.; Feng, S. Molecular Identification and Phylogenetic Analysis of Cymbidium Species (Orchidaceae) Based on the Potential DNA Barcodes matK, rbcL, psbA-trnH, and Internal Transcribed Spacer. Agronomy 2024, 14, 933. https://doi.org/10.3390/agronomy14050933
Chen Z, Gao L, Wang H, Feng S. Molecular Identification and Phylogenetic Analysis of Cymbidium Species (Orchidaceae) Based on the Potential DNA Barcodes matK, rbcL, psbA-trnH, and Internal Transcribed Spacer. Agronomy. 2024; 14(5):933. https://doi.org/10.3390/agronomy14050933
Chicago/Turabian StyleChen, Zhenming, Ling Gao, Huizhong Wang, and Shangguo Feng. 2024. "Molecular Identification and Phylogenetic Analysis of Cymbidium Species (Orchidaceae) Based on the Potential DNA Barcodes matK, rbcL, psbA-trnH, and Internal Transcribed Spacer" Agronomy 14, no. 5: 933. https://doi.org/10.3390/agronomy14050933
APA StyleChen, Z., Gao, L., Wang, H., & Feng, S. (2024). Molecular Identification and Phylogenetic Analysis of Cymbidium Species (Orchidaceae) Based on the Potential DNA Barcodes matK, rbcL, psbA-trnH, and Internal Transcribed Spacer. Agronomy, 14(5), 933. https://doi.org/10.3390/agronomy14050933






