HPPEM: A High-Precision Blueberry Cluster Phenotype Extraction Model Based on Hybrid Task Cascade
Abstract
:1. Introduction
- (1)
- Aiming at the problem of low accuracy of blueberry detection under severe occlusion, we adopt the ConvNeXt [27] backbone, hybrid cascade structure, and multi-scale training to strengthen the feature extraction capability and realize the accurate segmentation of blueberry fruits.
- (2)
- Combining contour detection, convex packet algorithm, and rotary caliper technology to design algorithm modules to realize the automatic extraction of the number of fruits, ripeness, and compactness within blueberry clusters.
- (3)
- Use HPPEM to extract cluster phenotypes of four varieties of blueberries, compare the trait differences between different varieties, and analyze the detection error and quantity extraction error.
2. Materials and Methods
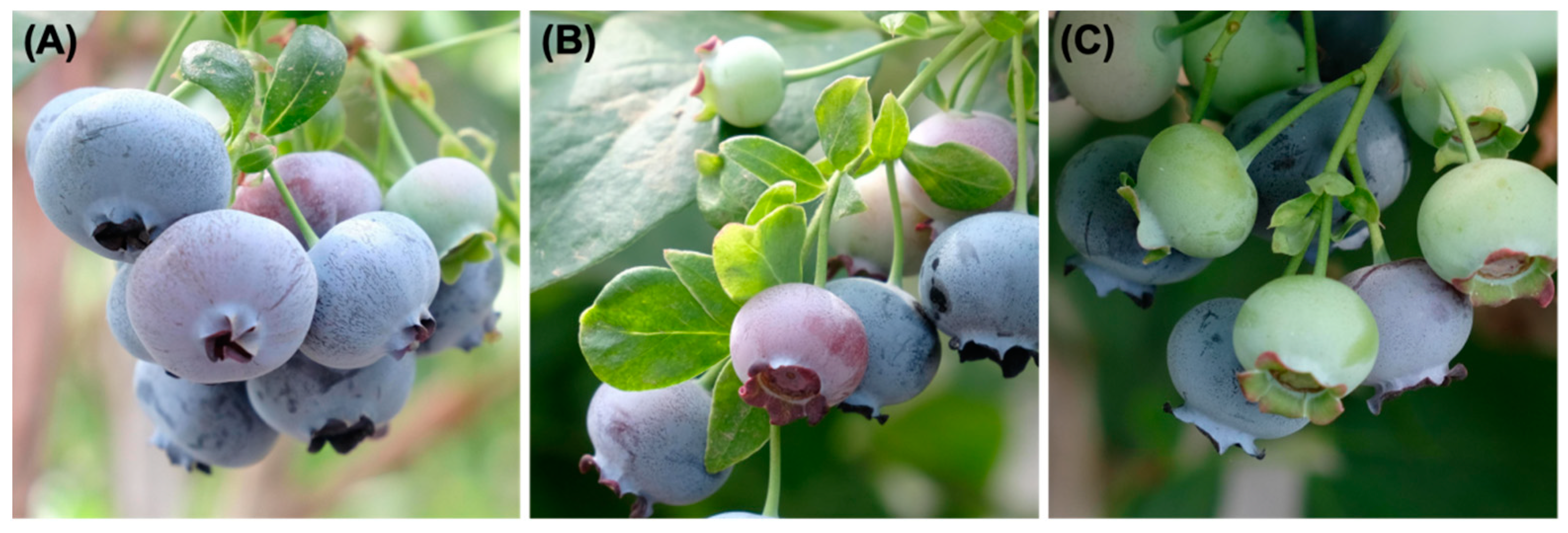
2.1. Data Collection
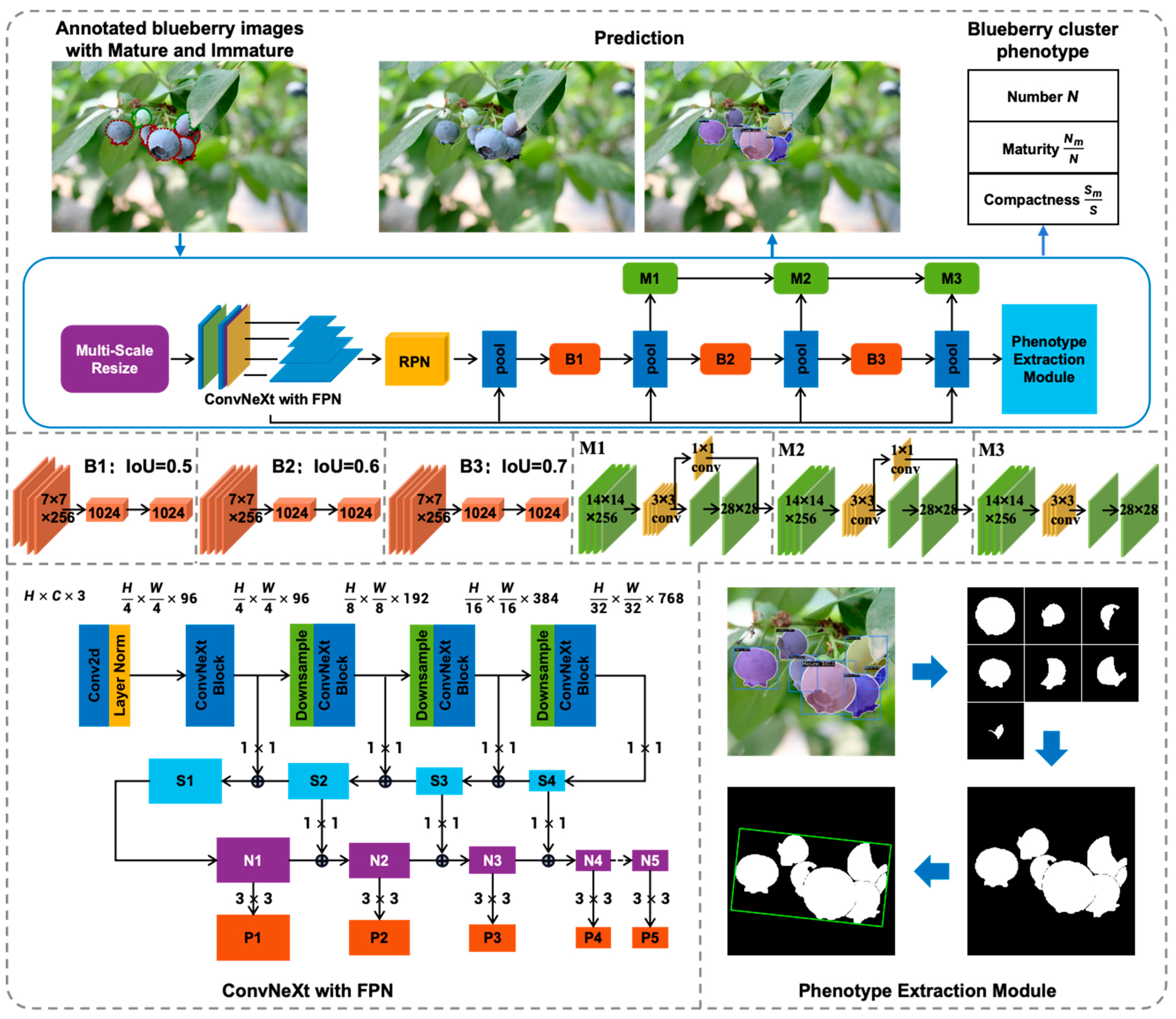
2.2. HPPEM
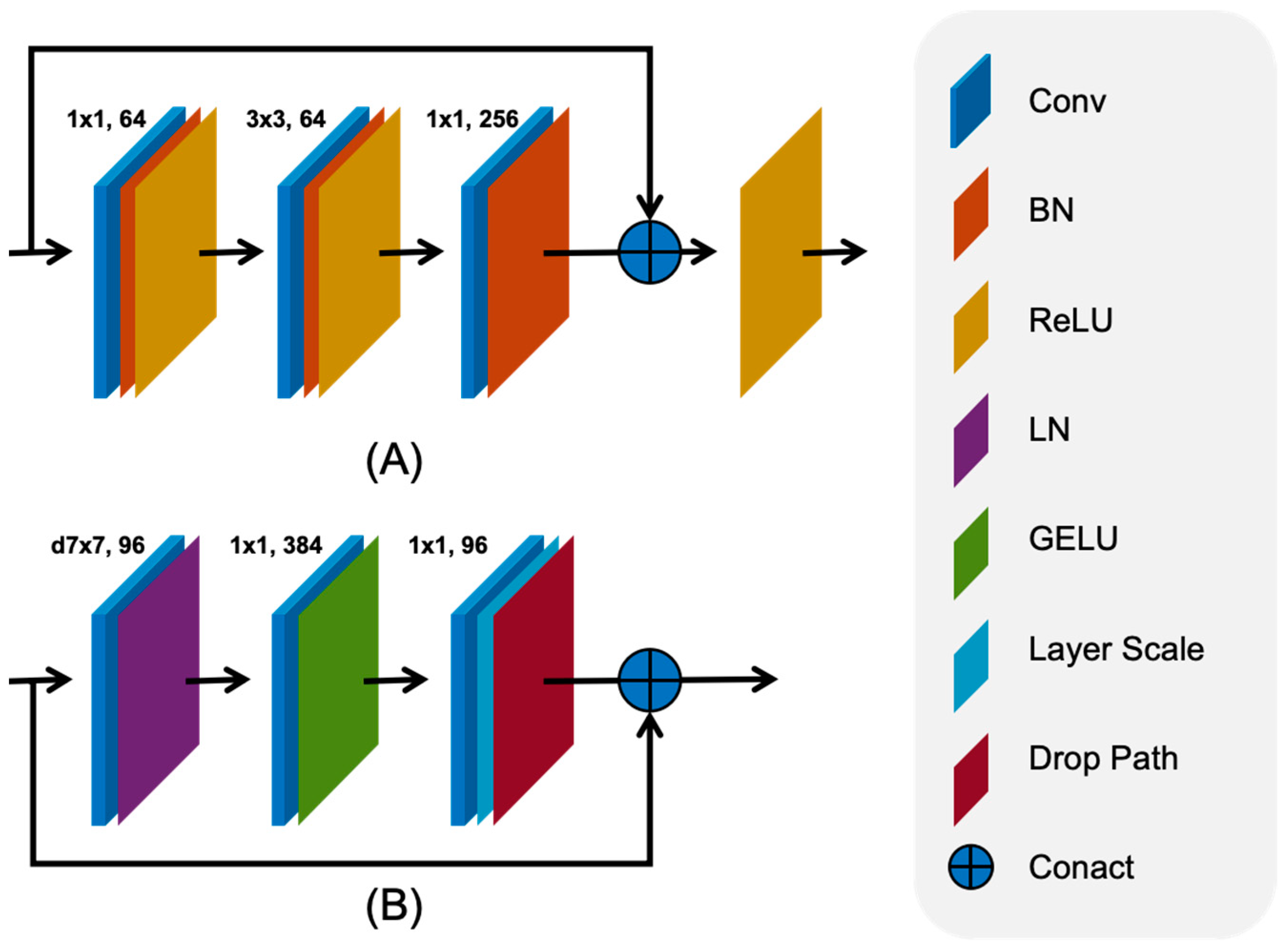
2.2.1. Feature Extraction
2.2.2. Multi-Scale Training
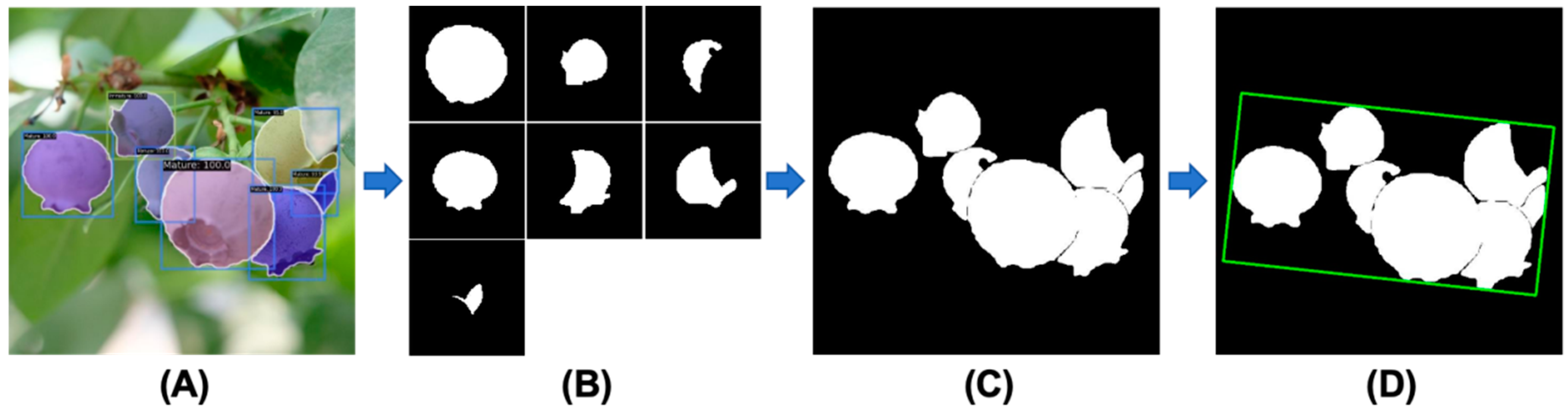
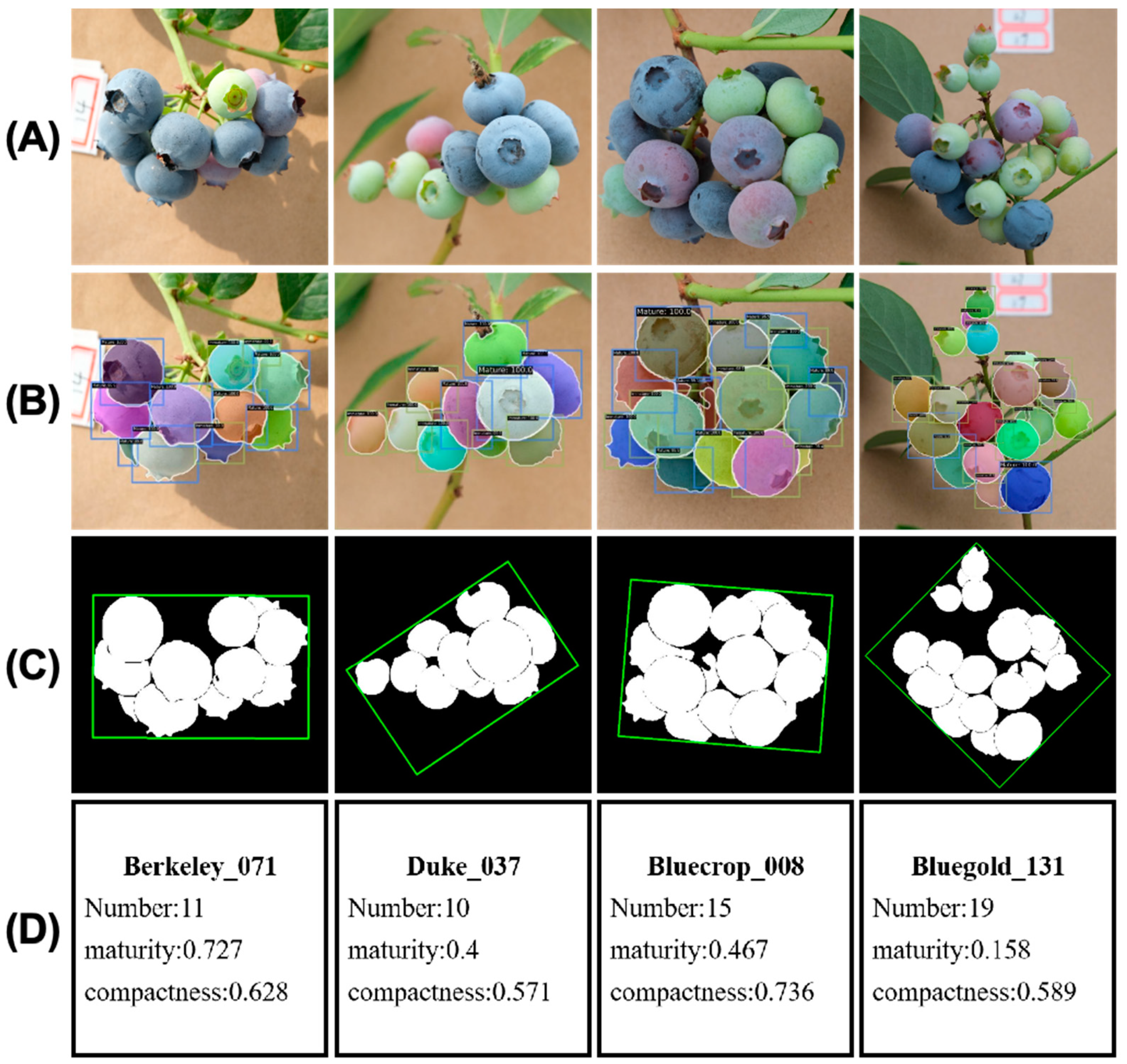
2.2.3. Phenotype Extraction Module
2.3. Evaluation Metrics
3. Results
3.1. Experimental Results
3.2. Ablation Study
3.3. Comparative Study
4. Discussion
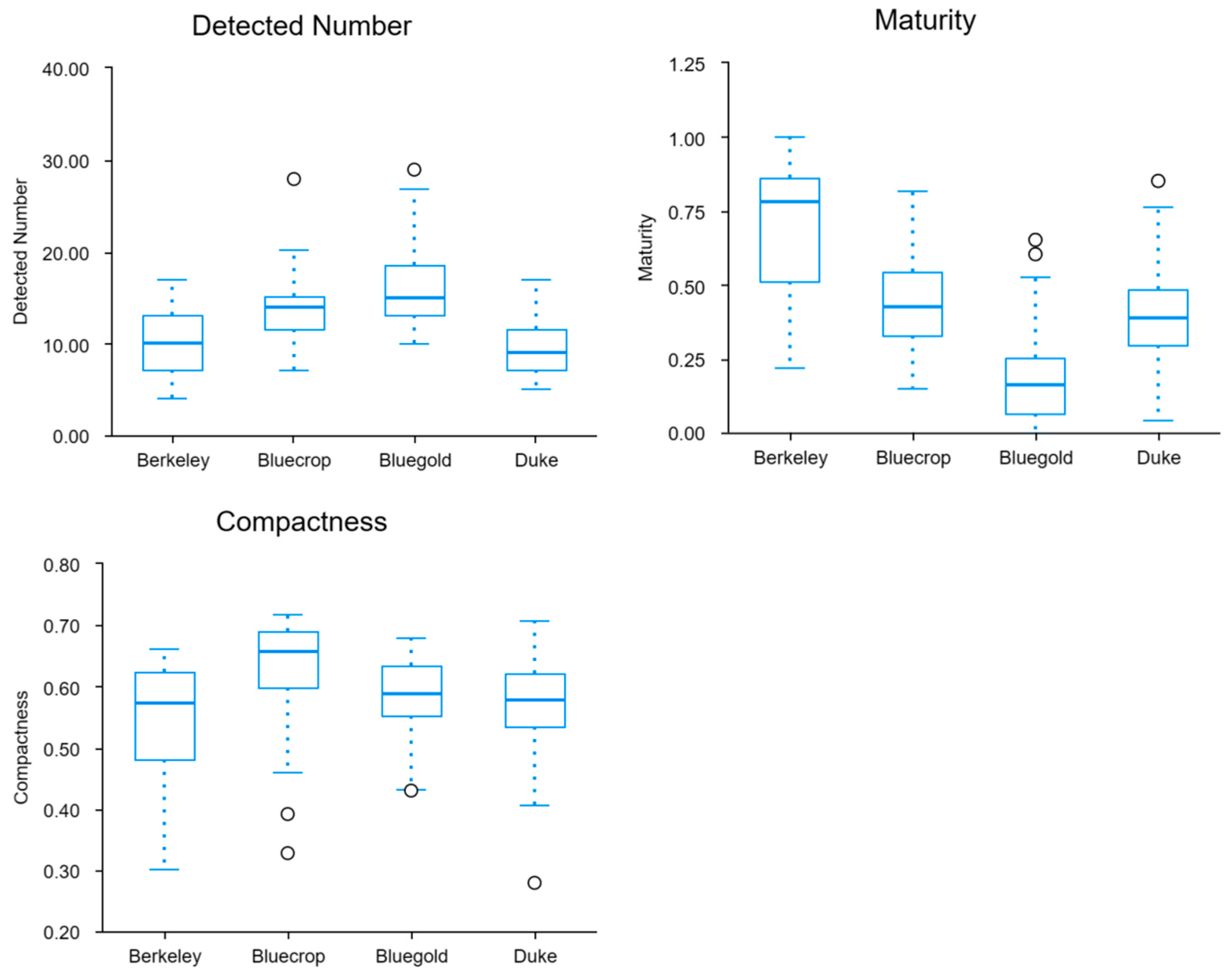
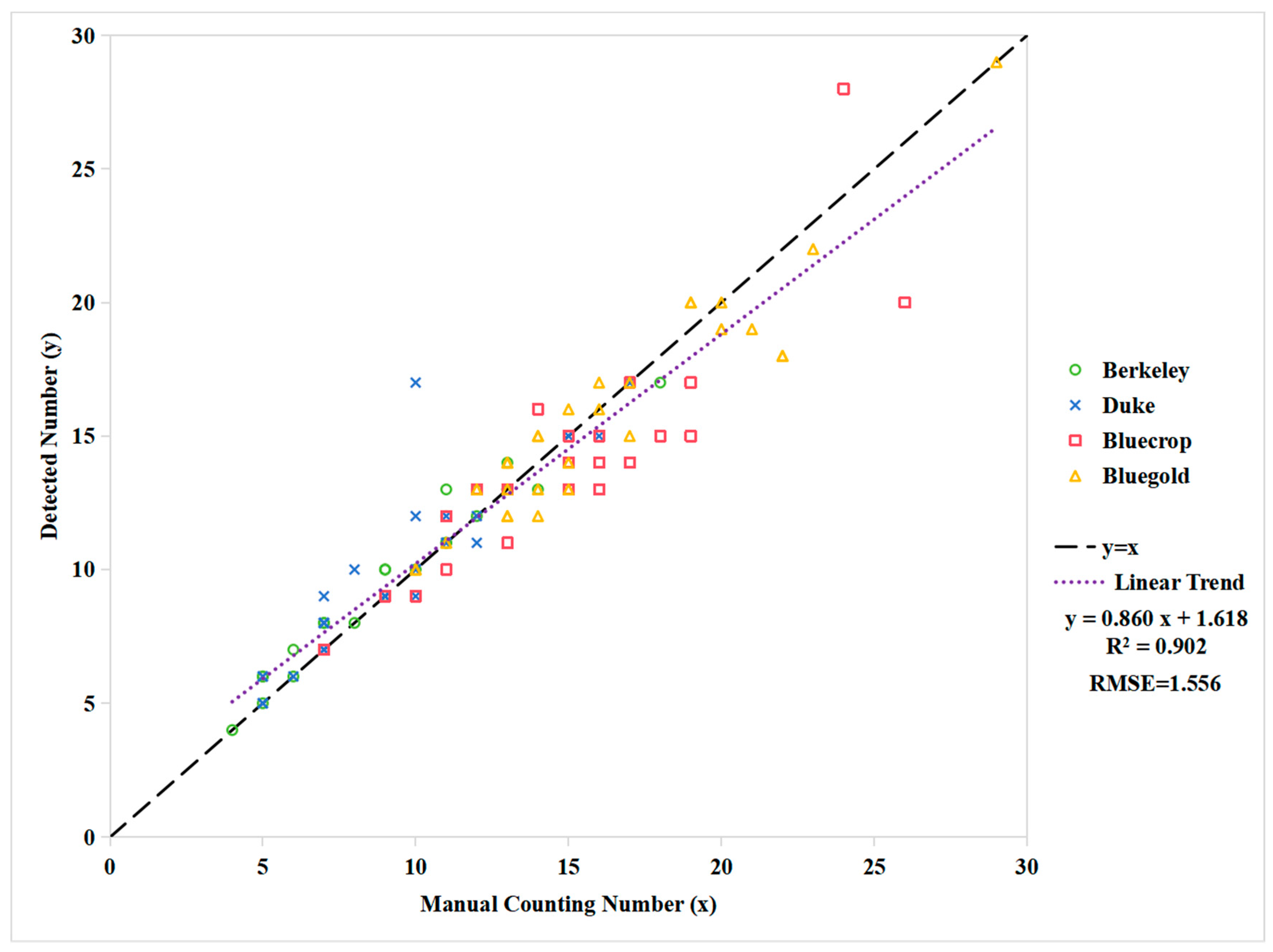
4.1. Detection Error
4.2. Quantity Extraction Error
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Qian, Y.P.L.; Zhou, Q.; Magana, A.A.; Qian, M.C. Comparative study of volatile composition of major Northern Highbush blueberry (Vaccinium corymbosum) varieties. J. Food Compos. Anal. 2022, 110, 104538. [Google Scholar] [CrossRef]
- Yang, W.; Guo, Y.; Liu, M.; Chen, X.; Xiao, X.; Wang, S.; Gong, P.; Ma, Y.; Chen, F. Structure and function of blueberry anthocyanins: A review of recent advance. J. Funct. Foods 2022, 88, 104864. [Google Scholar] [CrossRef]
- Duan, Y.; Tarafdar, A.; Chaurasia, D.; Singh, A.; Bhargava, P.C.; Yang, J.; Li, Z.; Ni, X.; Tian, Y.; Li, H.; et al. Blueberry fruit valorization and valuable constituents: A review. Int. J. Food Microbiol. 2022, 381, 109890. [Google Scholar] [CrossRef]
- Yang, H.; Wu, Y.; Zhang, C.; Wu, W.; Lyu, L.; Li, W. Growth and physiological characteristics of four blueberry cultivars under different high soil pH treatments. Environ. Exp. Bot. 2022, 197, 104842. [Google Scholar] [CrossRef]
- Sargent, S.A.; Takeda, F.; Williamson, J.G.; Berry, A.D. Harvest of Southern Highbush Blueberry with a Modified, Over-the-Row Mechanical Harvester: Use of Soft-Catch Surfaces to Minimize Impact Bruising. Agronomy 2021, 11, 1412. [Google Scholar] [CrossRef]
- Brondino, L.; Borra, D.; Giuggioli, N.R.; Massaglia, S. Mechanized Blueberry Harvesting: Preliminary Results in the Italian Context. Agriculture 2021, 11, 1197. [Google Scholar] [CrossRef]
- Wang, H.; Guo, Y.; Bao, Y.; Gen, L. Mechanism analysis and simulation of blueberry harvest by vibration mode. Trans. Chin. Soc. Agric. Eng. (Trans. CSAE) 2013, 29, 40–46. [Google Scholar] [CrossRef]
- Xu, G.; An, Q.; Zhao, L.; Liu, G.; Lou, X.; Wang, H. A new blueberry cultivar‘Morning Snow’suitable for cluster harvesting. Acta Hortic. Sin. 2021, 48, 2795–2796. [Google Scholar] [CrossRef]
- Xu, G.; Lei, L.; An, Q.; Luo, L.; Wang, H. Utilization and development trend analysis of Vaccinium of America in blueberry breeding. J. Fruit Sci. 2021, 38, 1173–1189, (In Chinese with English Abstract). [Google Scholar]
- Brightwell, W.T. A Comparison of the Ethel and Walker Varieties as Parents in Blueberry Breeding; The Ohio State University: Columbus, OH, USA, 1956. [Google Scholar]
- Ni, X.; Li, C.; Jiang, H.; Takeda, F. Deep learning image segmentation and extraction of blueberry fruit traits associated with harvestability and yield. Hortic. Res. 2020, 7, 110. [Google Scholar] [CrossRef]
- Shi, J.; Xiao, Y.; Jia, C.; Zhang, H.; Gan, H.; Li, X.; Yang, M.; Yin, Y.; Zhang, G.; Hao, J.; et al. Physiological and biochemical changes during fruit maturation and ripening in highbush blueberry (Vaccinium corymbosum L.). Food Chem. 2023, 410, 135299. [Google Scholar] [CrossRef] [PubMed]
- Lobos, T.E.; Retamales, J.B.; Hanson, E.J. Early preharvest calcium sprays improve postharvest fruit quality in “Liberty” highbush blueberries. Sci. Hortic. 2021, 277, 109790. [Google Scholar] [CrossRef]
- Ma, H.; Zhang, K.; Jin, X.; Ji, J.; Zhu, X. Identification of Blueberry Fruit Maturity Using Hyperspectral Images Technology. J. Agric. Sci. Technol. 2020, 22, 80–90. [Google Scholar] [CrossRef]
- Wu, Z.; Jiang, X. Extraction of Pine Wilt Disease Regions Using UAV RGB Imagery and Improved Mask R-CNN Models Fused with ConvNeXt. Forests 2023, 14, 1672. [Google Scholar] [CrossRef]
- Dong, S.; Wang, P.; Abbas, K. A survey on deep learning and its applications. Comput. Sci. Rev. 2021, 40, 100379. [Google Scholar] [CrossRef]
- Yazdinejad, A.; Dehghantanha, A.; Parizi, R.M.; Epiphaniou, G. An optimized fuzzy deep learning model for data classification based on NSGA-II. Neurocomputing 2023, 522, 116–128. [Google Scholar] [CrossRef]
- Wang, L.; Qin, M.; Lei, J.; Wang, X.; Tan, K. Blueberry maturity recognition method based on improved YOLOv4-Tiny. Trans. Chin. Soc. Agric. Eng. (Trans. CSAE) 2021, 37, 170–178. [Google Scholar] [CrossRef]
- Gonzalez, S.; Arellano, C.; Tapia, J.E. Deepblueberry: Quantification of Blueberries in the Wild Using Instance Segmentation. IEEE Access 2019, 7, 105776–105788. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Yazdinejad, A.; Dehghantanha, A.; Srivastava, G.; Karimipour, H.; Parizi, R.M. Hybrid Privacy Preserving Federated Learning Against Irregular Users in Next-Generation Internet of Things. J. Syst. Archit. 2024, 148, 103088. [Google Scholar] [CrossRef]
- Yazdinejad, A.; Dehghantanha, A.; Srivastava, G. AP2FL: Auditable Privacy-Preserving Federated Learning Framework for Electronics in Healthcare. IEEE Trans. Consum. Electron. 2024, 70, 2527–2535. [Google Scholar] [CrossRef]
- Namakshenas, D.; Yazdinejad, A.; Dehghantanha, A.; Srivastava, G. Federated Quantum-Based Privacy-Preserving Threat Detection Model for Consumer Internet of Things. IEEE Trans. Consum. Electron. 2024, 1. [Google Scholar] [CrossRef]
- Zhang, J.; Min, A.; Steffenson, B.J.; Su, W.; Hirsch, C.D.; Anderson, J.; Wei, J.; Ma, Q.; Yang, C. Wheat-Net: An Automatic Dense Wheat Spike Segmentation Method Based on an Optimized Hybrid Task Cascade Model. Front. Plant Sci. 2022, 13, 834938. [Google Scholar] [CrossRef]
- Deng, R.; Zhou, M.; Huang, Y.; Tu, W. Hybrid Task Cascade-Based Building Extraction Method in Remote Sensing Imagery. Remote Sens. 2023, 15, 4907. [Google Scholar] [CrossRef]
- Chen, K.; Pang, J.; Wang, J.; Xiong, Y.; Li, X.; Sun, S.; Feng, W.; Liu, Z.; Shi, J.; Ouyang, W.; et al. Hybrid Task Cascade for Instance Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 4974–4983. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 11976–11986. [Google Scholar]
- Russell, B.C.; Torralba, A.; Murphy, K.P.; Freeman, W.T. LabelMe: A Database and Web-Based Tool for Image Annotation. Int. J. Comput. Vis. 2007, 77, 157–173. [Google Scholar] [CrossRef]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common Objects in Context. In Computer Vision—ECCV 2014; Springer: Cham, Switzerland, 2014; Volume 8693, pp. 740–755. [Google Scholar] [CrossRef]
- Bello, I.; Fedus, W.; Du, X.; Cubuk, E.D.; Srinivas, A.; Lin, T.-Y.; Shlens, J.; Zoph, B. Revisiting resnets: Improved training and scaling strategies. Adv. Neural Inf. Process. Syst. 2021, 34, 22614–22627. [Google Scholar]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-end object detection with transformers. In Computer Vision—ECCV 2020; Springer International Publishing: Cham, Switzerland, 2020; pp. 213–229. [Google Scholar]
- Cai, Z.; Vasconcelos, N. Cascade R-CNN: High Quality Object Detection and Instance Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 43, 1483–1498. [Google Scholar] [CrossRef]
- Bolya, D.; Zhou, C.; Xiao, F.; Lee, Y.J. YOLACT: Real-Time Instance Segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 9157–9166. [Google Scholar]
- Wang, X.; Zhang, R.; Kong, T.; Li, L.; Shen, C. SOLOv2: Dynamic and Fast Instance Segmentation. Neural Inf. Process. Syst. 2020, 33, 17721–17732. [Google Scholar]
- Wang, X.; Kong, T.; Shen, C.; Jiang, Y.; Li, L. SOLO: Segmenting Objects by Locations. In Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, August 23–28, 2020, Proceedings, Part XVIII 16; Springer International Publishing: Cham, Switzerland, 2020; pp. 649–665. [Google Scholar] [CrossRef]
- Tian, Z.; Shen, C.; Chen, H. Conditional Convolutions for Instance Segmentation. In Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, August 23–28, 2020, Proceedings, Part I 16; Springer International Publishing: Cham, Switzerland, 2020; pp. 282–298. [Google Scholar] [CrossRef]
- Fang, Y.; Yang, S.; Wang, X.; Li, Y.; Fang, C.; Shan, Y.; Feng, B.; Liu, W. Instances As Queries. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 6910–6919. [Google Scholar]
- Cheng, B.; Misra, I.; Schwing, A.G.; Kirillov, A.; Girdhar, R. Masked-Attention Mask Transformer for Universal Image Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 1290–1299. [Google Scholar]





| Scales |
|---|
| (480, 1333), (512, 1333), (544, 1333), (576, 1333), (608, 1333), (640, 1333),(672, 1333), (704, 1333), (736, 1333), (768, 1333), (800, 1333) |
| Cultivar | Average Number of Fruits in Cluster | Average Cluster Maturity | Average Cluster Compactness |
|---|---|---|---|
| Berkeley | 9.828 | 0.696 | 0.548 |
| Duke | 9.655 | 0.407 | 0.567 |
| Bluecrop | 13.690 | 0.439 | 0.626 |
| Bluegold | 15.759 | 0.180 | 0.584 |
| ConvNeXt | Mulit-Scale | AP (IOU = 0.5) | AP (IOU = 0.5~0.95) | Params (M) | ||
|---|---|---|---|---|---|---|
| Bbox | Mask | Bbox | Mask | |||
| - | - | 0.948 | 0.949 | 0.842 | 0.845 | 96.148 |
| √ 1 | - | 0.96 | 0.961 | 0.846 | 0.849 | 80.855 |
| √ 1 | √ 1 | 0.974 | 0.975 | 0.877 | 0.855 | 80.855 |
| Type | F1-Score | AP (IOU = 0.5) | AP (IOU = 0.5~0.95) | ||
|---|---|---|---|---|---|
| Bbox | Mask | Bbox | Mask | ||
| Mask R-CNN | 0.889 | 0.940 | 0.937 | 0.785 | 0.790 |
| Cascade Mask R-CNN | 0.919 | 0.942 | 0.933 | 0.814 | 0.793 |
| Hybrid Task Cascade | 0.931 | 0.948 | 0.949 | 0.842 | 0.845 |
| Yolact | 0.837 | 0.869 | 0.850 | 0.295 | 0.497 |
| SOLOv2 | - | - | 0.907 | - | 0.700 |
| CondInst | 0.906 | 0.905 | 0.911 | 0.811 | 0.781 |
| QueryInst | 0.883 | 0.797 | 0.810 | 0.620 | 0.664 |
| Mask2Former | 0.915 | 0.902 | 0.914 | 0.791 | 0.836 |
| YOLO V8 | 0.898 | 0.911 | 0.911 | 0.789 | 0.752 |
| HPPEM | 0.938 | 0.974 | 0.975 | 0.877 | 0.855 |
| Cultivar | Linear Model | R2 | RMSE |
|---|---|---|---|
| Berkeley | y = 1.023 + 0.932x | 0.959 | 0.627 |
| Duke | y = 1.311 + 0.920x | 0.806 | 1.429 |
| Bluecrop | y = 1.406 + 0.838x | 0.798 | 1.759 |
| Bluegold | y = 1.412 + 0.885x | 0.895 | 1.313 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gai, R.; Gao, J.; Xu, G. HPPEM: A High-Precision Blueberry Cluster Phenotype Extraction Model Based on Hybrid Task Cascade. Agronomy 2024, 14, 1178. https://doi.org/10.3390/agronomy14061178
Gai R, Gao J, Xu G. HPPEM: A High-Precision Blueberry Cluster Phenotype Extraction Model Based on Hybrid Task Cascade. Agronomy. 2024; 14(6):1178. https://doi.org/10.3390/agronomy14061178
Chicago/Turabian StyleGai, Rongli, Jin Gao, and Guohui Xu. 2024. "HPPEM: A High-Precision Blueberry Cluster Phenotype Extraction Model Based on Hybrid Task Cascade" Agronomy 14, no. 6: 1178. https://doi.org/10.3390/agronomy14061178





