Qfhb.yzu.3B.1 and Qfhb.yzu.6B.3 Are Stable Quantitative Trait Loci for Wheat Resistance to Fusarium Head Blight with Diverse Genetic Backgrounds
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. FHB Inoculation and Phenotypic Evaluation
2.3. DNA Extraction and Genotyping
2.4. RNA Extraction and Genotyping
2.5. Statistical Analysis
2.6. GWAS Analysis
2.7. Linkage Disequilibrium and Haplotype Analysis
2.8. Development of Derived Cleaved Amplified Polymorphic Sequence (dCAPS) Markers and Validation
2.9. Experimental Design
3. Results
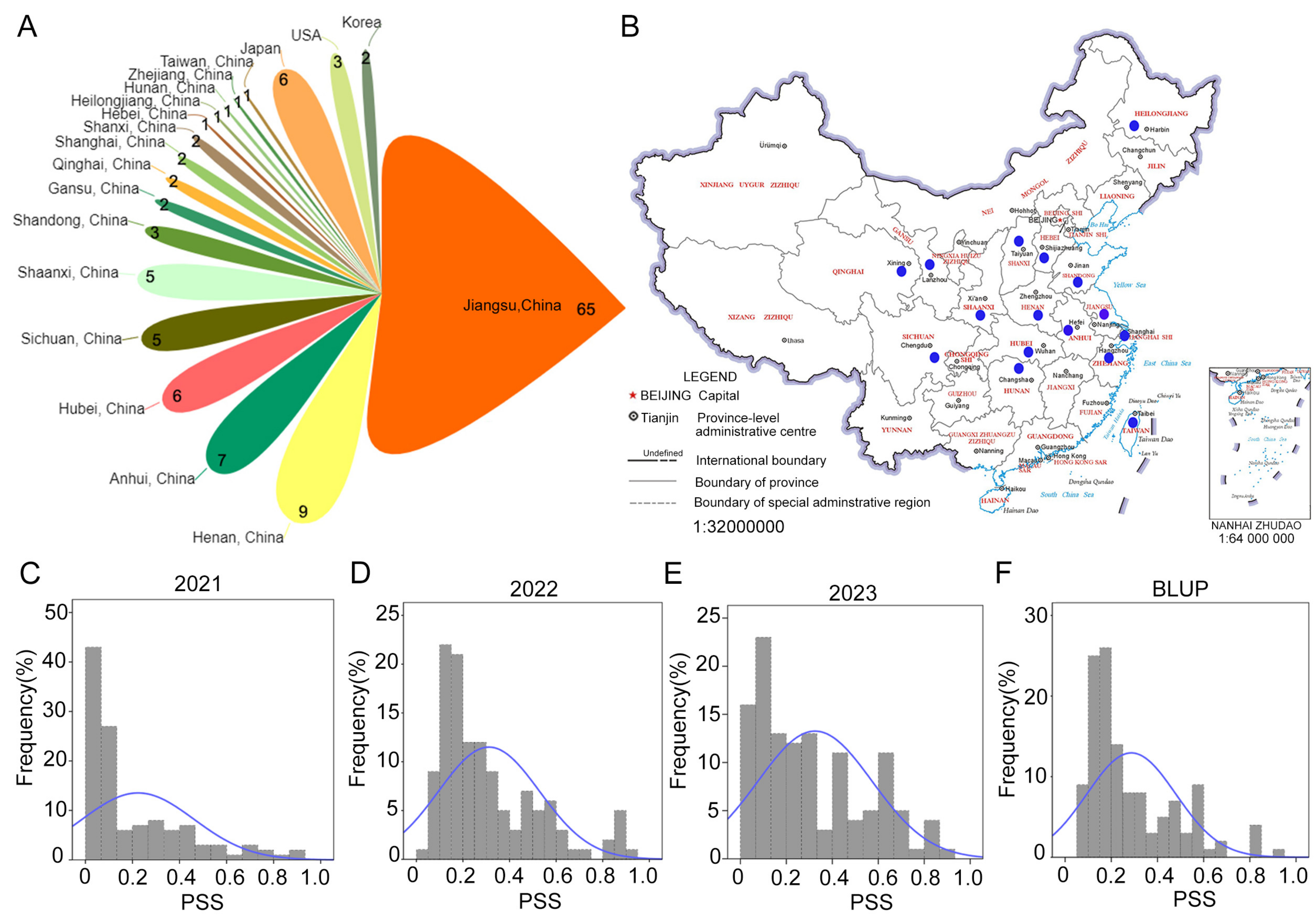
3.1. Phenotypic Variations of Two Independent Populations of Different Origins and Levels of Resistance to FHB
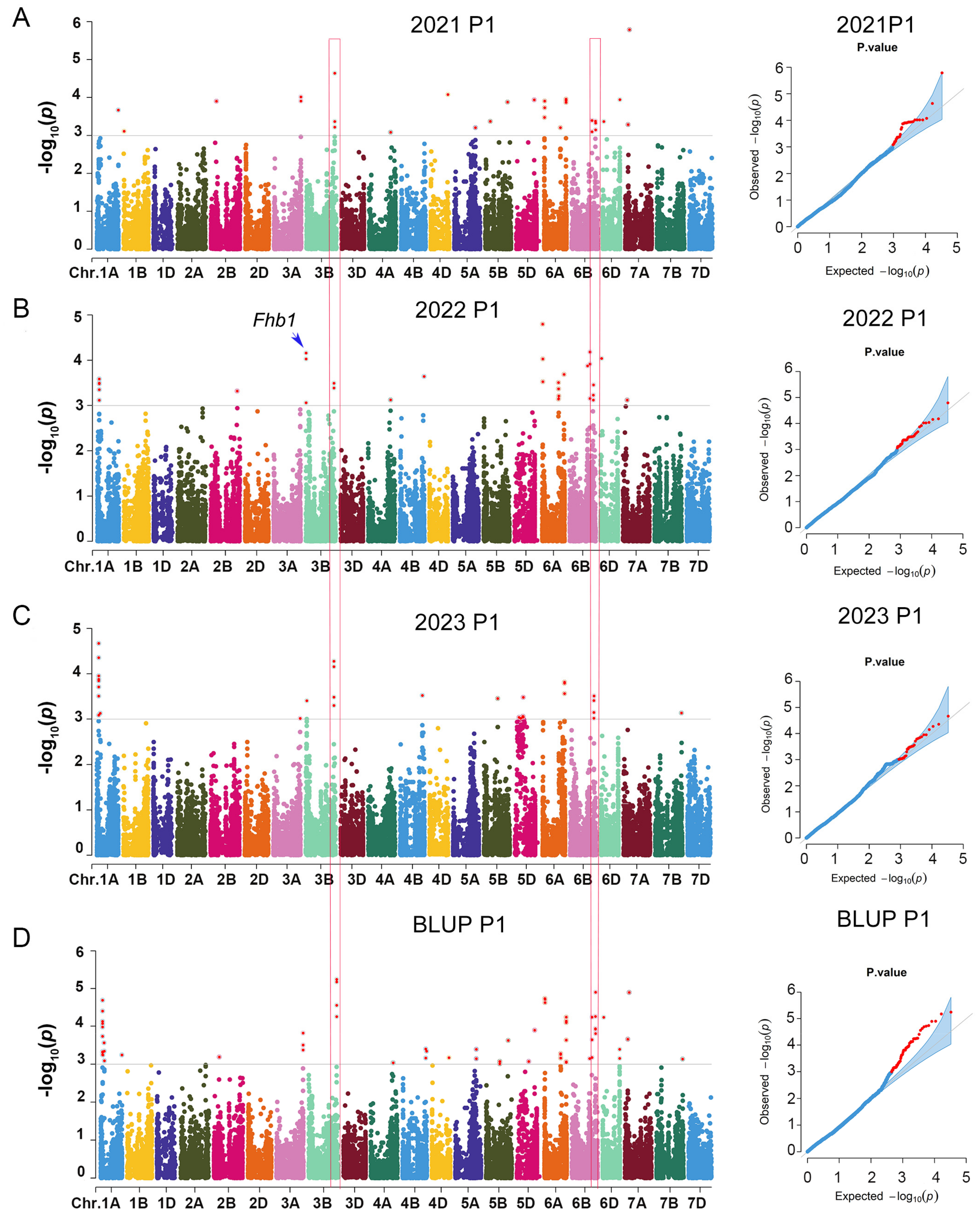
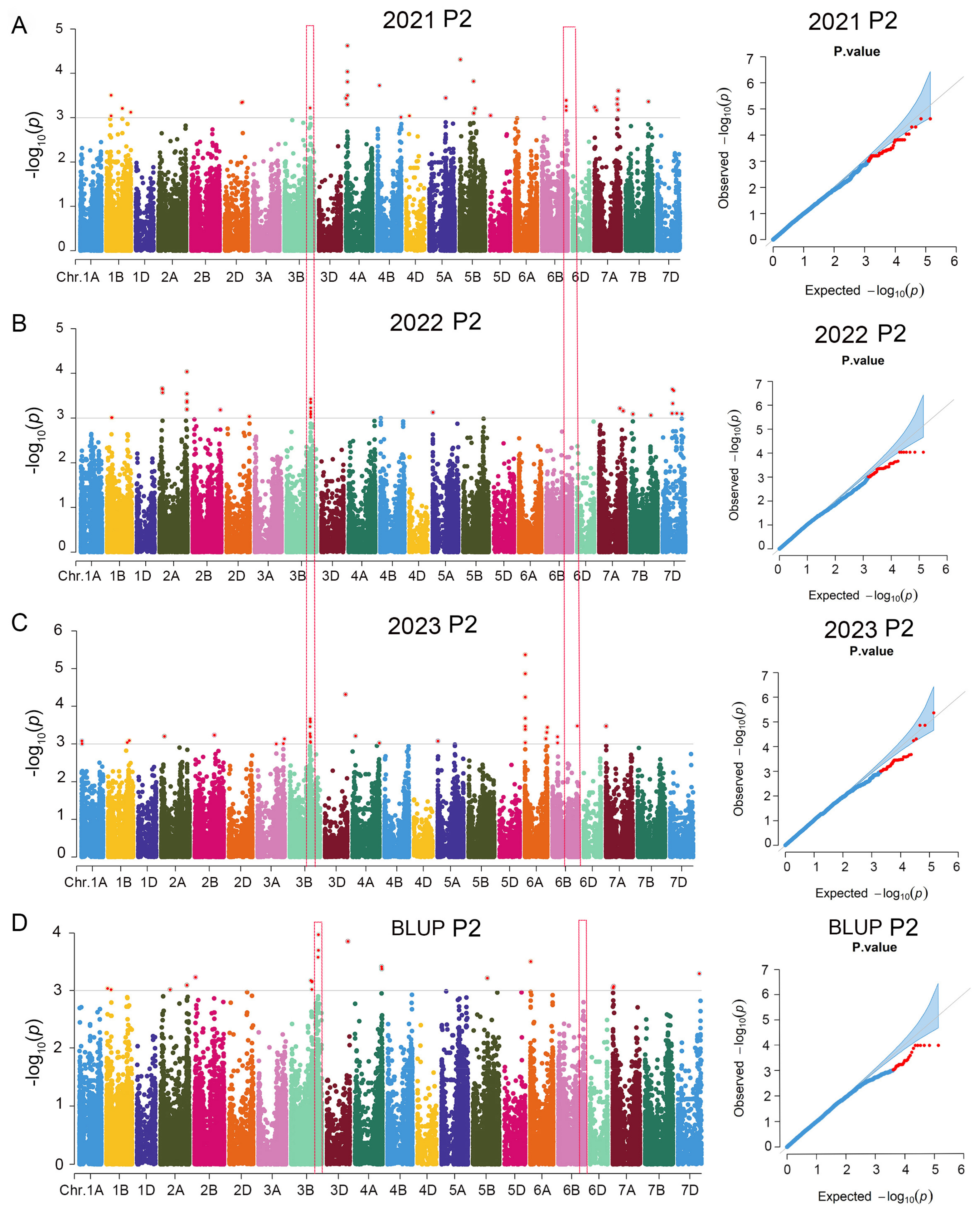
3.2. Significant SNPs for Marker–Trait Associations
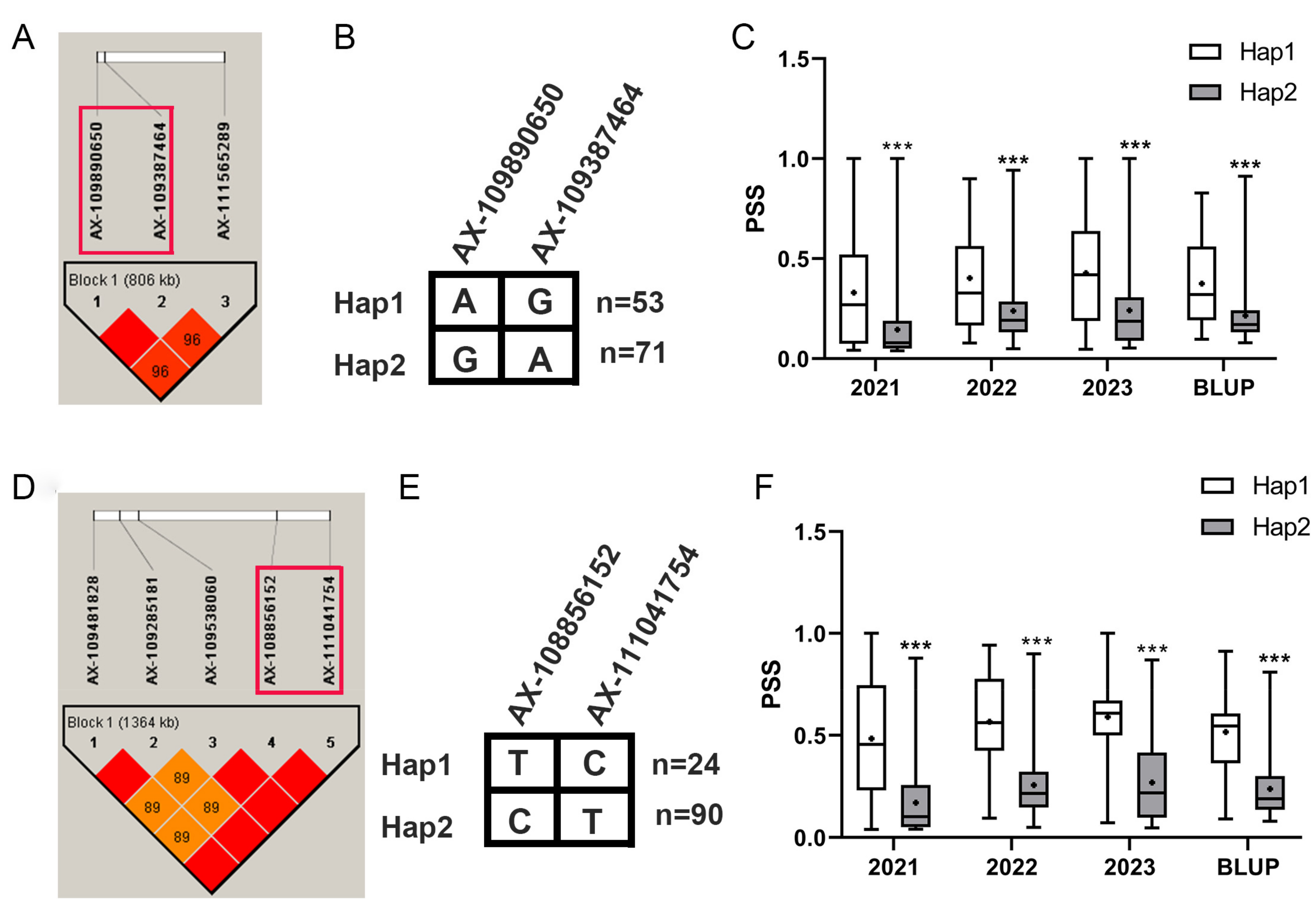
3.3. Favorable Alleles at Qfhb.yzu.3B.1 and Qfhb.yzu.6B.3 Significantly Decreased the Disease Severity
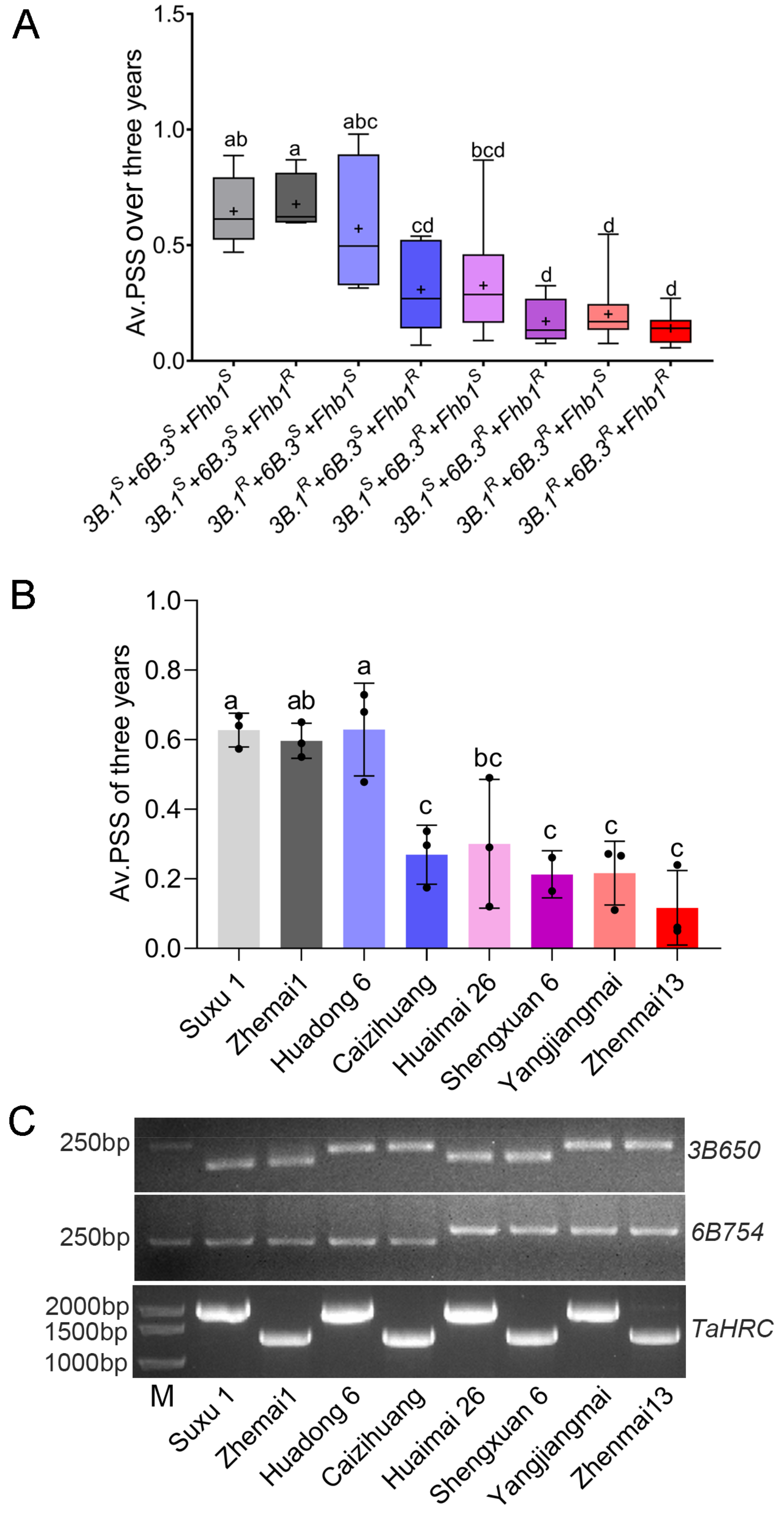
3.4. Pyramiding Favorable Alleles of Qfhb.yzu.3B.1, Qfhb.yzu.6B.3 and Fhb1 Significantly Improved FHB Resistance
4. Discussion
4.1. Qfhb.yzu.3B.1 and Qfhb.yzu.6B.3 Are Two Reliable QTL Associated with Resistance to FHB
4.2. Qfhb.yzu.3B.1 Represents a Newly Identified QTL
4.3. High Frequencies of Resistance Haplotypes at Qfhb.yzu.6B.3 in the Two Panels
4.4. Qfhb.yzu.3B.1 and Qfhb.yzu.6B.3 Were Additive to Fhb1 in Different Genetic Backgrounds
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Q.; Men, X.; Hui, C.; Ge, F.; Ouyang, F. Wheat Yield Losses from Pests and Pathogens in China. Agr. Ecosyst. Environ. 2022, 326, 107821. [Google Scholar] [CrossRef]
- Schroeder, H.W.; Christensen, J.J.; Christensen, J.D.; Platz-Christensen, J.; Schroeder, H.W.J.P. Factors Affecting Resistance of Wheat to Scab Caused by Gibberella zeae. Phytopathology 1963, 53, 831–838. [Google Scholar]
- Mesterhazy, A. Types and Components of Resistance to Fusarium Head Blight of Wheat. Plant Breed. 1995, 114, 377–386. [Google Scholar] [CrossRef]
- Mesterhazy, A. Updating the Breeding Philosophy of Wheat to Fusarium Head Blight (FHB): Resistance Components, QTL Identification, and Phenotyping—A Review. Plants 2020, 9, 1702. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; He, X.; Zhang, Y.; Li, L.; Sun, Z.; Bai, G.; Singh, P.K.; Li, T. Development of an Evaluation System for Fusarium Resistance in Wheat Grains and Its Application in Assessment of the Corresponding Effects of Fhb1. Plant Dis. 2020, 104, 2210–2216. [Google Scholar] [CrossRef] [PubMed]
- Windels, C.E. Economic and Social Impacts of Fusarium Head Blight: Changing Farms and Rural Communities in the Northern Great Plains. Phytopathology 2000, 90, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Shaner, G. Management and Resistance in Wheat and Barley to Fusarium Head Blight. Annu. Rev. Phytopathol. 2004, 42, 135–161. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Hua, C.; Li, L.; Sun, Z.; Yuan, M.; Bai, G.; Humphreys, G.; Li, T. Integration of Meta-QTL Discovery with Omics: Towards a Molecular Breeding Platform for Improving Wheat Resistance to Fusarium Head Blight. Crop J. 2020, 9, 739–749. [Google Scholar] [CrossRef]
- Cai, J.; Wang, S.; Su, Z.; Li, T.; Zhang, X.; Bai, G. Meta-analysis of QTL for Fusarium Head Blight Resistance in Chinese Wheat Landraces. Crop J. 2019, 7, 784–798. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, X.; Pumphrey, M.O.; Stack, R.W.; Gill, B.S.; Anderson, J.A. Complex Microcolinearity among Wheat, Rice, and Barley Revealed by Fine Mapping of the Genomic Region Harboring a Major QTL for Resistance to Fusarium Head Blight in Wheat. Funct. Integr. Genom. 2006, 6, 83–89. [Google Scholar] [CrossRef]
- Schweiger, W.; Steiner, B.; Vautrin, S.; Nussbaumer, T.; Siegwart, G.; Zamini, M.; Jungreithmeier, F.; Gratl, V.; Lemmens, M.; Mayer, K.F.; et al. Suppressed Recombination and Unique Candidate Genes in the Divergent Haplotype Encoding Fhb1, a Major Fusarium Head Blight Resistance Locus in Wheat. Theor. Appl. Genet. 2016, 129, 1607–1623. [Google Scholar] [CrossRef] [PubMed]
- Waldron, B.L.; Moreno-Sevilla, B.; Anderson, J.A.; Stack, R.W.; Frohberg, R.C. RFLP Mapping of QTL for Fusarium Head Blight Resistance in Wheat. Crop. Sci. 1999, 39, 805–811. [Google Scholar] [CrossRef]
- Su, Z.; Bernardo, A.; Tian, B.; Chen, H.; Wang, S.; Ma, H.; Cai, S.; Liu, D.; Zhang, D.; Li, T.; et al. A Deletion Mutation in TaHRC Confers Fhb1 Resistance to Fusarium Head Blight in Wheat. Nat. Genet. 2019, 51, 1099–1105. [Google Scholar] [CrossRef]
- Li, G.; Zhou, J.; Jia, H.; Gao, Z.; Fan, M.; Luo, Y.; Zhao, P.; Xue, S.; Li, N.; Yuan, Y.; et al. Mutation of a Histidine-Rich Calcium-Binding-Protein Gene in Wheat Confers Resistance to Fusarium Head Blight. Nat. Genet. 2019, 51, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Rawat, N.; Pumphrey, M.O.; Liu, S.; Zhang, X.; Tiwari, V.K.; Ando, K.; Trick, H.N.; Bockus, W.W.; Akhunov, E.; Anderson, J.A.; et al. Wheat Fhb1 Encodes a Chimeric Lectin with Agglutinin Domains and a Pore-Forming Toxin-Like Domain Conferring Resistance to Fusarium Head Blight. Nat. Genet. 2016, 48, 1576–1580. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Brown-Guedira, G.; Van Sanford, D.; Ohm, H.; Dong, Y.; McKendry, A.L. Novel QTL Associated with the Fusarium Head Blight Resistance in Truman Soft Red Winter Wheat. Euphytica 2015, 207, 571–592. [Google Scholar] [CrossRef]
- Zhang, X.; Pan, H.; Bai, G. Quantitative Trait Loci Responsible for Fusarium Head Blight Resistance in Chinese Landrace Baishanyuehuang. Theor. Appl. Genet. 2012, 125, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Kolb, F.L.; Yu, J.; Bai, G.; Boze, L.K.; Domier, L.L. Molecular Characterization of Fusarium Head Blight Resistance in Wangshuibai with Simple Sequence Repeat and Amplified Fragment Length Polymorphism Markers. Genome 2004, 47, 1137–1143. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Griffey, C.A.; Hall, M.D.; McKendry, A.L.; Chen, J.; Brooks, W.S.; Brown-Guedira, G.; Van Sanford, D.; Schmale, D.G. Molecular Characterization of Field Resistance to Fusarium Head Blight in Two US Soft Red Winter Wheat Cultivars. Theor. Appl. Genet. 2013, 126, 2485–2498. [Google Scholar] [CrossRef] [PubMed]
- Paillard, S.; Schnurbusch, T.; Tiwari, R.; Messmer, M.; Winzeler, M.; Keller, B.; Schachermayr, G. QTL Analysis of Resistance to Fusarium Head Blight in Swiss Winter Wheat (Triticum aestivum L.). Theor. Appl. Genet. 2004, 109, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Holzapfel, J.; Voss, H.H.; Miedaner, T.; Korzun, V.; Haberle, J.; Schweizer, G.; Mohler, V.; Zimmermann, G.; Hartl, L. Inheritance of Resistance to Fusarium Head Blight in Three European Winter Wheat Populations. Theor. Appl. Genet. 2008, 117, 1119–1128. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.B.; Bai, G.H.; Zhou, W.C.; Dong, Y.H.; Kolb, F.L. Quantitative Trait Loci for Fusarium Head Blight Resistance in a Recombinant Inbred Population of Wangshuibai/Wheaton. Phytopathology 2008, 98, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, H.; Huang, Y.; Su, Z.; Deng, Y.; Liu, H.; Mai, C.; Yu, G.; Li, H.; Yu, L.; et al. Effects of the Fhb1 Gene on Fusarium Head Blight Resistance and Agronomic Traits of Winter Wheat. Crop J. 2019, 7, 799–808. [Google Scholar] [CrossRef]
- Li, G.; Yuan, Y.; Zhou, J.; Cheng, R.; Chen, R.; Luo, X.; Shi, J.; Wang, H.; Xu, B.; Duan, Y.; et al. FHB Resistance Conferred by Fhb1 is under Inhibitory Regulation of Two Genetic Loci in Wheat (Triticum aestivum L.). Theor. Appl. Genet. 2023, 136, 134. [Google Scholar] [CrossRef] [PubMed]
- Korte, A.; Farlow, A. The Advantages and Limitations of Trait Analysis with GWAS: A Review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Gaire, R.; Brown-Guedira, G.; Dong, Y.; Ohm, H.; Mohammadi, M. Genome-Wide Association Studies for Fusarium Head Blight Resistance and Its Trade-Off with Grain Yield in Soft Red Winter Wheat. Plant Dis. 2021, 105, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Arruda, M.P.; Brown, P.; Brown-Guedira, G.; Krill, A.M.; Thurber, C.; Merrill, K.R.; Foresman, B.J.; Kolb, F.L. Genome-Wide Association Mapping of Fusarium Head Blight Resistance in Wheat Using Genotyping-by-Sequencing. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Tessmann, E.W.; Dong, Y.; Van Sanford, D.A. GWAS for Fusarium Head Blight Traits in a Soft Red Winter Wheat Mapping Panel. Crop Sci. 2019, 59, 1823–1837. [Google Scholar] [CrossRef]
- Bai, G.; Kolb, F.L.; Shaner, G.; Domier, L.L. Amplified Fragment Length Polymorphism Markers Linked to a Major Quantitative Trait Locus Controlling Scab Resistance in Wheat. Phytopathology 1999, 89, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA Spacer-length Polymorphisms in Barley: Mendelian Inheritance, Chromosomal Location, and Population Dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome Association and Prediction Integrated Tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, J.; Xue, X.; Li, C.; Yang, Z.; Li, T. CAPS/dCAPS Designer: A Web-Based High-Throughput dCAPS Marker Design Tool. Sci. China. Life. Sci. 2018, 61, 992–995. [Google Scholar] [CrossRef] [PubMed]
- Cardon, L.R.; Palmer, L.J. Population Stratification and Spurious Allelic Association. Lancet 2003, 361, 598–604. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Dreisigacker, S.; Singh, R.P.; Singh, P.K. Genetics for Low Correlation Between Fusarium Head Blight Disease and Deoxynivalenol (DON) Content in a Bread Wheat Mapping Population. Theor. Appl. Genet. 2019, 132, 2401–2411. [Google Scholar] [CrossRef]
- Ágnes, S.H.; Szabolcs, L.K.; Mónika, V.; László, P.; János, P.; Csaba, L.; Ákos, M. Differential Influence of QTL Linked to Fusarium Head Blight, Fusarium-Damaged Kernel, Deoxynivalenol Contents and Associated Morphological Traits in a Frontana-Derived Wheat Population. Euphytica 2014, 200, 9–26. [Google Scholar] [CrossRef]
- Basnet, B.R.; Glover, K.D.; Ibrahim, A.M.H.; Yen, Y.; Chao, S. A QTL on Chromosome 2DS of ‘Sumai 3’ Increases Susceptibility to Fusarium Head Blight in Wheat. Euphytica 2011, 186, 91–101. [Google Scholar] [CrossRef]
- Ghavami, F.; Elias, E.M.; Mamidi, S.; Ansari, O.; Sargolzaei, M.; Adhikari, T.; Mergoum, M.; Kianian, S.F. Mixed Model Association Mapping for Fusarium Head Blight Resistance in Tunisian-Derived Durum Wheat Populations. G3 2011, 1, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.J.; Sarti-Dvorjak, D.; Brown-Guedira, G.; Dong, Y.; Baik, B.K.; Van Sanford, D.A. Identifying Rare FHB-Resistant Segregants in Intransigent Backcross and F2 Winter Wheat Populations. Front. Microbiol. 2016, 7, 277. [Google Scholar] [CrossRef] [PubMed]
| QTL | Marker | Chr. | Pos. (Mb) | −LOG10(P) | Effect (%) | Meta-QTL | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2021 | 2022 | 2023 | BLUP | 2021 | 2022 | 2023 | BLUP | |||||
| Qfhb.yzu.1A.1 | AX-110971755 | 1A | 49.57 | 3.49 | 3.95 | 4.13 | 9.36 | 12.91 | 9.73 | No | ||
| AX-111073329 | 1A | 49.62 | 3.48 | 3.84 | 4.07 | 9.20 | 12.51 | 9.46 | ||||
| AX-108725829 | 1A | 49.90 | 3.49 | 3.95 | 4.13 | 9.36 | 12.91 | 9.73 | ||||
| AX-108835010 | 1A | 49.96 | 3.59 | 3.71 | 3.73 | 9.94 | 13.02 | 9.50 | ||||
| AX-111674822 | 1A | 50.29 | 3.35 | 3.88 | 3.98 | 9.41 | 13.16 | 9.78 | ||||
| AX-110464570 | 1A | 51.68 | 3.50 | 4.67 | 4.69 | 10.28 | 15.67 | 11.52 | ||||
| AX-110960234 | 1A | 51.96 | 3.12 | 4.36 | 4.40 | 9.74 | 15.10 | 11.28 | ||||
| AX-109433977 | 1A | 52.94 | 3.09 | 3.24 | 11.09 | 8.30 | ||||||
| Qfhb.yzu.1A.2 | AX-110670478 | 1A | 92.47 | 3.13 | 3.56 | 12.72 | 9.67 | No | ||||
| Qfhb.yzu.2B.1 | AX-109369175 | 2B | 144.59 | 3.90 | 3.19 | 16.77 | 10.24 | No | ||||
| Qfhb.yzu.3A.1 | AX-108757584 | 3A | 737.18 | 3.01 | 3.37 | 9.73 | 7.24 | No | ||||
| AX-108730958 | 3A | 738.80 | 4.02 | 3.82 | 8.29 | 6.02 | ||||||
| AX-109318717 | 3A | 738.82 | 4.02 | 3.82 | 8.29 | 6.02 | ||||||
| AX-110021843 | 3A | 738.86 | 3.91 | 3.50 | 8.16 | 5.73 | ||||||
| AX-110581293 | 3A | 738.90 | 4.02 | 3.82 | 8.29 | 6.02 | ||||||
| AX-111653891 | 3A | 738.94 | 4.02 | 3.82 | 8.29 | 6.02 | ||||||
| Qfhb.yzu.3B.1 | AX-111069466 | 3B | 779.97 | 3.30 | 4.25 | 7.89 | 6.57 | No | ||||
| AX-109890650 * | 3B | 780.00 | 3.22 | 3.49 | 4.28 | 5.18 | 7.53 | 6.19 | 8.95 | 7.25 | ||
| AX-109387464 * | 3B | 780.05 | 3.37 | 3.39 | 4.16 | 5.24 | 8.07 | 6.33 | 9.13 | 7.61 | ||
| AX-111565289 | 3B | 780.81 | 4.64 | 3.48 | 4.55 | 10.45 | 8.30 | 7.23 | ||||
| Qfhb.yzu.4A.1 | AX-111479307 | 4A | 621.78 | 3.13 | 3.03 | 6.48 | 5.97 | No | ||||
| Qfhb.yzu.4B.1 | AX-108781390 | 4B | 661.68 | 3.64 | 3.34 | 14.99 | 13.22 | No | ||||
| Qfhb.yzu.4D.1 | AX-111921300 | 4D | 469.76 | 4.08 | 3.17 | 13.80 | 8.75 | No | ||||
| Qfhb.yzu.5A.1 | AX-111535520 | 5A | 576.10 | 3.20 | 3.39 | 12.44 | 9.09 | No | ||||
| Qfhb.yzu.5B.1 | AX-110577474 | 5B | 379.72 | 3.46 | 3.00 | 13.79 | 9.42 | No | ||||
| Qfhb.yzu.5B.2 | AX-110077080 | 5B | 611.68 | 3.88 | 3.62 | 16.34 | 10.98 | Yes | ||||
| Qfhb.yzu.5D.1 | AX-110639062 | 5D | 495.06 | 3.94 | 3.89 | 32.85 | 23.05 | No | ||||
| AX-111481379 | 5D | 495.06 | 3.94 | 3.89 | 16.42 | 11.53 | ||||||
| Qfhb.yzu.6A.1 | AX-111507391 | 6A | 10.50 | 3.48 | 4.80 | 4.74 | 25.01 | 23.87 | 21.73 | Yes | ||
| AX-109333155 | 6A | 11.59 | 3.73 | 4.03 | 4.71 | 14.17 | 11.13 | 11.46 | ||||
| AX-110942399 | 6A | 11.87 | 3.91 | 3.53 | 4.63 | 14.45 | 10.26 | 11.32 | ||||
| Qfhb.yzu.6A.2 | AX-110985951 | 6A | 447.42 | 3.37 | 3.16 | 10.92 | 9.93 | Yes | ||||
| AX-110686572 | 6A | 447.56 | 3.51 | 3.28 | 11.06 | 10.02 | ||||||
| AX-109983752 | 6A | 447.60 | 3.37 | 3.16 | 10.92 | 9.93 | ||||||
| AX-108946683 | 6A | 447.62 | 3.20 | 3.26 | 17.34 | 12.16 | ||||||
| AX-111539462 | 6A | 447.81 | 3.37 | 3.16 | 10.92 | 9.93 | ||||||
| AX-111701725 | 6A | 448.19 | 3.37 | 3.16 | 10.92 | 9.93 | ||||||
| AX-108956212 | 6A | 448.66 | 3.37 | 3.16 | 10.92 | 9.93 | ||||||
| AX-111695448 | 6A | 453.12 | 3.37 | 3.16 | 10.92 | 9.93 | ||||||
| Qfhb.yzu.6A.3 | AX-110532405 | 6A | 599.83 | 3.96 | 3.69 | 4.24 | 21.97 | 15.29 | 15.80 | No | ||
| AX-108944873 | 6A | 601.26 | 3.92 | 3.82 | 4.14 | 15.40 | 15.13 | 11.17 | ||||
| AX-89773191 | 6A | 601.26 | 3.87 | 3.79 | 4.10 | 15.17 | 14.86 | 11.00 | ||||
| AX-111463277 | 6A | 601.48 | 3.56 | 3.63 | 14.37 | 10.25 | ||||||
| Qfhb.yzu.6B.1 | AX-108826673 | 6B | 491.96 | 3.87 | 3.15 | 10.63 | 8.80 | No | ||||
| Qfhb.yzu.6B.2 | AX-110480933 | 6B | 559.36 | 3.10 | 3.92 | 3.64 | 9.28 | 8.49 | 7.52 | Yes | ||
| AX-109393129 | 6B | 559.46 | 3.15 | 3.17 | 8.33 | 7.90 | ||||||
| AX-109816926 | 6B | 560.98 | 3.39 | 4.18 | 4.24 | 11.19 | 9.43 | 8.93 | ||||
| Qfhb.yzu.6B.3 | AX-109481828 | 6B | 654.80 | 3.12 | 3.02 | 3.94 | 19.21 | 24.36 | 20.52 | Yes | ||
| AX-109285181 | 6B | 654.95 | 3.12 | 3.02 | 3.94 | 19.21 | 24.36 | 20.52 | ||||
| AX-109538060 | 6B | 655.06 | 3.37 | 3.15 | 3.92 | 12.55 | 12.01 | 9.79 | ||||
| AX-108856152 | 6B | 655.86 | 3.23 | 3.41 | 4.26 | 10.22 | 13.66 | 11.22 | ||||
| AX-111041754 * | 6B | 656.16 | 3.15 | 3.45 | 3.51 | 4.90 | 13.91 | 13.09 | 16.97 | 14.72 | ||
| AX-108834761 | 6B | 656.19 | 3.33 | 3.80 | 18.68 | 14.05 | ||||||
| Qfhb.yzu.6D.1 | AX-111379959 | 6D | 25.03 | 3.37 | 4.04 | 4.24 | 12.49 | 10.24 | 9.92 | No | ||
| Qfhb.yzu.6D.2 | AX-109985214 | 6D | 465.51 | 3.94 | 3.39 | 17.06 | 10.60 | No | ||||
| Qfhb.yzu.7A.1 | AX-110505644 | 7A | 79.74 | 3.29 | 3.66 | 15.05 | 11.29 | No | ||||
| Qfhb.yzu.7A.2 | AX-109449565 | 7A | 117.27 | 5.79 | 3.12 | 4.90 | 19.33 | 10.11 | 12.32 | Yes | ||
| Qfhb.yzu.7B.1 | AX-109915196 | 7B | 725.37 | 3.13 | 3.13 | 11.43 | 8.23 | Yes | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Z.; Ye, H.; Chen, X.; Cheng, J.; Zhu, F.; Yang, D.; Hu, S.; Li, L.; Li, T. Qfhb.yzu.3B.1 and Qfhb.yzu.6B.3 Are Stable Quantitative Trait Loci for Wheat Resistance to Fusarium Head Blight with Diverse Genetic Backgrounds. Agronomy 2024, 14, 1230. https://doi.org/10.3390/agronomy14061230
Sun Z, Ye H, Chen X, Cheng J, Zhu F, Yang D, Hu S, Li L, Li T. Qfhb.yzu.3B.1 and Qfhb.yzu.6B.3 Are Stable Quantitative Trait Loci for Wheat Resistance to Fusarium Head Blight with Diverse Genetic Backgrounds. Agronomy. 2024; 14(6):1230. https://doi.org/10.3390/agronomy14061230
Chicago/Turabian StyleSun, Zhengxi, Hongyan Ye, Xiang Chen, Jiale Cheng, Fanfan Zhu, Dapeng Yang, Sijia Hu, Lei Li, and Tao Li. 2024. "Qfhb.yzu.3B.1 and Qfhb.yzu.6B.3 Are Stable Quantitative Trait Loci for Wheat Resistance to Fusarium Head Blight with Diverse Genetic Backgrounds" Agronomy 14, no. 6: 1230. https://doi.org/10.3390/agronomy14061230
APA StyleSun, Z., Ye, H., Chen, X., Cheng, J., Zhu, F., Yang, D., Hu, S., Li, L., & Li, T. (2024). Qfhb.yzu.3B.1 and Qfhb.yzu.6B.3 Are Stable Quantitative Trait Loci for Wheat Resistance to Fusarium Head Blight with Diverse Genetic Backgrounds. Agronomy, 14(6), 1230. https://doi.org/10.3390/agronomy14061230





