Enrichment of Grain Anthocyanin Content through Marker-Assisted Breeding for Ant1, Ant2 or HvMyc2 Genes in Barley (Hordeum vulgare L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and the Breeding Strategy
2.2. Molecular Markers
2.3. GBS
2.4. Total Phenolic and Anthocyanin Contents
2.5. AOA
2.6. Phenotyping
2.7. Statistical Analysis
3. Results
3.1. MAS of the Anthocyanin-Rich Barley NILs
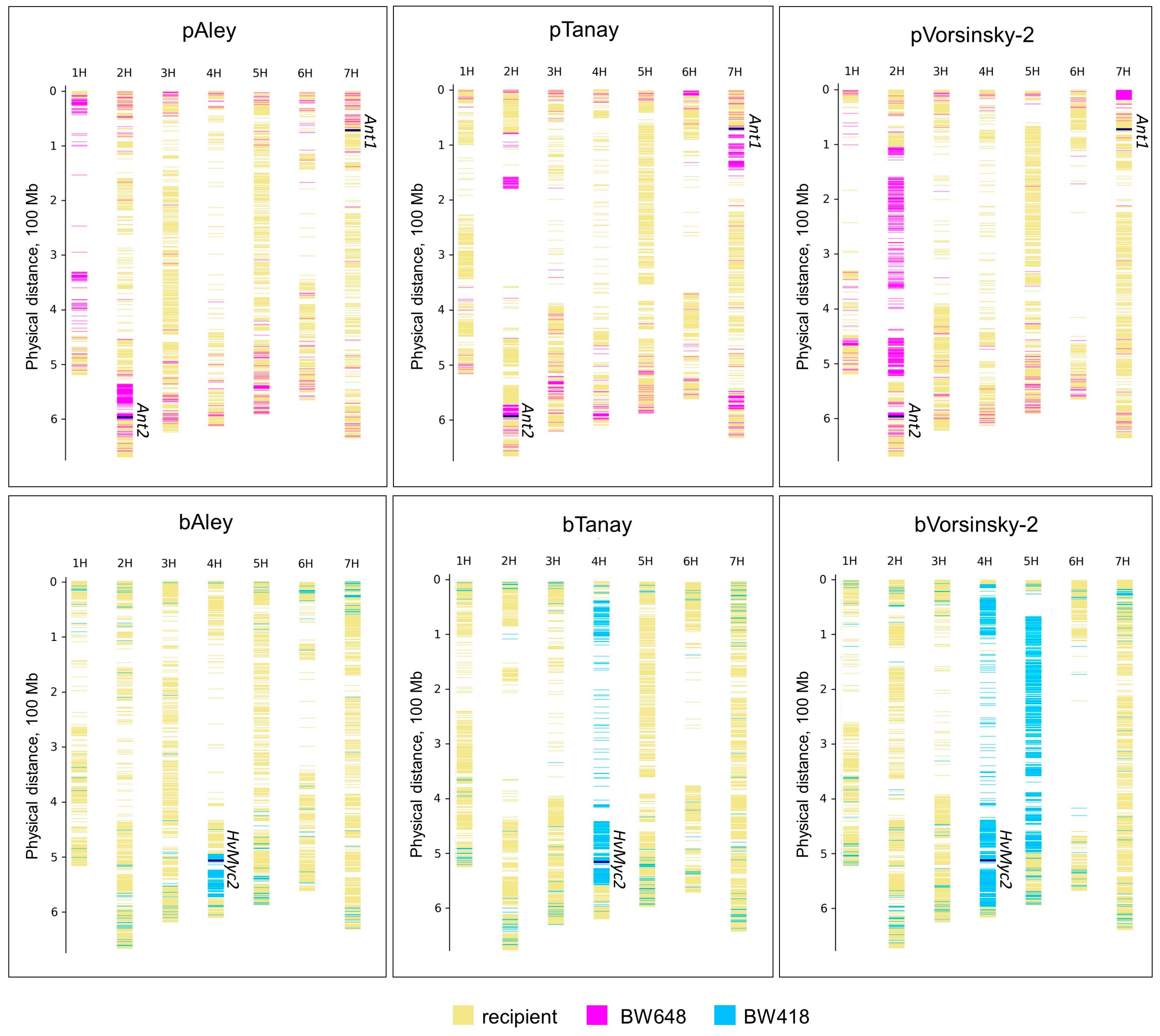
3.2. GBS of the NILs with Different Color of Grain
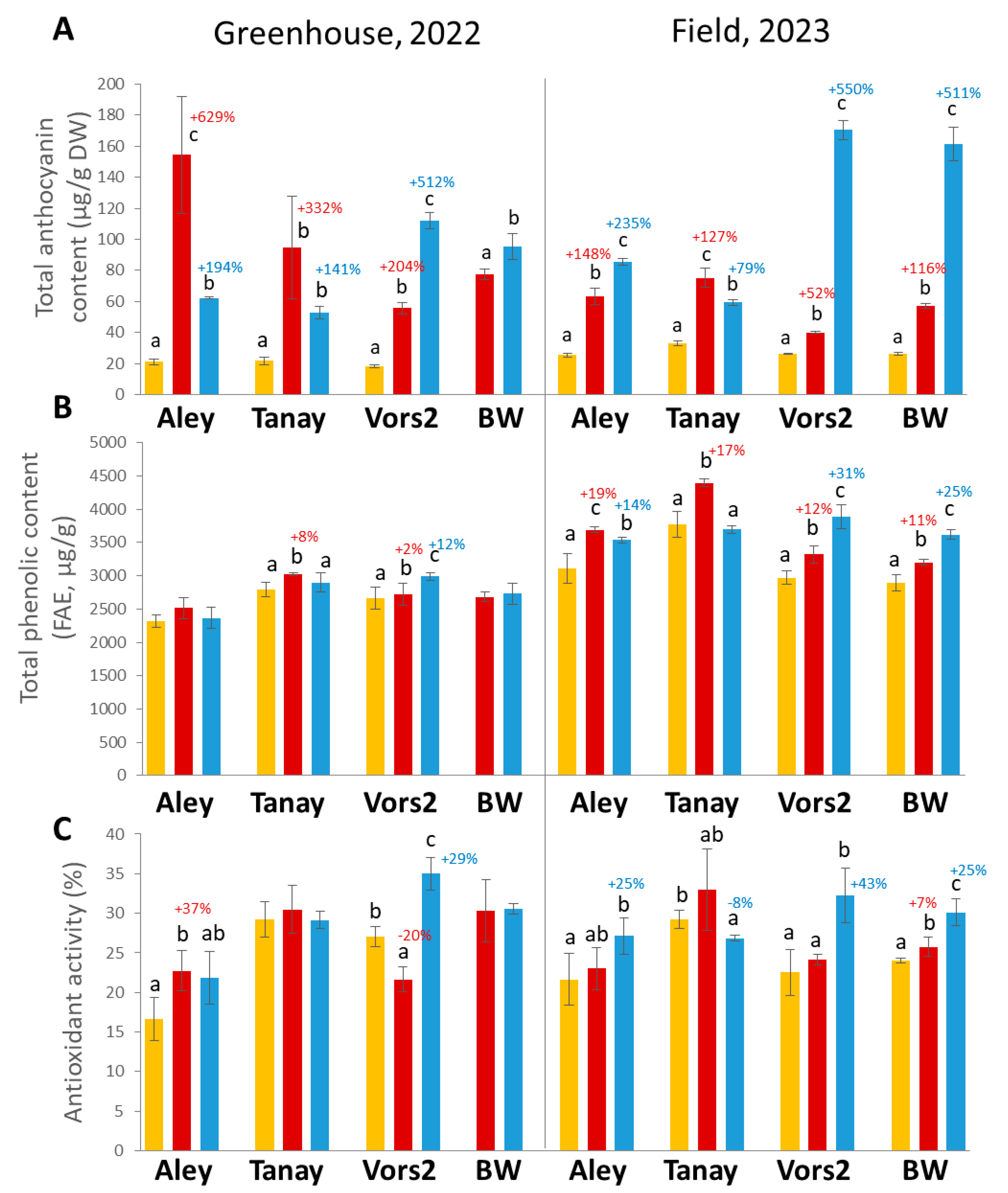
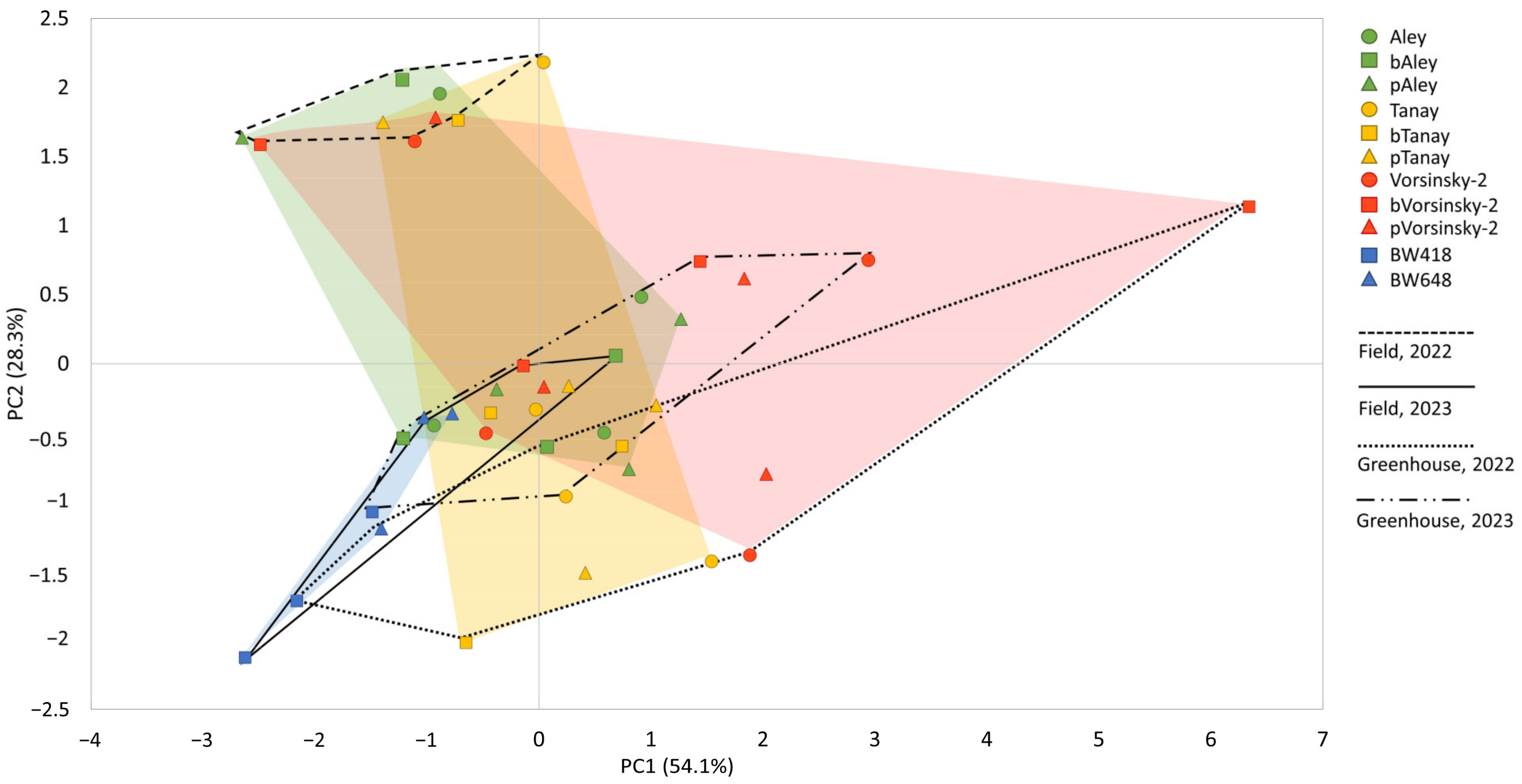
3.3. Comparative Study of Biochemical Characteristics of the Grain of the NILs
3.3.1. TAC
3.3.2. TPC
3.3.3. AOA
3.4. Comparative Study of Productivity of the NILs
4. Discussion
4.1. Marker-Assisted Breeding of Anthocyanin-Rich Barley NILs
4.2. Comparative Study of Biochemical Parameters
4.3. Comparative Study of Yield-Related Parameters
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Geng, L.; Li, M.; Zhang, G.; Ye, L. Barley: A Potential Cereal for Producing Healthy and Functional Foods. Food Qual. Saf. 2022, 6, fyac012. [Google Scholar] [CrossRef]
- Sullivan, P.; Arendt, E.; Gallagher, E. The Increasing Use of Barley and Barley By-Products in the Production of Healthier Baked Goods. Trends Food Sci. Technol. 2013, 29, 124–134. [Google Scholar] [CrossRef]
- Baik, B.-K.; Ullrich, S.E. Barley for Food: Characteristics, Improvement, and Renewed Interest. J. Cereal Sci. 2008, 48, 233–242. [Google Scholar] [CrossRef]
- Iannucci, A.; Suriano, S.; Codianni, P. Genetic Diversity for Agronomic Traits and Phytochemical Compounds in Coloured Naked Barley Lines. Plants 2021, 10, 1575. [Google Scholar] [CrossRef]
- Abdel-Aal, E.-S.M.; Young, J.C.; Rabalski, I. Anthocyanin Composition in Black, Blue, Pink, Purple, and Red Cereal Grains. J. Agric. Food Chem. 2006, 54, 4696–4704. [Google Scholar] [CrossRef]
- Kim, M.-J.; Hyun, J.-N.; Kim, J.-A.; Park, J.-C.; Kim, M.-Y.; Kim, J.-G.; Lee, S.-J.; Chun, S.-C.; Chung, I.-M. Relationship between Phenolic Compounds, Anthocyanins Content and Antioxidant Activity in Colored Barley Germplasm. J. Agric. Food Chem. 2007, 55, 4802–4809. [Google Scholar] [CrossRef]
- Li, D.; Wang, P.; Luo, Y.; Zhao, M.; Chen, F. Health Benefits of Anthocyanins and Molecular Mechanisms: Update from Recent Decade. Crit. Rev. Food Sci. Nutr. 2017, 57, 1729–1741. [Google Scholar] [CrossRef]
- Loskutov, I.G.; Khlestkina, E.K. Wheat, Barley, and Oat Breeding for Health Benefit Components in Grain. Plants 2021, 10, 86. [Google Scholar] [CrossRef]
- Francavilla, A.; Joye, I.J. Anthocyanins in Whole Grain Cereals and Their Potential Effect on Health. Nutrients 2020, 12, 2922. [Google Scholar] [CrossRef]
- Suriano, S.; Savino, M.; Codianni, P.; Iannucci, A.; Caternolo, G.; Russo, M.; Pecchioni, N.; Troccoli, A. Anthocyanin Profile and Antioxidant Capacity in Coloured Barley. Int. J. Food Sci. Tech. 2019, 54, 2478–2486. [Google Scholar] [CrossRef]
- Gryaznov, A.A.; Letjago, J.A.; Belkina, R.I.; Ponomareva, E.I. Obtaining of bread with the use of mixtures of wheat flour of the highest grade and wholemeal from the grain pigmented barley varieties Granal 32. Vestn. VGUIT Proc. VSUET 2019, 81, 196–200. [Google Scholar] [CrossRef]
- King, C. New Possibilities with Purple Wheat; Top Crop Manager: Saskatoon, ON, Canada, 2017. [Google Scholar]
- Khlestkina, E. Molecular Markers in Genetic Studies and Breeding. Russ. J. Genet. Appl. Res. 2014, 4, 236–244. [Google Scholar] [CrossRef]
- Shoeva, O.Y.; Strygina, K.V.; Khlestkina, E.K. Genes determining the synthesis of flavonoid and melanin pigments in barley. Vestn. VOGiS 2018, 22, 333–342. [Google Scholar] [CrossRef]
- Shoeva, O.Y.; Kukoeva, T.V.; Khlestkina, E.K. Set of DNA Markers for the Selection of Barley with a High Content of Anthocyanins in the Pericarp of the Caryopsis. Russian Patent RU 2 774 444 C1, 21 June 2022. [Google Scholar]
- Shoeva, O.Y.; Kukoeva, T.V. Relationship between the anthocyanin content values in the leaf sheath base of barley cultivars and in the grain of the hybrids derived from them. Proc. Appl. Bot. Genet. Breed. 2022, 183, 152–162. [Google Scholar] [CrossRef]
- Strygina, K.V.; Börner, A.; Khlestkina, E.K. Identification and Characterization of Regulatory Network Components for Anthocyanin Synthesis in Barley Aleurone. BMC Plant Biol. 2017, 17, 184. [Google Scholar] [CrossRef]
- Druka, A.; Franckowiak, J.; Lundqvist, U.; Bonar, N.; Alexander, J.; Houston, K.; Radovic, S.; Shahinnia, F.; Vendramin, V.; Morgante, M.; et al. Genetic Dissection of Barley Morphology and Development. Plant Physiol. 2011, 155, 617–627. [Google Scholar] [CrossRef]
- Plaschke, J.; Ganal, M.W.; Röder, M.S. Detection of Genetic Diversity in Closely Related Bread Wheat Using Microsatellite Markers. Theor. Appl. Genet. 1995, 91, 1001–1007. [Google Scholar] [CrossRef]
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannink, J.-L. Development of High-Density Genetic Maps for Barley and Wheat Using a Novel Two-Enzyme Genotyping-by-Sequencing Approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef]
- Pronozin, A.Y.; Salina, E.A.; Afonnikov, D.A. GBS-DP: A Bioinformatics Pipeline for Processing Data Coming from Genotyping by Sequencing. Vavilovskii Zhurnal Genet. Sel. Vavilov J. Genet. Breed. 2023, 27, 737–745. [Google Scholar] [CrossRef]
- Sharma, P.; Gujral, H.S. Antioxidant and Polyphenol Oxidase Activity of Germinated Barley and Its Milling Fractions. Food Chem. 2010, 120, 673–678. [Google Scholar] [CrossRef]
- Abdel-Aal, E.-S.M.; Hucl, P. A Rapid Method for Quantifying Total Anthocyanins in Blue Aleurone and Purple Pericarp Wheats. Cereal Chem. J. 1999, 76, 350–354. [Google Scholar] [CrossRef]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Jayakodi, M.; Padmarasu, S.; Haberer, G.; Bonthala, V.S.; Gundlach, H.; Monat, C.; Lux, T.; Kamal, N.; Lang, D.; Himmelbach, A.; et al. The Barley Pan-Genome Reveals the Hidden Legacy of Mutation Breeding. Nature 2020, 588, 284–289. [Google Scholar] [CrossRef]
- Hossain, F.; Jaiswal, S.K.; Muthusamy, V.; Zunjare, R.U.; Mishra, S.J.; Chand, G.; Bhatt, V.; Bhat, J.S.; Das, A.K.; Chauhan, H.S.; et al. Enhancement of Nutritional Quality in Maize Kernel through Marker-Assisted Breeding for Vte4, crtRB1, and Opaque2 Genes. J. Appl. Genet. 2023, 64, 431–443. [Google Scholar] [CrossRef]
- Zhang, T.; Ma, J.; Wu, X.; Hao, Z.; Dun, C.; Chen, C. Qualitative and Semi-Quantitative Assessment of Anthocyanins in Tibetan Hulless Barley from Different Geographical Locations by UPLC-QTOF-MS and Their Antioxidant Capacities. Open Chem. 2021, 19, 1–8. [Google Scholar] [CrossRef]
- Diczházi, I.; Kursinszki, L. Anthocyanin Content and Composition in Winter Blue Barley Cultivars and Lines. Cereal Chem. 2014, 91, 195–200. [Google Scholar] [CrossRef]
- Jin, H.-M.; Dang, B.; Zhang, W.-G.; Zheng, W.-C.; Yang, X.-J. Polyphenol and Anthocyanin Composition and Activity of Highland Barley with Different Colors. Molecules 2022, 27, 3411. [Google Scholar] [CrossRef]
- Shoeva, O.Y.; Mursalimov, S.R.; Gracheva, N.V.; Glagoleva, A.Y.; Börner, A.; Khlestkina, E.K. Melanin Formation in Barley Grain Occurs within Plastids of Pericarp and Husk Cells. Sci. Rep. 2020, 10, 179. [Google Scholar] [CrossRef]
- Lee, C.; Han, D.; Kim, B.; Baek, N.; Baik, B.-K. Antioxidant and Anti-Hypertensive Activity of Anthocyanin-Rich Extracts from Hulless Pigmented Barley Cultivars. Int. J. Food Sci. Technol. 2013, 48, 984–991. [Google Scholar] [CrossRef]
- Vasko, N.; Mykhailenko, E. Anthocyanins in Naked Pigmented Barley Grain as a Source of Antioxidant Activity. Food Sci. Nutr. Tech. 2023, 8, 000301. [Google Scholar] [CrossRef]
- Zhang, Y.; Yin, L.; Huang, L.; Tekliye, M.; Xia, X.; Li, J.; Dong, M. Composition, Antioxidant Activity, and Neuroprotective Effects of Anthocyanin-Rich Extract from Purple Highland Barley Bran and Its Promotion on Autophagy. Food Chem. 2021, 339, 127849. [Google Scholar] [CrossRef]
- Sumina, A.V.; Polonskiy, V.I.; Shaldaeva, T.M.; Shulbaeva, M.T. The content of antioxidants in the products of Khakass national cuisine based on barley grain. Vestn. Krasn. SAU 2019, 12, 125–131. [Google Scholar] [CrossRef]
- Gordeeva, E.; Shoeva, O.; Mursalimov, S.; Adonina, I.; Khlestkina, E. Fine Points of Marker-Assisted Pyramiding of Anthocyanin Biosynthesis Regulatory Genes for the Creation of Black-Grained Bread Wheat (Triticum aestivum L.) Lines. Agronomy 2022, 12, 2934. [Google Scholar] [CrossRef]
- Poljsak, B.; Kovač, V.; Milisav, I. Antioxidants, Food Processing and Health. Antioxidants 2021, 10, 433. [Google Scholar] [CrossRef]
- Scalbert, A.; Williamson, G. Dietary Intake and Bioavailability of Polyphenols. J. Nutr. 2000, 130, 2073S–2085S. [Google Scholar] [CrossRef]
- Pratyusha, S. Phenolic Compounds in the Plant Development and Defense: An Overview. In Plant Stress Physiology-Perspectives in Agriculture; IntechOpen: Rijeka, Croatia, 2022; ISBN 978-1-83969-867-5. [Google Scholar]
- Tena, N.; Martín, J.; Asuero, A.G. State of the Art of Anthocyanins: Antioxidant Activity, Sources, Bioavailability, and Therapeutic Effect in Human Health. Antioxidants 2020, 9, 451. [Google Scholar] [CrossRef]
- Treutter, D. Significance of Flavonoids in Plant Resistance: A Review. Environ. Chem. Lett. 2006, 4, 147–157. [Google Scholar] [CrossRef]
- Wang, X.; Ji, Z.; Cai, J.; Ma, L.; Li, X.; Yang, C. Construction of Near Isogenic Lines for Pericarp Color and Evaluation on Their Near Isogenicity in Rice. Rice Sci. 2009, 16, 261–266. [Google Scholar] [CrossRef]
- Garg, M.; Chawla, M.; Chunduri, V.; Kumar, R.; Sharma, S.; Sharma, N.K.; Kaur, N.; Kumar, A.; Mundey, J.K.; Saini, M.K.; et al. Transfer of Grain Colors to Elite Wheat Cultivars and Their Characterization. J. Cereal Sci. 2016, 71, 138–144. [Google Scholar] [CrossRef]
- Ji, Z.-J.; Wang, X.-G.; Zeng, Y.-X.; Ma, L.-Y.; Li, X.-M.; Liu, B.-X.; Yang, C.-D. Comparison of Physiological and Yield Traits between Purple- and White-Pericarp Rice Using SLs. Breed. Sci. 2012, 62, 71–77. [Google Scholar] [CrossRef]
- Zhou, H.; Pan, D.; Fan, Z. Research and application of Guangdong special rice and investigation on breeding problems. J. Guangdong Agric. Sci. 2005, 6, 19–21. [Google Scholar] [CrossRef]
- Gordeeva, E.; Shamanin, V.; Shoeva, O.; Kukoeva, T.; Morgounov, A.; Khlestkina, E. The Strategy for Marker-Assisted Breeding of Anthocyanin-Rich Spring Bread Wheat (Triticum aestivum L.) Cultivars in Western Siberia. Agronomy 2020, 10, 1603. [Google Scholar] [CrossRef]


| Cultivar/Line | Pedigree | Characterization | Origin |
|---|---|---|---|
| cv. Aley | Signal/Karina//Dina/Bagan///Vorsinsky-2 | Medium ripening, moderately resistant to stone and loose smut | Federal Altai Research Center of Agrobiotechnology (Barnaul, Russia) |
| cv. Tanay | G-20275/G-20191 | Mid-early ripening, resistant to stone and loose smut | Siberian Research Institute of Plant Production and Breeding (Krasnoobsk, Russia) |
| cv. Vorsinsky-2 | Signal//Irene/Gusar | Medium ripening, moderately tolerant to drought, melting cultivar, valuable in terms of quality | Altai Scientific Research Institute of Agriculture (Barnaul, Russia) |
| Grain Color | Gene | Chr | Primers (5′→3′), Restriction Enzymes | PCR Product Length (Restriction Fragment Length) in Donor/Recurrent Cultivar, bp | Reference |
|---|---|---|---|---|---|
| Purple | Ant1 | 7H | F: GCGGCTTGATTTGTTTCATA R: TTTAAATGGCGAGGTAAGGT | 488/455 | [15,16] |
| Ant2 | 2H | F: GCTGGAACACACGTACAAGA R: CTTTGAGCTATGGAGACCAAGAA | 514/719 | [15,16] | |
| Blue | HvMyc2 | 4H | F: CAAGTAGGTCCGAAGGCTCT R: CGGGCACTTTACCTCCAACA Restriction enzyme: Bse1I | 610/611 (485 + 125/611) | [17] |
| Crossing Combinations | Total Number of F2 Hybrids | Number of F2 Hybrids with Red Leaf Sheath Bases | Number of F2 Hybrids Homozygous for Donor-Like Alleles of Ant1 and Ant2 or HvMyc2 | Number of F2 (BC1F1-BC6F1) Hybrids Chosen for Backcrossing | Number of Hybrids Chosen in BC6F2 and the Resultant BC6F6 Lines |
|---|---|---|---|---|---|
| Breeding of purple-grained lines | |||||
| cv. Aley × BW648 | 65 | 49 | 5 | 4 (2–15) | 10 |
| cv. Tanay × BW648 | 89 | 64 | 14 | 6 (2–15) | 3 |
| cv. Vorsinsky-2 × BW648 | 61 | 43 | 7 | 6 (1–15) | 10 |
| Breeding of blue-grained lines | |||||
| cv. Aley × BW418 | 83 | n.a. | 31 | 6 (1–15) | 12 |
| cv. Tanay × BW418 | 72 | n.a. | 25 | 6 (5–15) | 14 |
| cv. Vorsinsky-2 × BW418 | 64 | n.a. | 29 | 6 (4–15) | 15 |
| NIL | Number of SNPs | SNP Density (Number of SNPs per Mbp) | Number of Donors’ SNPs | Frequency of Donors’ SNPs, % |
|---|---|---|---|---|
| pAley | 22,311 | 5.32 | 2025 | 9.08 |
| bAley | 23,858 | 5.69 | 1091 | 4.57 |
| pTanay | 26,481 | 6.31 | 2218 | 8.38 |
| bTanay | 29,649 | 7.07 | 1930 | 6.51 |
| pVorsinsky-2 | 25,769 | 6.14 | 3148 | 12.22 |
| bVorsinsky-2 | 30,289 | 7.22 | 5323 | 17.57 |
| Average | 26,393 | 6.29 | 2623 | 9.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kukoeva, T.V.; Molobekova, C.A.; Totsky, I.V.; Vasiliev, G.V.; Pronozin, A.Y.; Afonnikov, D.A.; Khlestkina, E.K.; Shoeva, O.Y. Enrichment of Grain Anthocyanin Content through Marker-Assisted Breeding for Ant1, Ant2 or HvMyc2 Genes in Barley (Hordeum vulgare L.). Agronomy 2024, 14, 1231. https://doi.org/10.3390/agronomy14061231
Kukoeva TV, Molobekova CA, Totsky IV, Vasiliev GV, Pronozin AY, Afonnikov DA, Khlestkina EK, Shoeva OY. Enrichment of Grain Anthocyanin Content through Marker-Assisted Breeding for Ant1, Ant2 or HvMyc2 Genes in Barley (Hordeum vulgare L.). Agronomy. 2024; 14(6):1231. https://doi.org/10.3390/agronomy14061231
Chicago/Turabian StyleKukoeva, Tatjana V., Camilla A. Molobekova, Igor V. Totsky, Gennady V. Vasiliev, Artem Yu. Pronozin, Dmitry A. Afonnikov, Elena K. Khlestkina, and Olesya Yu. Shoeva. 2024. "Enrichment of Grain Anthocyanin Content through Marker-Assisted Breeding for Ant1, Ant2 or HvMyc2 Genes in Barley (Hordeum vulgare L.)" Agronomy 14, no. 6: 1231. https://doi.org/10.3390/agronomy14061231
APA StyleKukoeva, T. V., Molobekova, C. A., Totsky, I. V., Vasiliev, G. V., Pronozin, A. Y., Afonnikov, D. A., Khlestkina, E. K., & Shoeva, O. Y. (2024). Enrichment of Grain Anthocyanin Content through Marker-Assisted Breeding for Ant1, Ant2 or HvMyc2 Genes in Barley (Hordeum vulgare L.). Agronomy, 14(6), 1231. https://doi.org/10.3390/agronomy14061231







