ZmD11 Gene Regulates Tobacco Plant Floral Development under Drought Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth
2.2. Gene Clone and Transformation
2.3. Relative Water Content
2.4. Enzyme Activity
2.5. BR, SA, and ABA Content Determination
2.6. BR Treatment
2.7. RNA Extraction, Reverse Transcription and qRT-PCR
2.8. Statistical Analysis
3. Results
3.1. The Expression of ZmD11
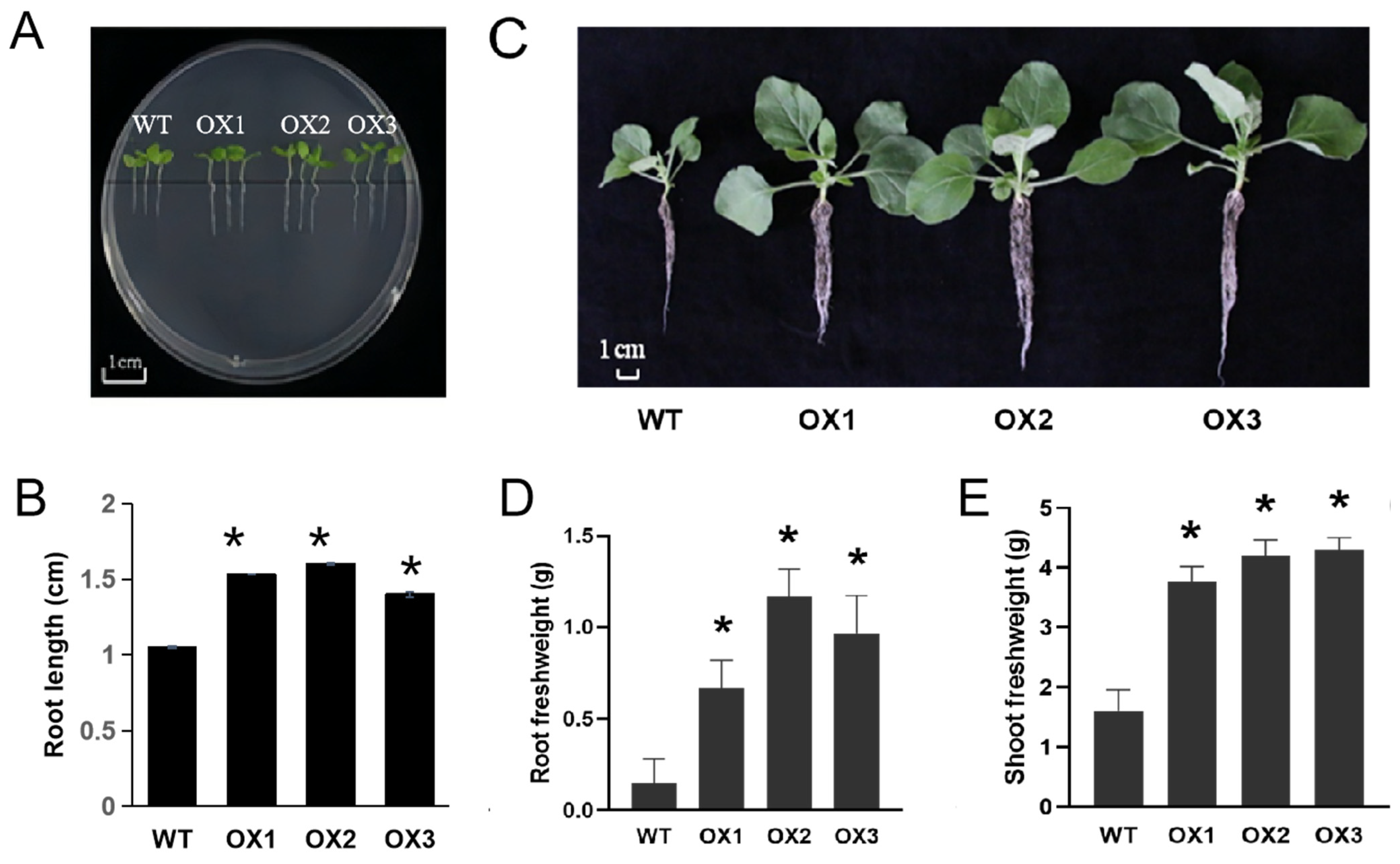
3.2. Over-expression of ZmD11 Increased Vegetative and Floral Organ Size in Tobacco
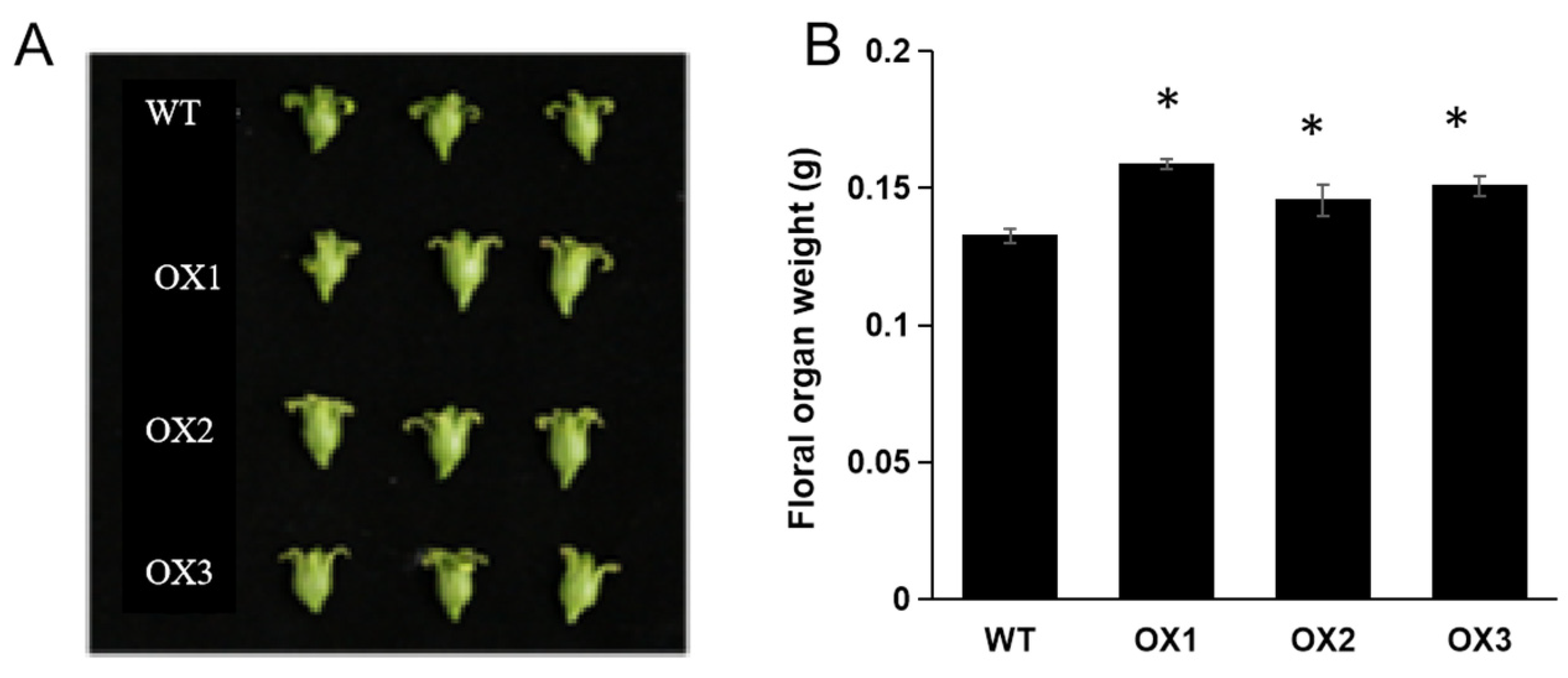
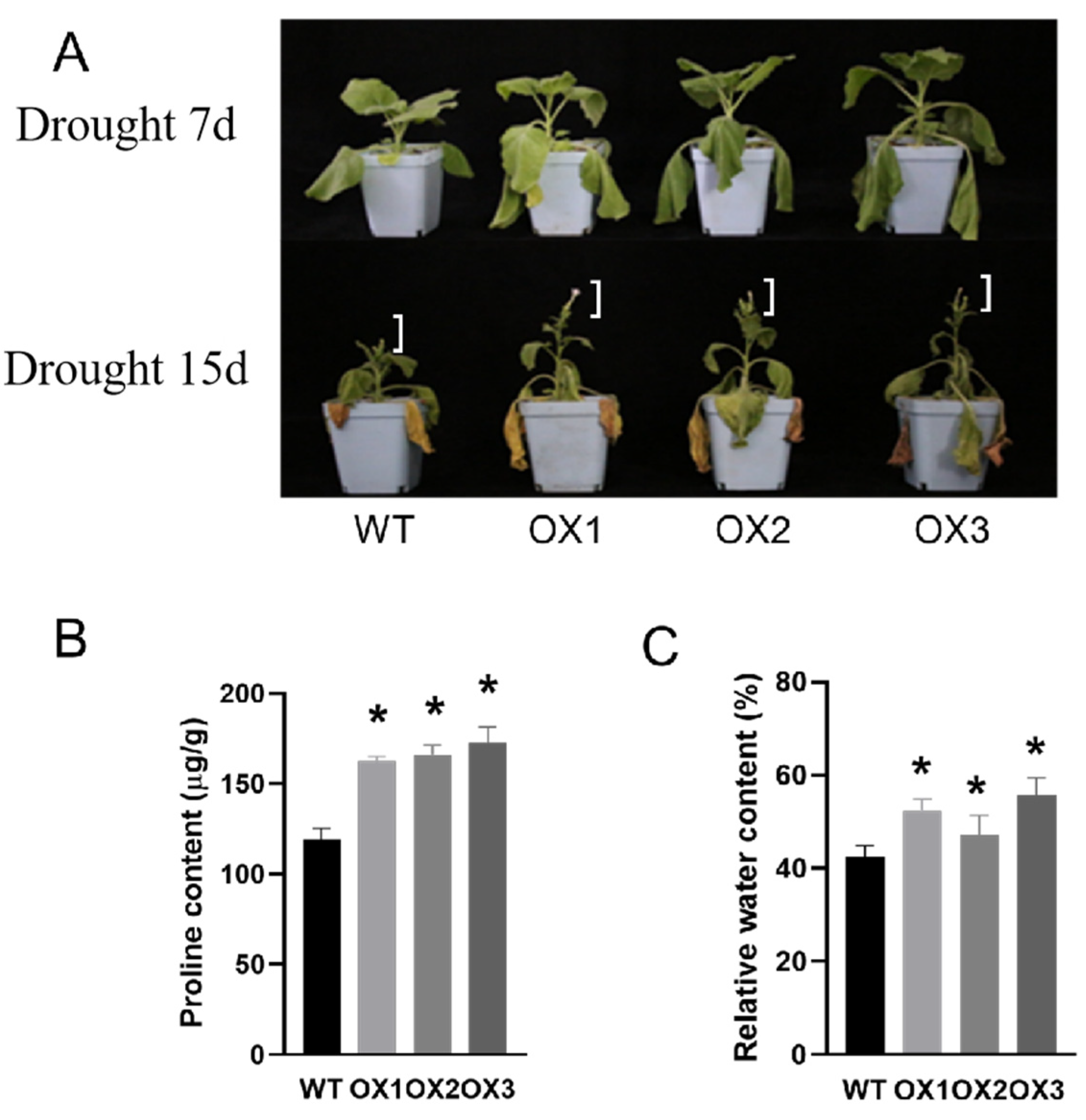
3.3. Over-Expression of ZmD11 Increases Floral Size under Drought Stress
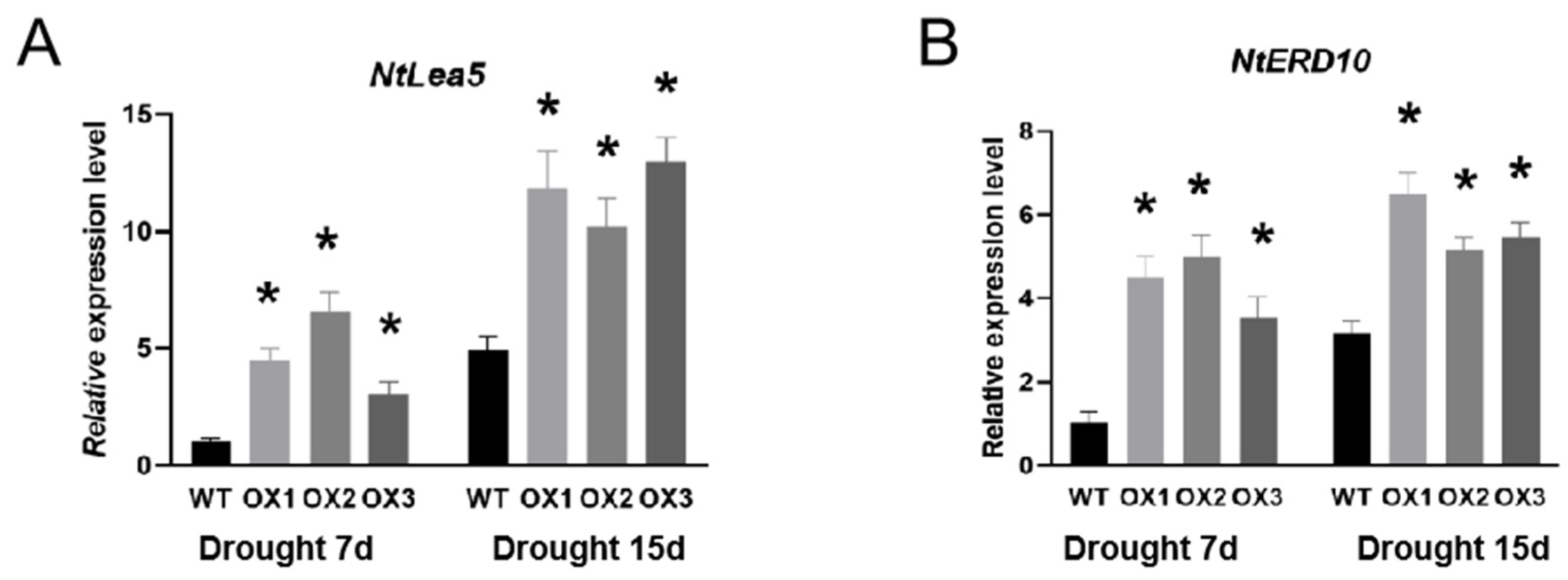
3.4. The Activity of Protective Enzymes and the Expression of Drought-Tolerant-Related Genes Are Changed
3.5. The Content of Hormones Have Been Affected by ZmD11
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nuccio, M.L.; Wu, J.; Mowers, R.; Zhou, H.P.; Meghji, M.; Primavesi, L.F.; Paul, M.J.; Chen, X.; Gao, Y.; Haque, E.; et al. Expression of Trehalose-6-Phosphate Phosphatase in Maize Ears Improves Yield in Well-Watered and Drought Conditions. Nat. Biotechnol. 2015, 33, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Liu, S.; Ferjani, A.; Li, J.; Yan, J.; Yang, X.; Qin, F. Genetic Variation in ZmVPP1 Contributes to Drought Tolerance in Maize Seedlings. Nat. Genet. 2016, 48, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Hao, Z.; Pang, J.; Zhang, M.; Wang, N.; Li, X.; Li, W.; Wang, L.; Xu, M. Effect of Water-Deficit on Tassel Development in Maize. Gene 2019, 681, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhang, B.; Yang, Z.; Liu, Y.; Yang, S.; Shi, Y.; Jiang, C.; Qin, F. Manipulating ZmEXPA4 Expression Ameliorates the Drought-Induced Prolonged Anthesis and Silking Interval in Maize. Plant Cell 2021, 33, 2058–2071. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.E.; Repetti, P.P.; Adams, T.R.; Creelman, R.A.; Wu, J.; Warner, D.C.; Anstrom, D.C.; Bensen, R.J.; Castiglioni, P.P.; Donnarummo, M.G.; et al. Plant Nuclear Factor Y (NF-Y) B Subunits Confer Drought Tolerance and Lead to Improved Corn Yields on Water-Limited Acres. Proc. Natl. Acad. Sci. USA 2007, 104, 16450–16455. [Google Scholar] [CrossRef]
- Virlouvet, L.; Jacquemot, M.P.; Gerentes, D.; Corti, H.; Bouton, S.; Gilard, F.; Valot, B.; Trouverie, J.; Tcherkez, G.; Falque, M.; et al. The ZmASR1 Protein Influences Branched-Chain Amino Acid Biosynthesis and Maintains Kernel Yield in Maize under Water-Limited Conditions. Plant Physiol. 2011, 157, 917–936. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 Variants Generated by CRISPR-Cas9 Improve Maize Grain Yield under Field Drought Stress Conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Cheng, M.; Chen, Y.; Liu, B.; Wang, X.; Li, G.; Zhou, Y.; Luo, P.; Xi, Z.; Yong, H.; et al. Natural Variations in the Non-Coding Region of ZmNAC080308 Contributes Maintaining Grain Yield under Drought Stress in Maize. BMC Plant Biol. 2021, 21, 305. [Google Scholar] [CrossRef]
- Planas-Riverola, A.; Gupta, A.; Betegoń-Putze, I.; Bosch, N.; Ibanḛs, M.; Cano-Delgado, A.I. Brassinosteroid Signaling in Plant Development and Adaptation to Stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Webster, S.; He, S.Y. Growth–Defense Trade-Offs in Plants. Curr. Biol. 2022, 32, R634–R639. [Google Scholar] [CrossRef]
- Nolan, T.M.; Brennan, B.; Yang, M.; Chen, J.; Zhang, M.; Li, Z.; Wang, X.; Bassham, D.C.; Walley, J.; Yin, Y. Selective Autophagy of BES1 Mediated by DSK2 Balances Plant Growth and Survival. Dev. Cell 2017, 41, 33–46. [Google Scholar] [CrossRef]
- Nolan, T.M.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional Regulators of Plant. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The Physiology of Plant Responses to Drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Iqbal, S.; Wang, X.; Mubeen, I.; Kamran, M.; Kanwal, I.; Díaz, G.A.; Abbas, A.; Parveen, A.; Atiq, M.N.; Alshaya, H.; et al. Phytohormones Trigger Drought Tolerance in Crop Plants: Outlook and Future Perspectives. Front. Plant Sci. 2022, 12, 799318. [Google Scholar] [CrossRef]
- Tian, G.; Wang, S.; Wu, J.; Wang, Y.; Wang, X.; Liu, S.; Han, D.; Xia, G.; Wang, M. Allelic variation of TaWD40-4B.1 contributes to drought tolerance by modulating catalase activity in wheat. Nat. Commun. 2023, 14, 1200. [Google Scholar] [CrossRef] [PubMed]
- Saed-Moucheshi, A.; Sohrabi, F.; Fasihfar, E.; Baniasadi, F.; Riasat, M.; Mozafari, A.A. Superoxide dismutase (SOD) as a selection criterion for triticale grain yield under drought stress: A comprehensive study on genomics and expression profiling, bioinformatics, heritability, and phenotypic variability. BMC Plant Biol. 2021, 21, 148. [Google Scholar] [CrossRef]
- Sofo, A.; Scopa, A.; Nuzzaci, M.; Vitti, A. Ascorbate Peroxidase and Catalase Activities and Their Genetic Regulation in Plants Subjected to Drought and Salinity Stresses. Int. J. Mol. Sci. 2015, 16, 13561–13578. [Google Scholar] [CrossRef]
- Xu, C.; Shan, J.; Liu, T.; Wang, Q.; Ji, Y.; Zhang, Y.; Wang, M.; Xia, N.; Zhao, L. CONSTANS-LIKE 1a positively regulates salt and drought tolerance in soybean. Plant Physiol. 2023, 191, 2427–2446. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, Y.; Koehler, G.; Blacklock, B.J.; Randall, S.K. Dehydrin expression in soybean. Plant Physiol. Biochem. 2013, 70, 213–220. [Google Scholar] [CrossRef]
- Decena, M.A.; Gálvez-Rojas, S.; Agostini, F.; Sancho, R.; Contreras-Moreira, B.; Des Marais, D.L.; Hernandez, P.; Catalán, P. Comparative Genomics, Evolution, and Drought-Induced Expression of Dehydrin Genes in Model Brachypodium Grasses. Plants 2021, 10, 2664. [Google Scholar] [CrossRef]
- Wu, Y.; Fu, Y.; Zhao, S.; Gu, P.; Zhu, Z.; Sun, C.; Tan, L. CLUSTERED PRIMARY BRANCH 1, a New Allele of DWARF11, Controls Panicle Architecture and Seed Size in Rice. Plant Biotechnol. J. 2016, 14, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Xu, H.; Li, B.; Shang, Y.; Wei, M.; Zhang, S.; Zhao, C.; Qin, R.; Cui, F.; Wu, Y. The Brassinosteroid Biosynthesis Gene, ZmD11, Increases Seed Size and Quality in Rice and Maize. Plant Physiol. Biochem. 2021, 160, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Sheng, J.; Xu, Y.; Xiong, F.; Wu, Y.; Wang, W.; Wang, Z.; Yang, J.; Zhang, J. Role of Brassinosteroids in Rice Spikelet Differentiation and Degeneration under Soil-Drying during Panicle Development. BMC Plant Biol. 2019, 19, 409. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiao, F.; Li, J.; Pei, Y.; Zhao, M.; Song, X.; Guo, X. Transcriptomic Analysis of the Maize Inbred Line Chang7-2 and a Large-Grain Mutant Tc19. BMC Genom. 2022, 23, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, N.; Li, H.; Zhang, S.; Zhang, X.; Yan, X.; Wang, Z.; Yang, Y.; Zhang, S. Overexpression of NtGCN2 improves drought tolerance in tobacco by regulating proline accumulation, ROS scavenging ability, and stomatal closure. Plant Physiol. Biochem. 2023, 198, 107665. [Google Scholar] [CrossRef]
- Ji, X.; Gao, Q.; Zhuang, Z.; Chang, F.; Peng, Y. WGCNA analysis of the effect of exogenous BR on leaf angle of maize mutant lpa1. Sci. Rep. 2024, 14, 5238. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Yong, R.; Zhang, J.; Cai, G.; Wang, R.; Li, J.; Wang, Y.; Zhang, H.; Gao, X.; Huang, J. OsBAK2/OsSERK2 Expression Is Repressed by OsBZR1 to Modulate Brassinosteroid Response and Grain Length in Rice. J. Exp. Bot. 2023, 74, 4978–4993. [Google Scholar] [CrossRef]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yina, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Responses. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Fàbregas, N.; Lozano-Elena, F.; Blasco-Escámez, D.; Tohge, T.; Martínez-Andújar, C.; Albacete, A.; Osorio, S.; Bustamante, M.; Riechmann, J.L.; Nomura, T.; et al. Overexpression of the Vascular Brassinosteroid Receptor BRL3 Confers Drought Resistance without Penalizing Plant Growth. Nat. Commun. 2018, 9, 4680. [Google Scholar] [CrossRef]
- Muhammad, I.; Yang, L.; Ahmad, S.; Farooq, S.; Khan, A.; Muhammad, N.; Ullah, S.; Adnan, M.; Ali, S.; Liang, Q.P.; et al. Melatonin-Priming Enhances Maize Seedling Drought Tolerance by Regulating the Antioxidant Defense System. Plant Physiol. 2023, 191, 2301–2315. [Google Scholar] [CrossRef]
- Zhu, D.; Chang, Y.; Pei, T.; Zhang, X.; Liu, L.; Li, Y.; Zhuang, J.; Yang, H.; Qin, F.; Song, C.; et al. MAPK-like Protein 1 Positively Regulates Maize Seedling Drought Sensitivity by Suppressing ABA Biosynthesis. Plant J. 2020, 102, 747–760. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Ma, X.; Hussain, Q.; Asim, M.; Iqbal, A.; Ren, X.; Shah, S.; Chen, K.; Shi, Y. Application of 2,4-Epibrassinolide Improves Drought Tolerance in Tobacco through Physiological and Biochemical Mechanisms. Biology 2022, 11, 1192. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Wang, S.; Yang, S.; Yang, Z.; Liu, S.; Wang, Y.; Gao, H.; Zhang, S.; Yang, X.; Jiang, C.; et al. Genome assembly and genetic dissection of a prominent drought-resistant maize germplasm. Nat. Genet. 2023, 55, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xiang, Y.; Yan, J.; Di, P.; Li, J.; Sun, X.; Han, G.; Ni, L.; Jiang, M.; Yuan, J.; et al. Brassinosteroid-Signaling Kinase 1 Phosphorylating Calcium/Calmodulin-Dependent Protein Kinase Functions in Drought Tolerance in Maize. New Phytol. 2021, 231, 695–712. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yan, Y.; Lu, W.; Lu, D. Application of Exogenous Phytohormones at Silking Stage Improve Grain Quality under Post-Silking Drought Stress in Waxy Maize. Plants 2021, 10, 48. [Google Scholar] [CrossRef]
- Liao, K.; Peng, Y.J.; Yuan, L.B.; Dai, Y.S.; Chen, Q.F.; Yu, L.J.; Bai, M.Y.; Zhang, W.Q.; Xie, L.J.; Xiao, S. Brassinosteroids Antagonize Jasmonate-Activated Plant Defense Responses through BRI1-EMS-Suppressor1 (BES1)1. Plant Physiol. 2020, 182, 1066–1082. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Jiao, F.; Sun, Z.; Zhang, E.; Song, X.; Pei, Y.; Li, J.; Cannon, N.; Chang, X.; Guo, X. ZmD11 Gene Regulates Tobacco Plant Floral Development under Drought Stress. Agronomy 2024, 14, 1381. https://doi.org/10.3390/agronomy14071381
Li Z, Jiao F, Sun Z, Zhang E, Song X, Pei Y, Li J, Cannon N, Chang X, Guo X. ZmD11 Gene Regulates Tobacco Plant Floral Development under Drought Stress. Agronomy. 2024; 14(7):1381. https://doi.org/10.3390/agronomy14071381
Chicago/Turabian StyleLi, Zhanfeng, Fuchao Jiao, Zhiyi Sun, Enying Zhang, Xiyun Song, Yuhe Pei, Jun Li, Nicola Cannon, Xianmin Chang, and Xinmei Guo. 2024. "ZmD11 Gene Regulates Tobacco Plant Floral Development under Drought Stress" Agronomy 14, no. 7: 1381. https://doi.org/10.3390/agronomy14071381





