Natural Variation in Chromium Accumulation and the Development of Related EST-SSR Molecular Markers in Miscanthus sinensis
Abstract
:1. Introduction
2. Material and Methods
2.1. Plant Material and Cr Stress Treatment
2.2. Determination of Chromium Content in Different Plant Tissues
2.3. Identification of EST-SSRs
2.4. DNA Extraction and Validation of EST-SSRs
2.5. Data Analysis
3. Results
3.1. The Cr Content in Root, Stem, and Leaf Tissues of Various M. sinensis Genotypes
3.2. Identification and Validation of EST-SSRs
3.3. Clustering Analysis and Population Structure
3.4. Marker–Trait Association Analysis for Cr Treatment
3.5. Expression and Phylogenetic Analysis of Associated Candidate Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Islam, M.K.; Alam, I.; Khanam, S.; Lee, S.Y.; Huh, M.R. Accumulation and tolerance characteristics of chromium in nine jute varieties (Corchorus spp. and Hibiscus spp.). Plant Omics 2014, 7, 392–402. [Google Scholar]
- Zhu, W.; Zhang, K.; Xu, D.; Liu, Z.; Gao, J. Statistical analysis on the effect of the utilization of mineral resources on the environmental impact in China. Sustainability 2021, 13, 8462. [Google Scholar] [CrossRef]
- Sharma, A.; Kapoor, D.; Wang, J.; Shahzad, B.; Kumar, V.; Bali, A.S.; Jasrotia, S.; Zheng, B.; Yuan, H.; Yan, D. Chromium bioaccumulation and its impacts on plants: An overview. Plants 2020, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Mohan, D.; Pittman, C.U. Activated carbons and low cost adsorbents for remediation of tri- and hexavalent chromium from water. J. Hazard. Mater. 2006, 137, 762–811. [Google Scholar] [CrossRef] [PubMed]
- Shanker, A.K.; Cervantes, C.; Loza-Tavera, H.; Avudainayagam, S. Chromium toxicity in plants. Environ. Int. 2005, 31, 739–753. [Google Scholar] [CrossRef] [PubMed]
- Ertani, A.; Mietto, A.; Borin, M.; Nardi, S. Chromium in Agricultural Soils and Crops: A Review. Water Air Soil Pollut. 2017, 228, 190. [Google Scholar] [CrossRef]
- Akinci, I.E.; Akinci, S. Effect of chromium toxicity on germination and early seedling growth in melon (Cucumis melo L.). Afr. J. Biotechnol. 2010, 9, 4589–4594. [Google Scholar]
- Sallah-Ud-Din, R.; Farid, M.; Saeed, R.; Ali, S.; Rizwan, M.; Tauqeer, H.M.; Bukhari, S.A.H. Citric acid enhanced the antioxidant defense system and chromium uptake by Lemna minor L. grown in hydroponics under Cr stress. Environ. Sci. Pollut. Res. 2017, 24, 17669–17678. [Google Scholar] [CrossRef] [PubMed]
- Goupil, P.; Souguir, D.; Ferjani, E.; Faure, O.; Hitmi, A.; Ledoigt, G. Expression of stress-related genes in tomato plants exposed to arsenic and chromium in nutrient solution. J. Plant Physiol. 2009, 166, 1446–1452. [Google Scholar] [CrossRef]
- Bah, A.M.; Sun, H.; Chen, F.; Zhou, J.; Dai, H.; Zhang, G.; Wu, F. Comparative proteomic analysis of Typha angustifolia leaf under chromium, cadmium and lead stress. J. Hazard. Mater. 2010, 184, 191–203. [Google Scholar] [CrossRef]
- Clifton-Brown, J.C.; Lewandowski, I. Screening Miscanthus genotypes in field trials to optimise biomass yield and quality in southern Germany. Eur. J. Agron. 2002, 16, 97–110. [Google Scholar] [CrossRef]
- Heaton, E.A.; Long, S.P.; Voigt, T.B.; Jones, M.B.; Clifton-Brown, J. Miscanthus for renewable energy generation: European Union expierence and projections for Illinois. Mitig. Adapt. Strateg. Glob. Chang. 2004, 9, 433–451. [Google Scholar] [CrossRef]
- Nie, G.; Zhong, M.; Cai, J.; Yang, X.; Zhang, X. Transcriptome characterization of candidate genes related to chromium uptake, transport and accumulation in Miscanthus sinensis. Ecotoxicol. Environ. Saf. 2021, 221, 112445. [Google Scholar] [CrossRef] [PubMed]
- Tubeileh, A.; Rennie, T.J.; Goss, M.J. A review on biomass production from C4 grasses: Yield and quality for end-use. Curr. Opin. Plant Biol. 2016, 31, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Nie, G.; Tang, L.; Zhang, Y.J.; Huang, L.K.; Ma, X.; Cao, X.; Pan, L.; Zhang, X.; Zhang, X.Q. Development of SSR markers based on transcriptome sequencing and association analysis with drought tolerance in perennial grass Miscanthus from China. Front. Plant Sci. 2017, 8, 801. [Google Scholar] [CrossRef] [PubMed]
- Fernando, A.; Duarte, P.; Oliveira, J.F.S. Bioremoval of heavy metals from soil by Miscanthus sinensis × giganteus. In Proceedings of the Biomass for Energy and the Environment, Proceedings of the 9th European Bioenergy Conference, Copenhagen, Denmark, 24–27 June 1996; pp. 531–536. [Google Scholar]
- Namasivayam, C.; Höll, W.H. Chromium (III) removal in tannery waste waters using Chinese Reed (Miscanthus sinensis), a fast growing plant. Eur. J. Wood Wood Prod. 2004, 62, 74–80. [Google Scholar] [CrossRef]
- Hung, K.H.; Chiang, T.Y.; Chiu, C.T.; Hsu, T.W.; Ho, C.W. Isolation and characterization of microsatellite loci from a potential biofuel plant Miscanthus sinensis (Poaceae). Conserv. Genet. 2009, 10, 1377–1380. [Google Scholar] [CrossRef]
- Clark, L.V.; Brummer, J.E.; Glowacka, K.; Hall, M.C.; Sacks, E.J. A footprint of past climate change on the diversity and population structure of Miscanthus sinensis. Ann. Bot. 2014, 114, 97–107. [Google Scholar] [CrossRef]
- Nie, G.; Zhang, X.Q.; Huang, L.K.; Xu, W.Z.; Wang, J.P.; Zhang, Y.W.; Ma, X.; Yan, H. Genetic variability and population structure of the potential bioenergy crop Miscanthus sinensis (Poaceae) in Southwest China based on SRAP markers. Molecules 2014, 19, 12881–12897. [Google Scholar] [CrossRef] [PubMed]
- Nie, G.; Huang, L.; Zhang, X.; Megan, T.; Jiang, Y.; Yu, X.; Lin, X.; Wang, X.; Zhang, Y. Marker-Trait Association for Biomass Yield of Potential Bio-fuel Feedstock Miscanthus sinensis from Southwest China. Front. Plant Sci. 2016, 7, 802. [Google Scholar] [CrossRef]
- Liu, W.; Xu, L.; Wang, Y.; Shen, H.; Zhu, X.; Zhang, K.; Chen, Y.; Yu, R.; Limera, C.; Liu, L. Transcriptome-wide analysis of chromium-stress responsive micrornas to explore mirna-mediated regulatory networks in radish (Raphanus sativus L.). Sci. Rep. 2015, 5, 14024. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Nakatani, M.; Mizuno, K.; Fujimura, T. Development and characterization of microsatellite markers in Sweetpotato. Breed. Sci. 2004, 54, 177–188. [Google Scholar] [CrossRef]
- Gupta, S.; Prasad, M. Development and characterization of genic SSR markers in Medicago truncatula and their transferability in leguminous and non-leguminous species. Genome 2009, 52, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K. Gene-based marker systems in plants: High throughput approaches for marker discovery and genotyping. In Molecular Techniques in Crop Improvement; Springer: Dordrecht, The Netherlands, 2010; pp. 119–142. [Google Scholar]
- Kane, N.C.; Rieseberg, L.H. Selective sweeps reveal candidate genes for adaptation to drought and salt tolerance in common sunflower, Helianthus annuus. Genetics 2007, 175, 1823–1834. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Bai, G.; Luo, N.; Chen, Z.; Liu, S.; Liu, J. Association of simple sequence repeat (SSR) markers with submergence tolerance in diverse populations of perennial ryegrass. Plant Sci. 2011, 180, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Nie, G.; Sun, M.; Huang, L.; Ma, X.; Zhang, X. Effect of moist pre-chill and dry pre-heat treatment on the germination of Miscanthus sinensis seed from southwest China. Grassl. Sci. 2017, 63, 93–100. [Google Scholar] [CrossRef]
- Arduini, I.; Masoni, A.; Ercoli, L. Effects of high chromium applications on Miscanthus during the period of maximum growth. Environ. Exp. Bot. 2006, 58, 234–243. [Google Scholar] [CrossRef]
- Sebastian, B.; Thomas, T.; Thomas, M.; Uwe, S.; Martin, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar]
- Liu, K.J.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Evanno, G.S.; Regnaut, S.J.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Wilkins, C. The uptake of copper, arsenic and zinc by Miscanthus-environmental implications for use as an energy crop. Asp. Appl. Biol. 1997, 49, 335–340. [Google Scholar]
- Adhikari, A.; Adhikari, S.; Ghosh, S.; Azahar, I.; Hossain, Z. Imbalance of redox homeostasis and antioxidant defense status in maize under chromium (VI) stress. Environ. Exp. Bot. 2019, 169, 103873. [Google Scholar] [CrossRef]
- Jia, H.; Wang, X.; Wei, T.; Zhou, R.; Ding, Y. Accumulation and fixation of Cd by tomato cell wall pectin under Cd stress. Environ. Exp. Bot. 2019, 167, 103829. [Google Scholar] [CrossRef]
- De Caroli, M.; Furini, A.; DalCorso, G.; Rojas, M.; Di Sansebastiano, G.P. Endomembrane Reorganization induced by heavy metals. Plants 2020, 9, 482. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Y.Y.; Zhang, Q.; Fu, B.S.; Cai, J.; Wu, J.Z.; Chen, Y.H. Genome-wide association analysis of quantitative trait loci for salinity-tolerance related morphological indices in bread wheat. Euphytica 2018, 214, 176. [Google Scholar] [CrossRef]
- Zhang, P.; Zhong, K.; Tong, H.; Shahid, M.Q.; Li, J. Association mapping for aluminum tolerance in a core collection of rice landraces. Front. Plant Sci. 2016, 7, 1415. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Tiwari, M.; Dutta, P.; Singh, P.; Chawda, K.; Kumari, M.; Chakrabarty, D. Chromium stress in plants: Toxicity, tolerance and phytoremediation. Sustainability 2021, 13, 4629. [Google Scholar] [CrossRef]
- Xu, Z.; Cai, M.; Chen, S.; Huang, X.; Wang, P. High-affinity sulfate transporter Sultr1; 2 is a major transporter for Cr (VI) uptake in plants. Environ. Sci. Technol. 2021, 55, 1576–1584. [Google Scholar] [CrossRef]
- Li, P.; Luo, T.; Pu, X. Plant transporters: Roles in stress responses and effects on growth and development. Plant Growth Regul. 2021, 93, 253–266. [Google Scholar] [CrossRef]
- Shewry, P.R.; Peterson, P.J. The Uptake and transport of chromium by barley seedlings (Hordeum vulgare L.). J. Exp. Bot. 1974, 25, 785–797. [Google Scholar] [CrossRef]
- Ferrari, M.; Cozza, R.; Marieschi, M.; Torelli, A. Role of sulfate transporters in chromium tolerance in Scenedesmus acutus M. (Sphaeropleales). Plants 2022, 11, 223. [Google Scholar] [CrossRef] [PubMed]
- Schiavon, M.; Pilon-Smits, E.A.; Wirtz, M.; Hell, R.; Malagoli, M. Interactions between chromium and sulfur metabolism in Brassica juncea. J. Environ. Qual. 2008, 37, 1536–1545. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Yuan, X.; Li, L.; Zeng, M.; Yang, J.; Tang, H.; Duan, C. Genome-Wide Analysis of the ATP-Binding Cassette (ABC) Transporter family in Zea mays L. and its response to heavy metal stresses. Int. J. Mol. Sci. 2022, 23, 2109. [Google Scholar] [CrossRef] [PubMed]
- Banasiak, J.; Jasiński, M. ATP-binding cassette (ABC) transporters in nonmodel plants. New Phytol. 2022, 233, 1597–1612. [Google Scholar] [CrossRef] [PubMed]
- Ghanizadeh, H.; Harrington, K.C. Non-target site mechanisms of resistance to herbicides. Crit. Rev. Plant Sci. 2017, 36, 24–34. [Google Scholar] [CrossRef]
- Thanh Ha Thi, D.; Enrico, M.; Youngsook, L. Functions of ABC transporters in plant growth and development. Curr. Opin. Plant Biol. 2018, 41, 32–38. [Google Scholar]
- Dahuja, A.; Kumar, R.R.; Sakhare, A. Role of ABC transporters in maintaining plant homeostasis under abiotic and biotic stresses. Physiol. Plant. 2020, 171, 785–801. [Google Scholar] [CrossRef]
- Dubey, S.; Saxena, S.; Chauhan, A.S.; Mathur, P.; Chakrabaroty, D. Identification and expression analysis of conserved micrornas during short and prolonged chromium stress in rice (Oryza sativa). Environ. Sci. Pollut. Res. 2020, 27, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Lally, D.; Ingmire, P.; Tong, H.; He, Z. Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell 2001, 13, 1317–1331. [Google Scholar]
- Anderson, C.M.; Wagner, T.A.; Perret, M. WAKs: Cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol. Biol. 2001, 47, 197–206. [Google Scholar] [CrossRef]
- Wang, D.; Qin, L.; Wu, M. Identification and characterization of WAK gene family in Saccharum and the negative roles of ScWAK1 under the pathogen stress. Int. J. Biol. Macromol. 2022, 224, 1–19. [Google Scholar] [CrossRef]
- Sivaguru, M.; Ezaki, B.; He, Z.; Tong, H.; Osawa, H.; Baluška, F. Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol. 2003, 132, 2256–2266. [Google Scholar] [CrossRef]
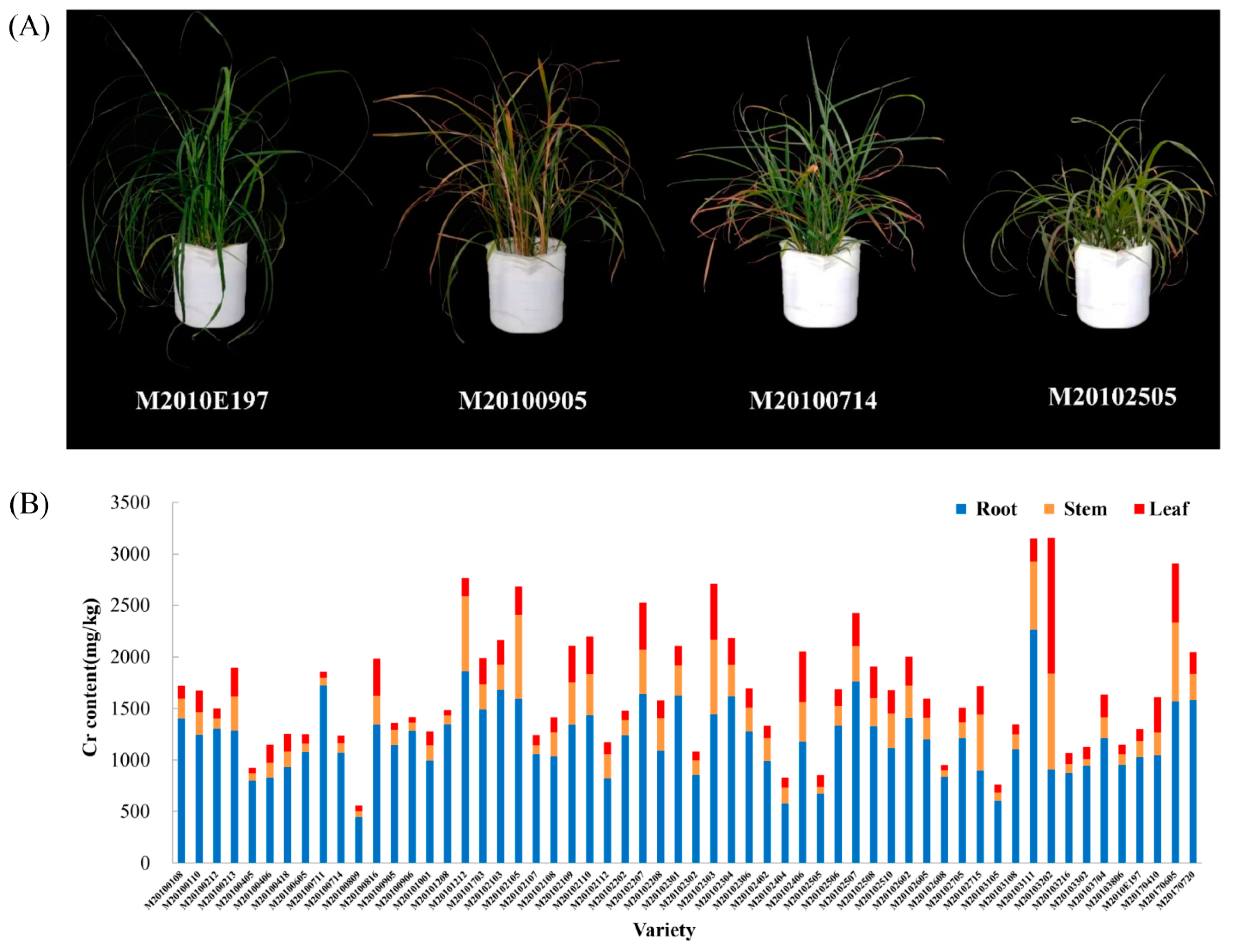

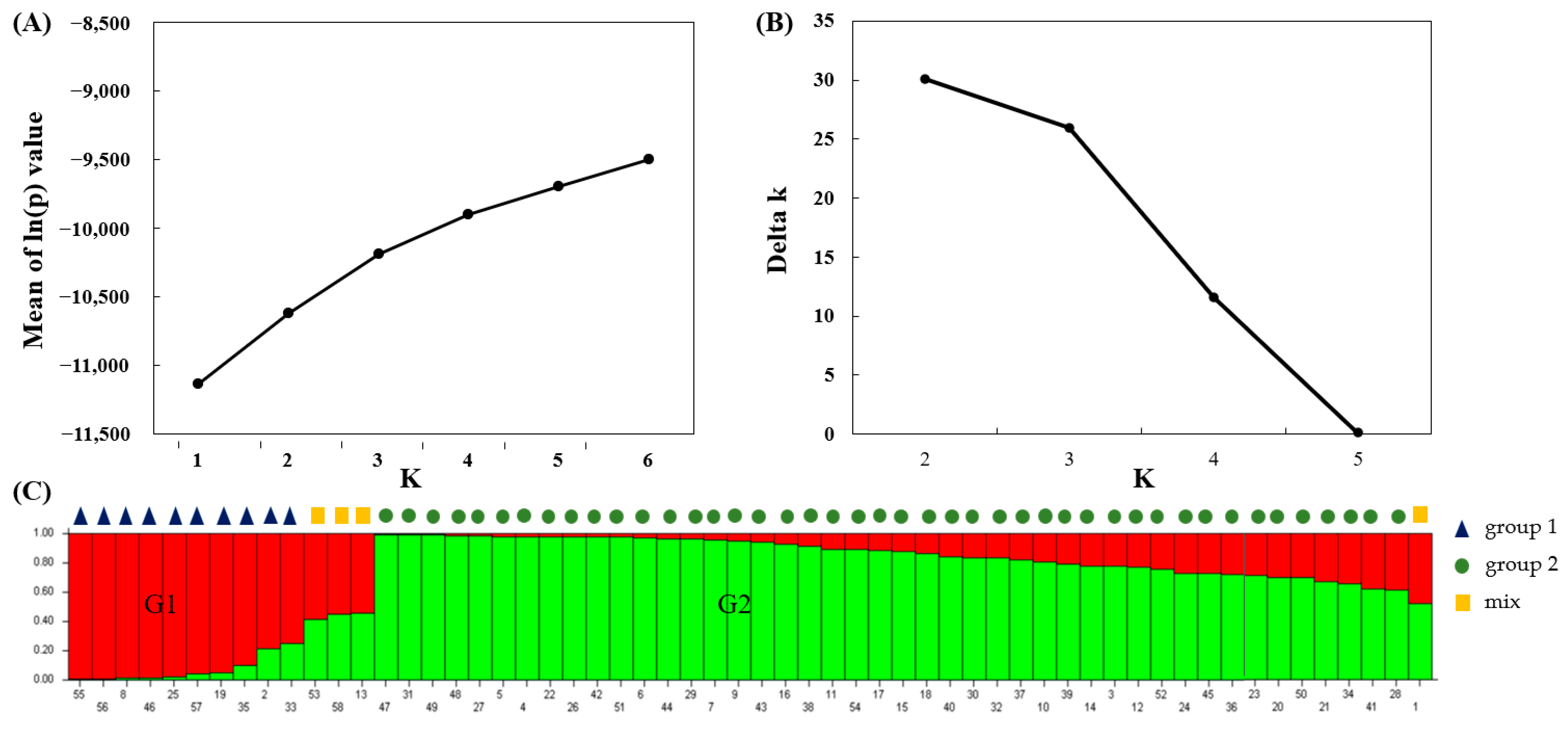
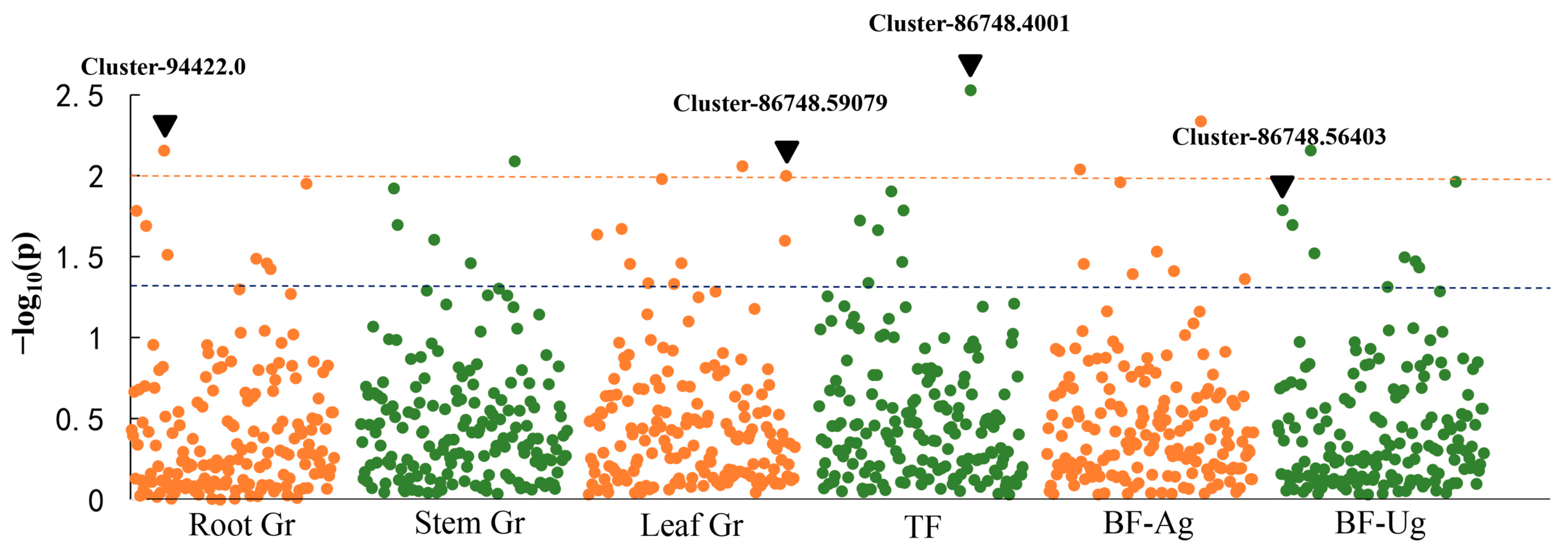
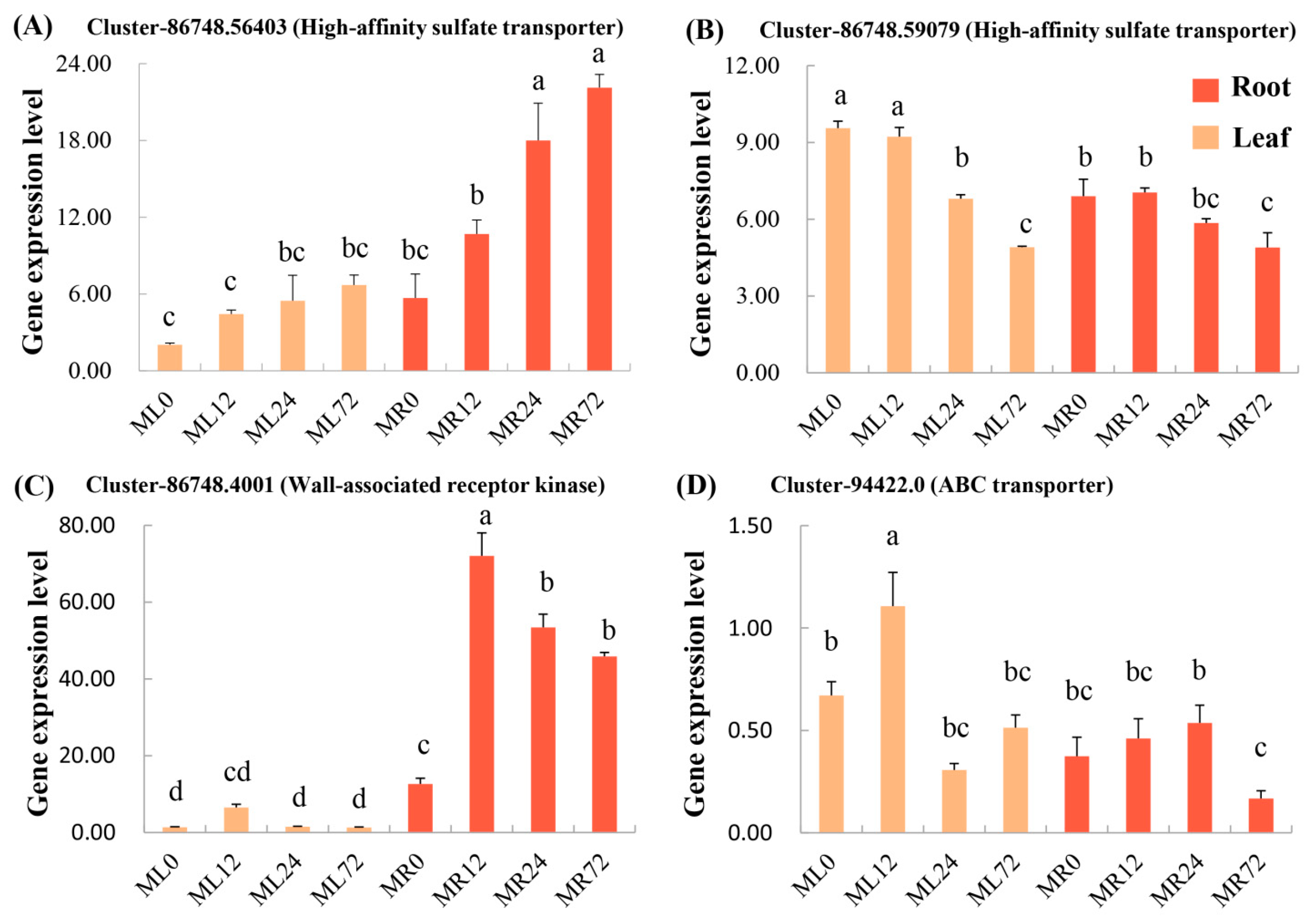

| Varieties | Cr Concentration/(mg/kg) | TF | BF | |||
|---|---|---|---|---|---|---|
| Root | Stem | Leaf | Aboveground | Underground | ||
| Mean | 1205.77 | 261.71 | 212.53 | 0.39 | 2.37 | 6.03 |
| Max | 2265.09 | 933.08 | 1317.93 | 2.48 | 11.26 | 11.33 |
| Min | 443.15 | 56.97 | 52.49 | 0.07 | 0.56 | 2.22 |
| SD | 343.22 | 205.33 | 191.69 | 0.33 | 1.86 | 1.72 |
| CV (%) | 28.46 | 78.46 | 90.19 | 85.13 | 78.53 | 28.46 |
| F | 13.32 *** | 32.78 *** | 83.48 *** | 1.41 | 63.15 *** | 13.31 *** |
| Primer Name | Gene Name | Total Number of Amplified Bands | Number of Polymorphic Bands | PPB (%) | PIC (%) |
|---|---|---|---|---|---|
| 1 | Cluster-86748.56403 | 10 | 10 | 100.00 | 26.53 |
| 2 | Cluster-99255.0 | 12 | 8 | 66.67 | 35.34 |
| 3 | Cluster-97571.0 | 10 | 5 | 50.00 | 14.84 |
| 4 | Cluster-94422.0 | 9 | 5 | 55.56 | 24.53 |
| 5 | Cluster-86748.83255 | 3 | 1 | 33.33 | 46.20 |
| 6 | Cluster-86748.82223 | 11 | 8 | 72.73 | 31.48 |
| 7 | Cluster-86748.81965 | 12 | 7 | 58.33 | 37.28 |
| 8 | Cluster-86748.81248 | 9 | 7 | 77.78 | 41.01 |
| 9 | Cluster-85378.1 | 12 | 10 | 83.33 | 23.39 |
| 10 | Cluster-66462.1 | 17 | 9 | 52.94 | 39.54 |
| 11 | Cluster-66335.1 | 15 | 11 | 73.33 | 34.72 |
| 12 | Cluster-65872.0 | 13 | 12 | 92.31 | 32.84 |
| 13 | Cluster-64532.0 | 3 | 1 | 33.33 | 49.76 |
| 14 | Cluster-45227.0 | 15 | 10 | 66.67 | 41.90 |
| 15 | Cluster-86748.54106 | 10 | 8 | 80.00 | 38.36 |
| 16 | Cluster-86748.20466 | 9 | 6 | 66.67 | 35.66 |
| 17 | Cluster-86748.4001 | 17 | 12 | 70.59 | 39.95 |
| 18 | Cluster-86748.55829 | 9 | 8 | 88.89 | 37.34 |
| 19 | Cluster-86748.33713 | 12 | 9 | 75.00 | 39.25 |
| 20 | Cluster-86748.16394 | 15 | 12 | 80.00 | 43.85 |
| 21 | Cluster-104317.0 | 16 | 10 | 62.50 | 37.08 |
| 22 | Cluster-86748.76270 | 15 | 8 | 53.33 | 28.05 |
| 23 | Cluster-86748.6834 | 10 | 8 | 80.00 | 26.33 |
| 24 | Cluster-86748.59079 | 8 | 5 | 62.50 | 20.10 |
| Total | - | 272 | 190 | - | - |
| Average | - | 11.33 | 7.92 | 68.16 | 34.39 |
| Primer | Gene ID | Associated Trait | NCBI Annotation |
|---|---|---|---|
| 1 | Cluster-86748.56403 | Root Cr, Leaf Cr, BF-Ug | High-affinity sulfate transporter |
| 24 | Cluster-86748.59079 | Leaf Cr, BF-Ag | High-affinity sulfate transporter |
| 4 | Cluster-94422.0 | Root Cr, Steam Cr, Leaf Cr, BF-Ug, BF-Ag | ABC transporter family |
| 5 | Cluster-86748.83255 | Root Cr, Steam Cr, Leaf Cr, BF-Ug, BF-Ag, TF | ABC transporter family |
| 7 | Cluster-86748.81965 | Leaf Cr, TF | ABC transporter family |
| 13 | Cluster-64532.0 | TF | ABC transporter family |
| 2 | Cluster-99255.0 | Root Cr, BF-Ug | ABC transporter family |
| 8 | Cluster-86748.81248 | Steam Cr, Leaf Cr, BF-Ag, TF | ABC transporter family |
| 9 | Cluster-85378.1 | Leaf Cr, TF, BF-Ag | ABC transporter family |
| 15 | Cluster-86748.54106 | Root Cr, BF-Ug, BF-Ag | Heavy metal ATPase (HMA) family |
| 16 | Cluster-86748.20466 | Root Cr, BF-Ug | Heavy metal ATPase (HMA) family |
| 19 | Cluster-86748.33713 | Root Cr, BF-Ug | Zinc/iron regulated transporter protein |
| 20 | Cluster-86748.16394 | Steam Cr, BF-Ag | Zinc/iron regulated transporter protein |
| 17 | Cluster-86748.4001 | Steam Cr, Leaf Cr, BF-Ag, TF | Wall-associated receptor kinase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, G.; Liu, A.; Ghanizadeh, H.; Wang, Y.; Tang, M.; He, J.; Feng, G.; Huang, L.; Zhang, X. Natural Variation in Chromium Accumulation and the Development of Related EST-SSR Molecular Markers in Miscanthus sinensis. Agronomy 2024, 14, 1458. https://doi.org/10.3390/agronomy14071458
Nie G, Liu A, Ghanizadeh H, Wang Y, Tang M, He J, Feng G, Huang L, Zhang X. Natural Variation in Chromium Accumulation and the Development of Related EST-SSR Molecular Markers in Miscanthus sinensis. Agronomy. 2024; 14(7):1458. https://doi.org/10.3390/agronomy14071458
Chicago/Turabian StyleNie, Gang, Aiyu Liu, Hossein Ghanizadeh, Yang Wang, Mingyu Tang, Jie He, Guangyan Feng, Linkai Huang, and Xinquan Zhang. 2024. "Natural Variation in Chromium Accumulation and the Development of Related EST-SSR Molecular Markers in Miscanthus sinensis" Agronomy 14, no. 7: 1458. https://doi.org/10.3390/agronomy14071458
APA StyleNie, G., Liu, A., Ghanizadeh, H., Wang, Y., Tang, M., He, J., Feng, G., Huang, L., & Zhang, X. (2024). Natural Variation in Chromium Accumulation and the Development of Related EST-SSR Molecular Markers in Miscanthus sinensis. Agronomy, 14(7), 1458. https://doi.org/10.3390/agronomy14071458







