Linking the Laboratory and the Field in Potato Early Dying Detection: From Spectral Signatures to Vegetation Indices Obtained with Multispectral Cameras Coupled to Drones
Abstract
:1. Introduction
2. Materials and Methods
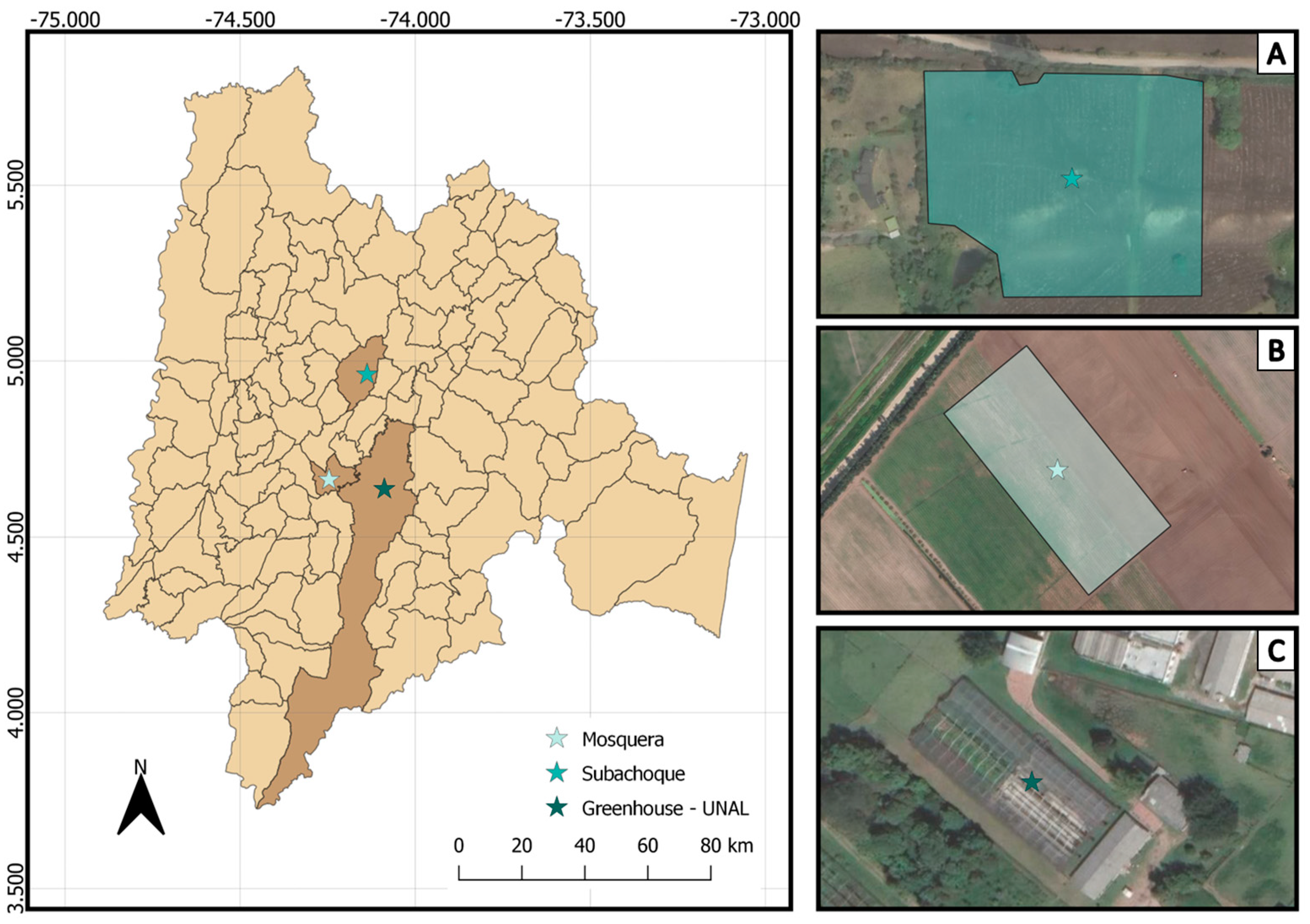
2.1. Location of the Greenhouse and Field for Experiments
2.2. Data Acquisition
2.2.1. Greenhouse Controlled Conditions
2.2.2. Field Conditions
2.3. Data Analysis
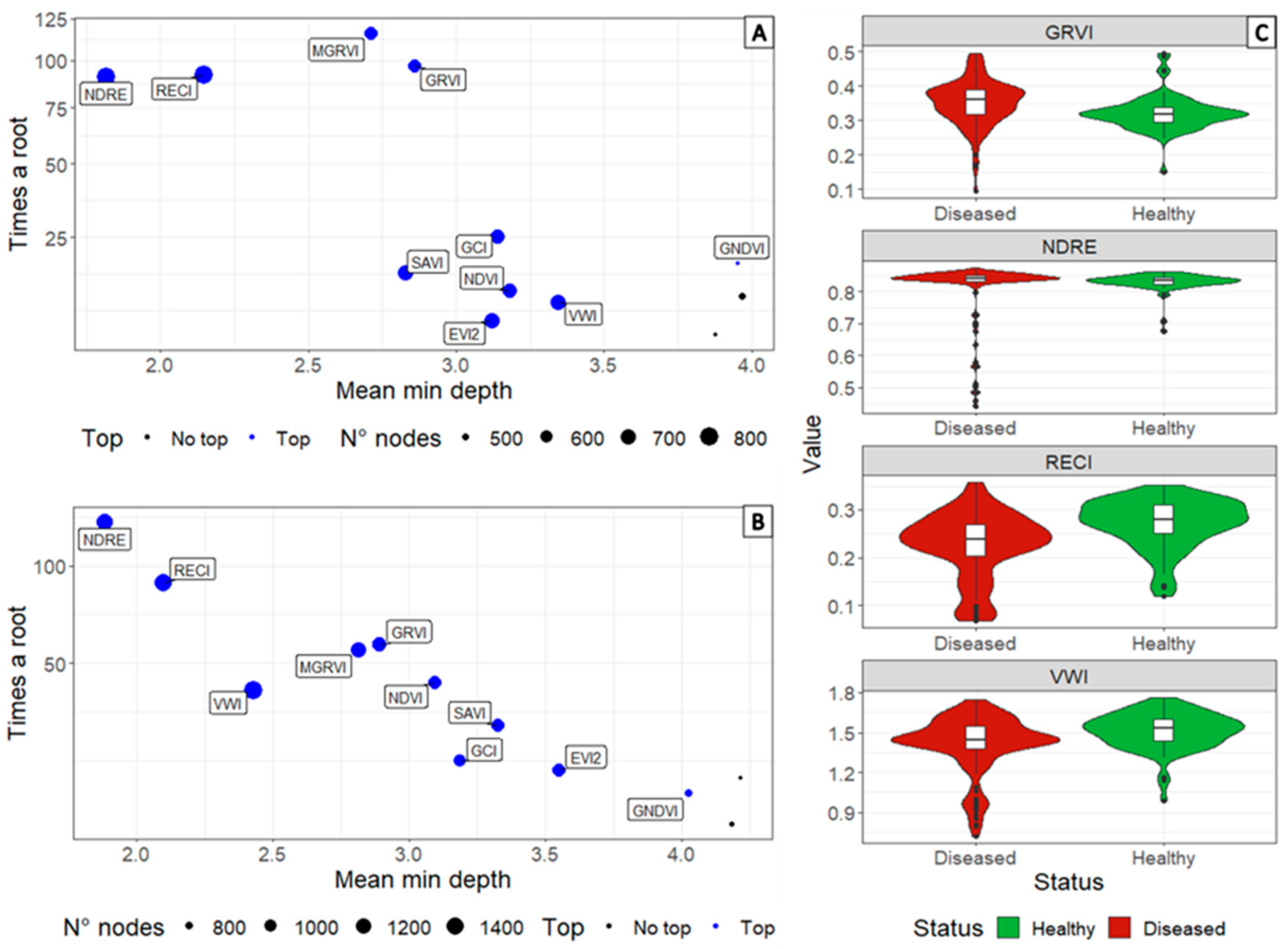
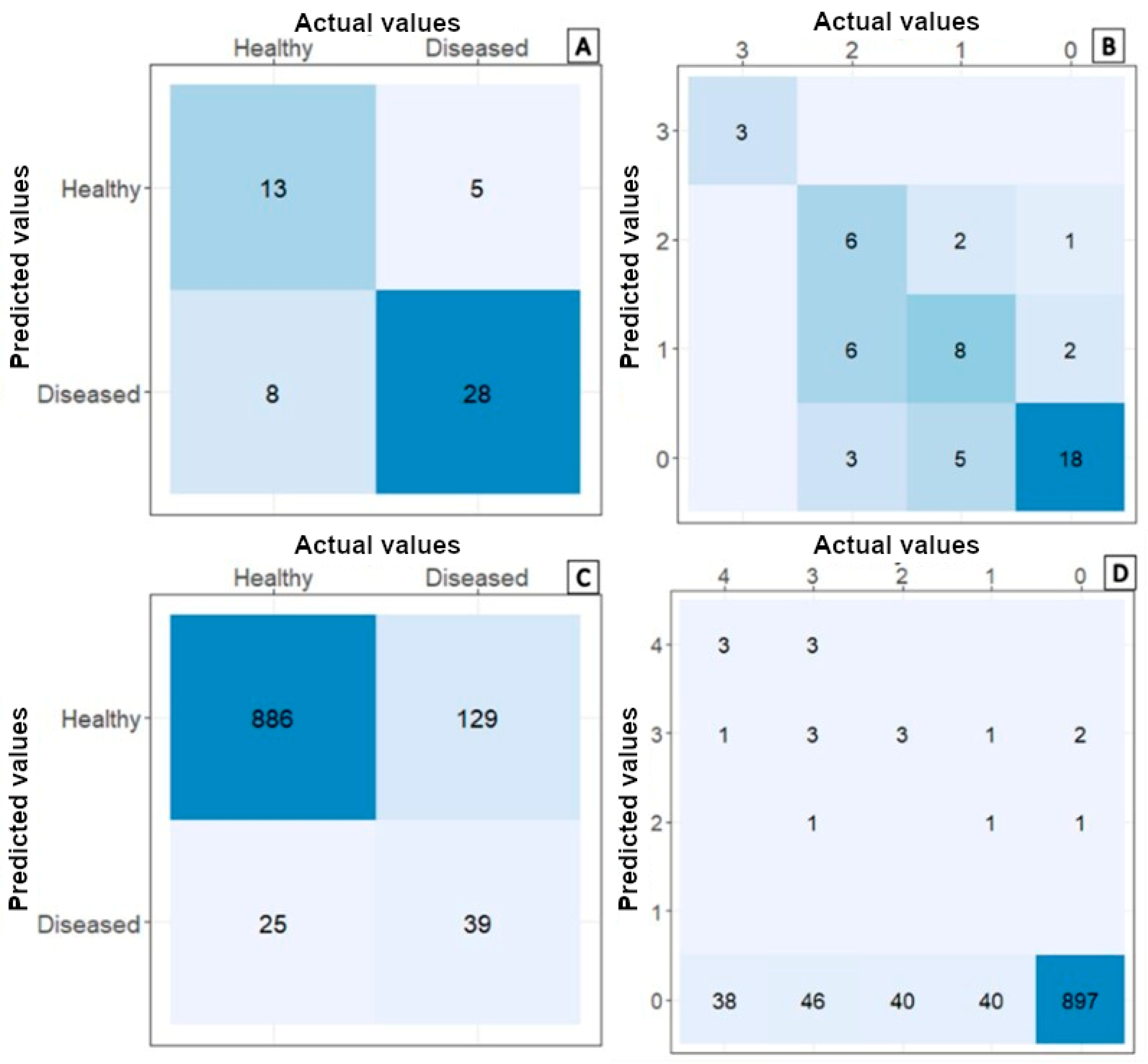
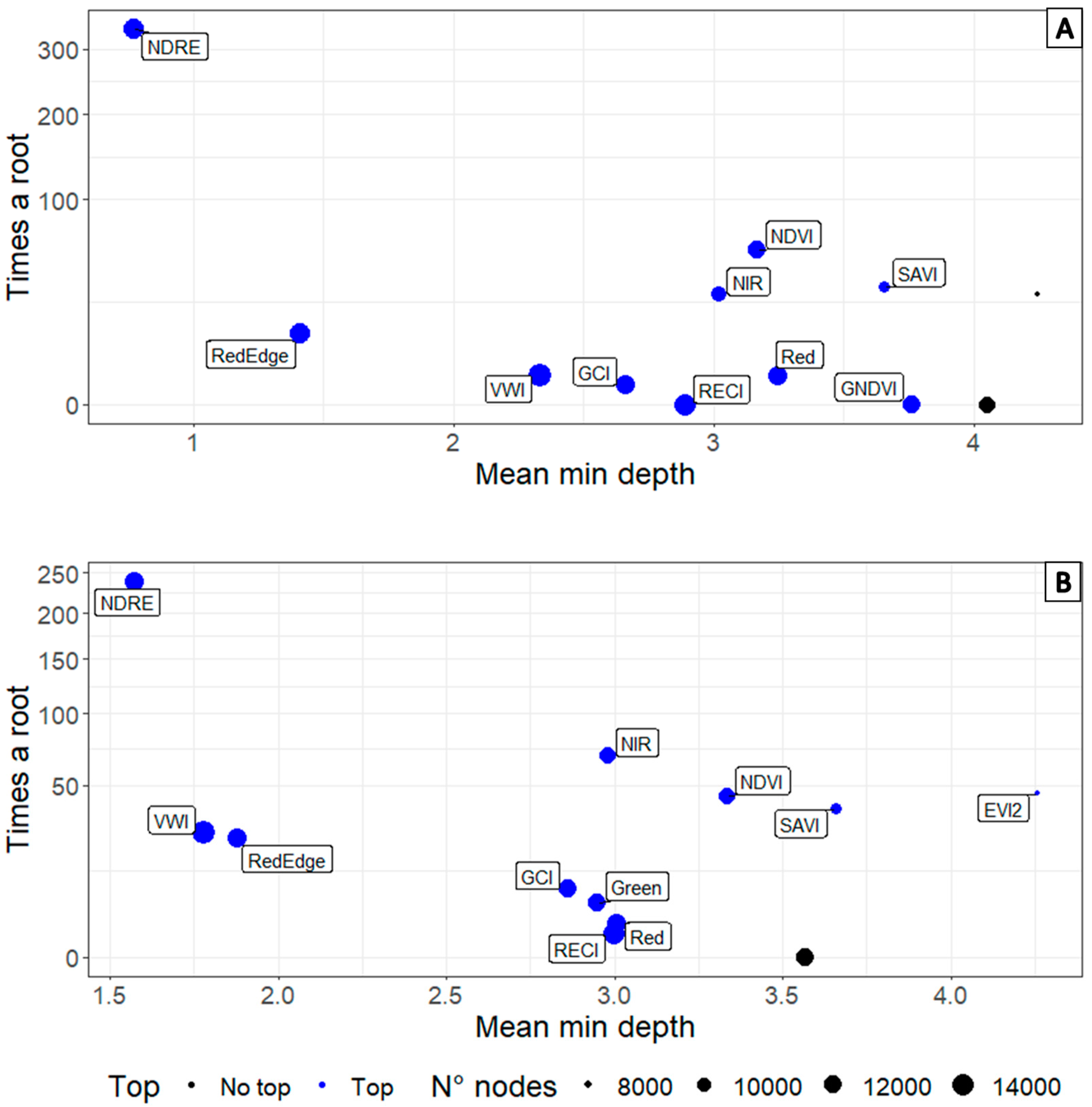
3. Results
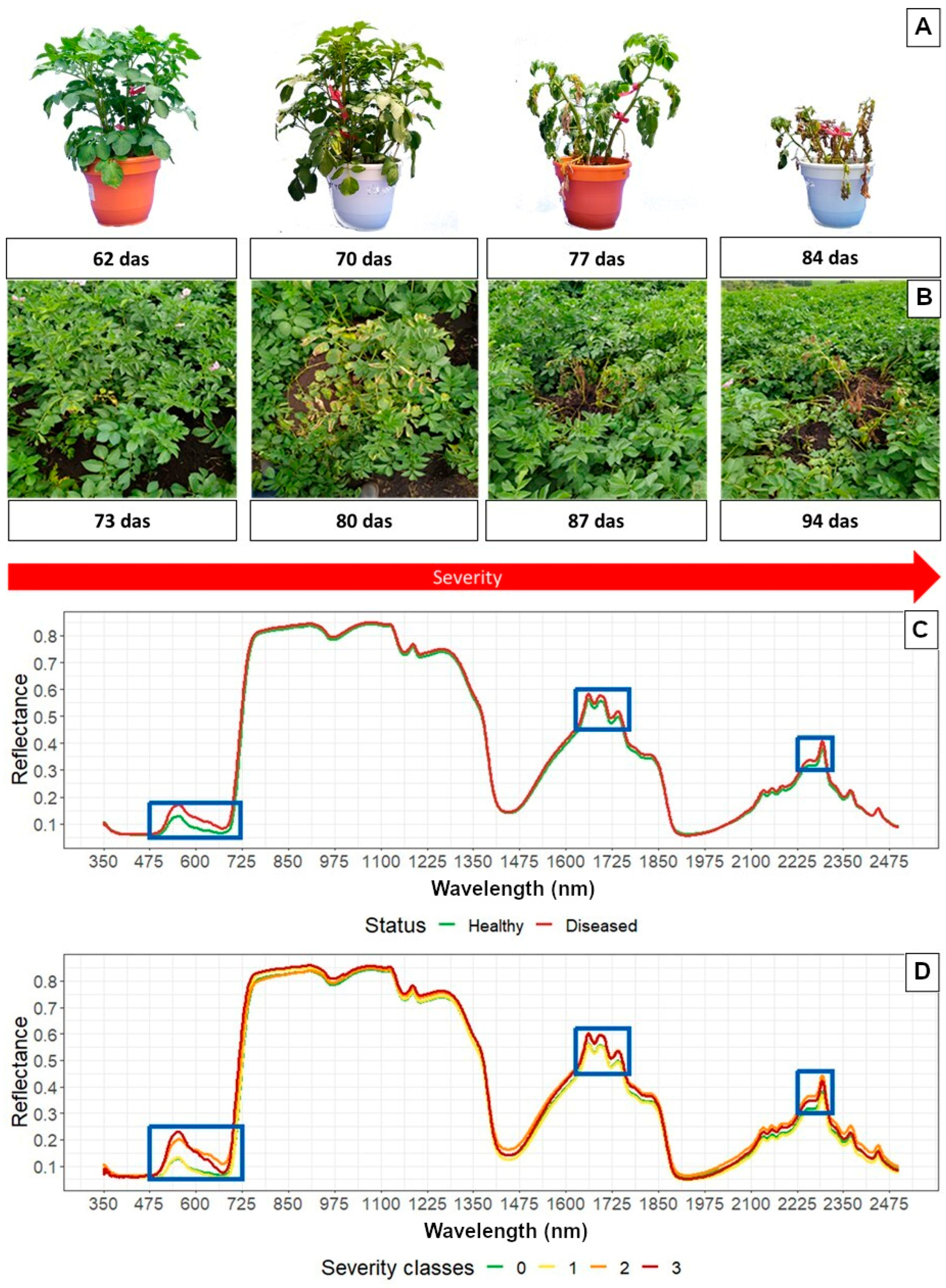
3.1. Symptoms under Greenhouse and Field Conditions Associated with PED
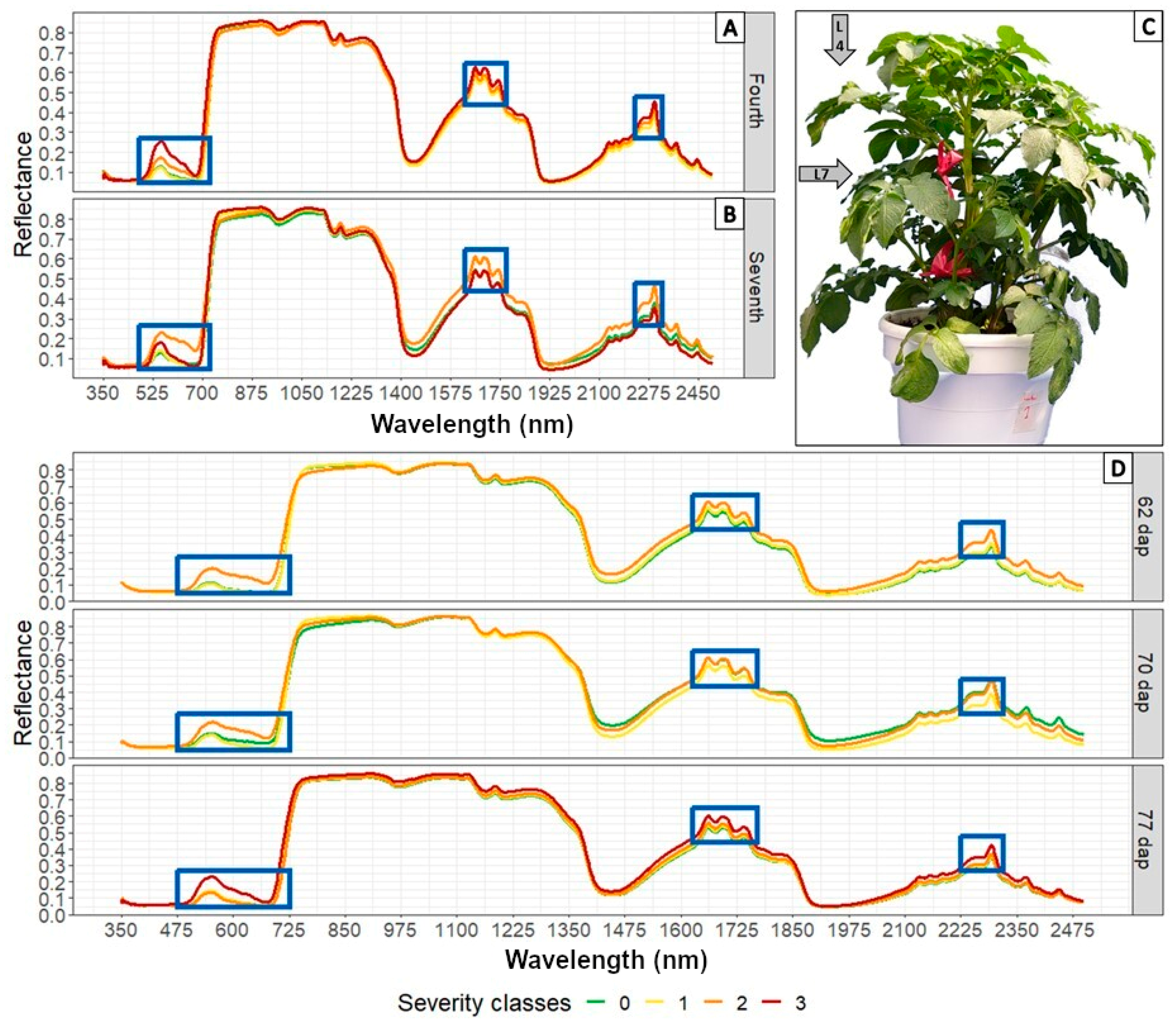
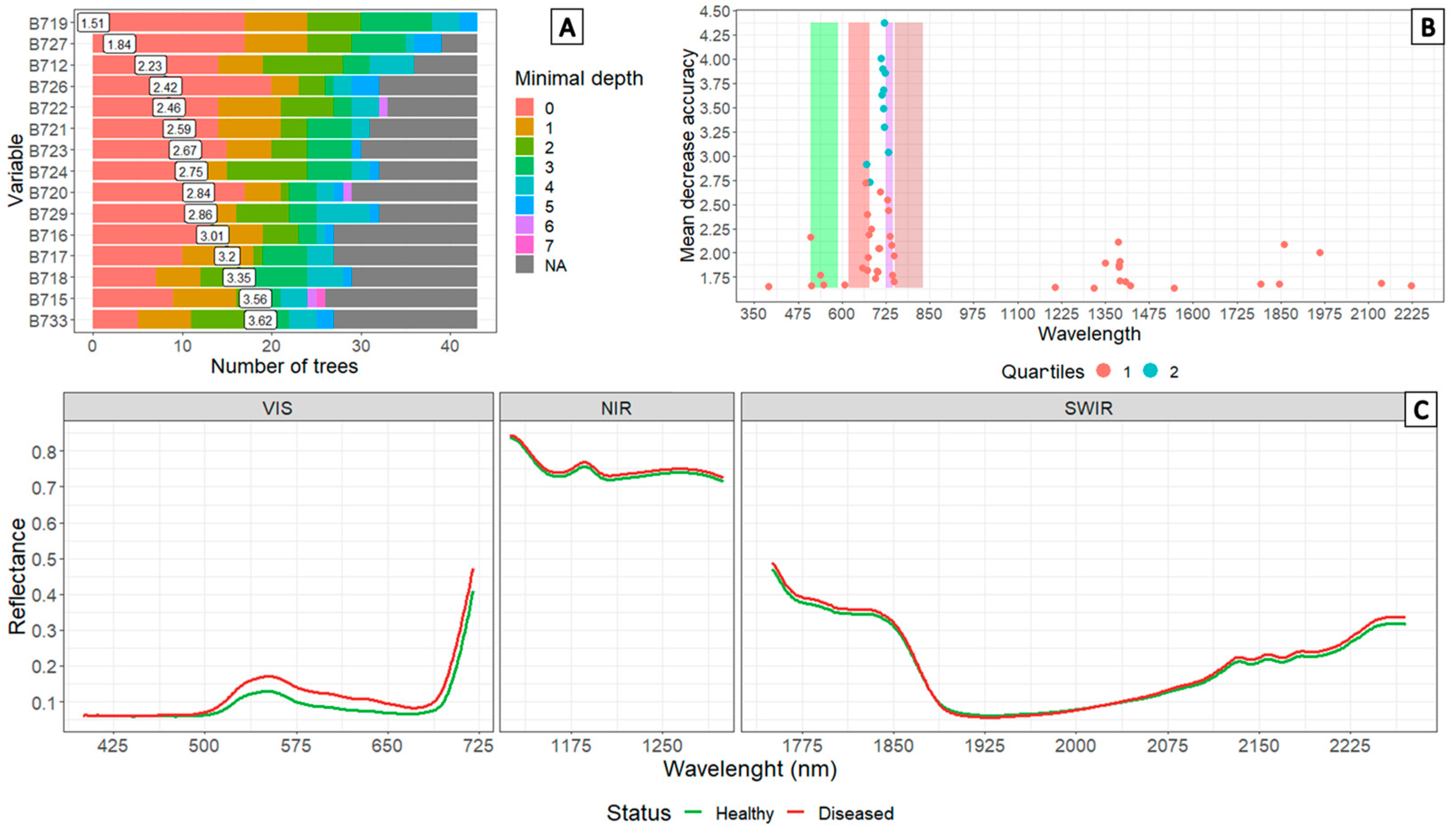
3.2. Analysis of Spectral Signatures Obtained in Plants Subjected to Artificial Infection in a Greenhouse
3.3. Determination of Informative Bands to Discriminate between Healthy and Diseased Plants
3.4. Comparison of Spectral Signatures and Multispectral Camera Data Obtained in Commercial Crops
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Index | Equation | References |
|---|---|---|
| Verticillium Wilt Index (VWI) | (A1) | Proposed Index |
| Soil Adjusted Vegetation Index (SAVI) * | (A2) | [49] |
| Enhanced Vegetation Index (EVI2) ** | (A3) | [49] |
| Green Normalized Difference Vegetation Index (GNDVI) | (A4) | [50] |
| Green–Red Vegetation Index (GRVI) | (A5) | [48,51] |
| Modified Green–Red Vegetation Index (MGRVI) | (A6) | [51] |
| Green Chlorophyll Index (GCI) | (A7) | [47] |
| Red Edge Chlorophyll Index (RECI) | (A8) | [47] |
| Normalized Difference Red Edge Index (NDRE) | (A9) | [51] |
| Chlorophyll Index Green (CIGreen) | (A10) | [52] |
| Anthocyanin Reflectance Index (ARI) | (A11) | [45] |
| Anthocyanin Reflectance Index (CARI) | (A12) | [53] |
| Normalized Difference Vegetation Index (NDVI) | (A13) | [54] |
References
- Del Mar Martínez-Prada, M.; Curtin, S.J.; Gutiérrez-González, J.J. Potato improvement through genetic engineering. GM Crop. Food 2022, 12, 479–496. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT FAOSTAT: Statistical Database. Available online: https://www.fao.org/faostat/es/#home (accessed on 17 April 2022).
- Li, H.; Wang, Z.; Hu, X.; Shang, W.; Shen, R.; Guo, C.; Guo, Q.; Subbarao, K.V. Assessment of Resistance in Potato Cultivars to Verticillium Wilt Caused by Verticillium Dahliae and Verticillium Nonalfalfae. Plant Dis. 2019, 103, 1357–1362. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.A.; Jeremiah, K.; Dung, S. Verticillium Wilt of Potato—The Pathogen, Disease and Management. Can. J. Plant Pathol. 2010, 32, 58–67. [Google Scholar] [CrossRef]
- Shattock, R. Compendium of Potato Diseases, Second Edition. W.R. Stevenson. Plant Pathol. 2002, 51, 520. [Google Scholar] [CrossRef]
- Ramegowda, V.; Senthil-Kumar, M. The Interactive Effects of Simultaneous Biotic and Abiotic Stresses on Plants: Mechanistic Understanding from Drought and Pathogen Combination. J. Plant Physiol. 2015, 176, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Kollist, H.; Zandalinas, S.I.; Sengupta, S.; Nuhkat, M.; Kangasjärvi, J.; Mittler, R. Rapid Responses to Abiotic Stress: Priming the Landscape for the Signal Transduction Network. Trends Plant Sci. 2019, 24, 25–37. [Google Scholar] [CrossRef]
- Galieni, A.; D’Ascenzo, N.; Stagnari, F.; Pagnani, G.; Xie, Q.; Pisante, M. Past and Future of Plant Stress Detection: An Overview From Remote Sensing to Positron Emission Tomography. Front. Plant Sci. 2021, 11, 1975. [Google Scholar] [CrossRef] [PubMed]
- Oerke, E.C. Remote Sensing of Diseases. Annu. Rev. Phytopathol. 2020, 58, 225–252. [Google Scholar] [CrossRef] [PubMed]
- Buja, I.; Sabella, E.; Monteduro, A.G.; Chiriacò, M.S.; De Bellis, L.; Luvisi, A.; Maruccio, G. Advances in Plant Disease Detection and Monitoring: From Traditional Assays to In-Field Diagnostics. Sensors 2021, 21, 2129. [Google Scholar] [CrossRef]
- Mahlein, A.K. Plant Disease Detection by Imaging Sensors—Parallels and Specific Demands for Precision Agriculture and Plant Phenotyping. Plant Dis. 2016, 100, 241–254. [Google Scholar] [CrossRef]
- Liu, L.; Dong, Y.; Huang, W.; Du, X.; Ren, B.; Huang, L.; Zheng, Q.; Ma, H. A Disease Index for Efficiently Detecting Wheat Fusarium Head Blight Using Sentinel-2 Multispectral Imagery. IEEE Access 2020, 8, 52181–52191. [Google Scholar] [CrossRef]
- Singh, V.; Sharma, N.; Singh, S. A Review of Imaging Techniques for Plant Disease Detection. Artif. Intell. Agric. 2020, 4, 229–242. [Google Scholar] [CrossRef]
- León-Rueda, W.A.; León, C.; Caro, S.G.; Ramírez-Gil, J.G. Identification of Diseases and Physiological Disorders in Potato via Multispectral Drone Imagery Using Machine Learning Tools. Trop. Plant Pathol. 2022, 47, 152–167. [Google Scholar] [CrossRef]
- Ramesh Reddy, D.; Naga Santhosh, K.; Kodali, P. Convolutional Neural Networks for the Intuitive Identification of Plant Diseases. In Proceedings of the 5th International Conference on Inventive Computation Technologies, ICICT 2022—Proceedings, Lalitpur, Nepal, 20–22 July 2022; Volume 10, p. 941. [Google Scholar]
- Baldi, P.; La Porta, N. Molecular Approaches for Low-Cost Point-of-Care Pathogen Detection in Agriculture and Forestry. Front. Plant Sci. 2020, 11, 570862. [Google Scholar] [CrossRef] [PubMed]
- Marín-Ortiz, J.C.; Gutierrez-Toro, N.; Botero-Fernández, V.; Hoyos-Carvajal, L.M. Linking Physiological Parameters with Visible/near-Infrared Leaf Reflectance in the Incubation Period of Vascular Wilt Disease. Saudi J. Biol. Sci. 2020, 27, 88. [Google Scholar] [CrossRef] [PubMed]
- Couture, J.J.; Singh, A.; Charkowski, A.O.; Groves, R.L.; Gray, S.M.; Bethke, P.C.; Townsend, P.A. Integrating Spectroscopy with Potato Disease Management. Plant Dis. 2018, 102, 2233–2240. [Google Scholar] [CrossRef] [PubMed]
- Imanian, K.; Pourdarbani, R.; Sabzi, S.; García-Mateos, G.; Arribas, J.I.; Molina-Martínez, J.M. Identification of Internal Defects in Potato Using Spectroscopy and Computational Intelligence Based on Majority Voting Techniques. Foods 2021, 10, 982. [Google Scholar] [CrossRef]
- Agilandeeswari, L.; Prabukumar, M.; Radhesyam, V.; Phaneendra, K.L.N.B.; Farhan, A. Crop Classification for Agricultural Applications in Hyperspectral Remote Sensing Images. Appl. Sci. 2022, 12, 1670. [Google Scholar] [CrossRef]
- Moshou, D.; Bravo, C.; Oberti, R.; West, J.S.; Ramon, H.; Vougioukas, S.; Bochtis, D. Intelligent Multi-Sensor System for the Detection and Treatment of Fungal Diseases in Arable Crops. Biosyst. Eng. 2011, 108, 311–321. [Google Scholar] [CrossRef]
- Wei, X.; Johnson, M.A.; Langston, D.B.; Mehl, H.L.; Li, S. Identifying Optimal Wavelengths as Disease Signatures Using Hyperspectral Sensor and Machine Learning. Remote Sens. 2021, 13, 2833. [Google Scholar] [CrossRef]
- Meng, R.; Lv, Z.; Yan, J.; Chen, G.; Zhao, F.; Zeng, L.; Xu, B. Development of Spectral Disease Indices for Southern Corn Rust Detection and Severity Classification. Remote Sens. 2020, 12, 3233. [Google Scholar] [CrossRef]
- Sun, W.; Du, Q. Hyperspectral Band Selection: A Review. IEEE Geosci. Remote Sens. Mag. 2019, 7, 118–139. [Google Scholar] [CrossRef]
- Junges, A.H.; Almança, M.A.K.; Fajardo, T.V.M.; Ducati, J.R. Leaf Hyperspectral Reflectance as a Potential Tool to Detect Diseases Associated with Vineyard Decline. Trop. Plant Pathol. 2020, 45, 522–533. [Google Scholar] [CrossRef]
- Mountrakis, G.; Im, J.; Ogole, C. Support Vector Machines in Remote Sensing: A Review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Rodríguez, J.; Lizarazo, I.; Prieto, F.; Angulo-Morales, V. Assessment of Potato Late Blight from UAV-Based Multispectral Imagery. Comput. Electron. Agric. 2021, 184, 106061. [Google Scholar] [CrossRef]
- Lizarazo, I.; Rodriguez, J.L.; Cristancho, O.; Olaya, F.; Duarte, M.; Prieto, F. Identification of Symptoms Related to Potato Verticillium Wilt from UAV-Based Multispectral Imagery Using an Ensemble of Gradient Boosting Machines. Smart Agric. Technol. 2023, 3, 100138. [Google Scholar] [CrossRef]
- Shin, M.Y.; Gonzalez Viejo, C.; Tongson, E.; Wiechel, T.; Taylor, P.W.J.; Fuentes, S. Early Detection of Verticillium Wilt of Potatoes Using Near-Infrared Spectroscopy and Machine Learning Modeling. Comput. Electron. Agric. 2023, 204, 107567. [Google Scholar] [CrossRef]
- Ashraf, A.; Rauf, A.; Fahim Abbas, M.; Rehman, R. Isolation and Identification of Verticillium Dahliae Causing Wilt on Potato in Pakistan. J. Phytopathol. 2012, 24, 112–116. [Google Scholar]
- Hunter, D.E.; Darling, H.M.; Stevenson, F.J.; Cunningham, C.E. Inheritance of Resistance to Verticillium Wilt in Wisconsin. Am. Potato J. 1968, 45, 72–78. [Google Scholar] [CrossRef]
- Mauromicale, G.; Ierna, A.; Marchese, M. Chlorophyll Fluorescence and Chlorophyll Content in Field-Grown Potato as Affected by Nitrogen Supply, Genotype, and Plant Age. Photosynthetica 2006, 44, 76–82. [Google Scholar] [CrossRef]
- Ren, Y.; Zhang, L.; Suganthan, P.N. Ensemble Classification and Regression-Recent Developments, Applications and Future Directions [Review Article]. IEEE Comput. Intell. Mag. 2016, 11, 41–53. [Google Scholar] [CrossRef]
- Gholami, R.; Fakhari, N. Support Vector Machine: Principles, Parameters, and Applications. In Handbook of Neural Computation; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 515–535. ISBN 9780128113196. [Google Scholar]
- Wang, J.; Lu, S.; Wang, S.H.; Zhang, Y.D. A Review on Extreme Learning Machine. Multimed. Tools Appl. 2021, 81, 41611–41660. [Google Scholar] [CrossRef]
- Gupta, S.; Saluja, K.; Goyal, A.; Vajpayee, A.; Tiwari, V. Comparing the Performance of Machine Learning Algorithms Using Estimated Accuracy. Meas. Sens. 2022, 24, 100432. [Google Scholar] [CrossRef]
- Kuhn, M. Building Predictive Models in R Using the Caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Hopkins, D.W. What Is a Norris Derivative? NIR News 2001, 12, 3–5. [Google Scholar] [CrossRef]
- Stevens, A.; Ramirez Lopez, L. R Package Vignette, Report No.: R Package, Version 0.1. In An Introduction to the Prospectr Package; 2014; Volume 3, pp. 1–22. Available online: https://www.researchgate.net/publication/255941339_An_introduction_to_the_prospectr_package (accessed on 15 July 2024).
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Gold, K.M.; Townsend, P.A.; Chlus, A.; Herrmann, I.; Couture, J.J.; Larson, E.R.; Gevens, A.J. Hyperspectral Measurements Enable Pre-Symptomatic Detection and Differentiation of Contrasting Physiological Effects of Late Blight and Early Blight in Potato. Remote Sens. 2020, 12, 286. [Google Scholar] [CrossRef]
- Singh, A.; Kaur, H. Potato Plant Leaves Disease Detection and Classification Using Machine Learning Methodologies. IOP Conf. Ser. Mater. Sci. Eng. 2021, 1022, 012121. [Google Scholar] [CrossRef]
- Calderón, R.; Navas-Cortés, J.A.; Zarco-Tejada, P.J. Early Detection and Quantification of Verticillium Wilt in Olive Using Hyperspectral and Thermal Imagery over Large Areas. Remote Sens. 2015, 7, 5584–5610. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Chivkunova, O.B. Optical Properties and Nondestructive Estimation of Anthocyanin Content in Plant Leaves. Photochem. Photobiol. 2001, 74, 38. [Google Scholar] [CrossRef]
- Li, H.; Yang, W.; Lei, J.; She, J.; Zhou, X. Estimation of Leaf Water Content from Hyperspectral Data of Different Plant Species by Using Three New Spectral Absorption Indices. PLoS ONE 2021, 16, e0249351. [Google Scholar] [CrossRef] [PubMed]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between Leaf Chlorophyll Content and Spectral Reflectance and Algorithms for Non-Destructive Chlorophyll Assessment in Higher Plant Leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Motohka, T.; Nasahara, K.N.; Oguma, H.; Tsuchida, S. Applicability of Green-Red Vegetation Index for Remote Sensing of Vegetation Phenology. Remote Sens. 2010, 2, 2369–2387. [Google Scholar] [CrossRef]
- Jiang, Z.; Huete, A.R.; Didan, K.; Miura, T. Development of a Two-Band Enhanced Vegetation Index without a Blue Band. Remote Sens. Environ. 2008, 112, 3833–3845. [Google Scholar] [CrossRef]
- Yang, C.M.; Cheng, C.H.; Chen, R.K. Changes in Spectral Characteristics of Rice Canopy Infested with Brown Planthopper and Leaffolder. Crop Sci. 2007, 47, 329–335. [Google Scholar] [CrossRef]
- Yeom, J.; Jung, J.; Chang, A.; Ashapure, A.; Maeda, M.; Maeda, A.; Landivar, J. Comparison of Vegetation Indices Derived from UAV Data for Differentiation of Tillage Effects in Agriculture. Remote Sens. 2019, 11, 1548. [Google Scholar] [CrossRef]
- Xie, Q.; Dash, J.; Huang, W.; Peng, D.; Qin, Q.; Mortimer, H.; Casa, R.; Pignatti, S.; Laneve, G.; Pascucci, S.; et al. Vegetation Indices Combining the Red and Red-Edge Spectral Information for Leaf Area Index Retrieval. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 1482–1492. [Google Scholar] [CrossRef]
- Zhou, X.; Huang, W.; Zhang, J.; Kong, W.; Casa, R.; Huang, Y. A Novel Combined Spectral Index for Estimating the Ratio of Carotenoid to Chlorophyll Content to Monitor Crop Physiological and Phenological Status. Int. J. Appl. Earth Obs. Geoinf. 2019, 76, 128–142. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS. NASA Spec. Publ. 1973, 351, 309. [Google Scholar]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
León-Rueda, W.A.; Gómez-Caro, S.; Mendoza-Vargas, L.A.; León-Sánchez, C.A.; Ramírez-Gil, J.G. Linking the Laboratory and the Field in Potato Early Dying Detection: From Spectral Signatures to Vegetation Indices Obtained with Multispectral Cameras Coupled to Drones. Agronomy 2024, 14, 1569. https://doi.org/10.3390/agronomy14071569
León-Rueda WA, Gómez-Caro S, Mendoza-Vargas LA, León-Sánchez CA, Ramírez-Gil JG. Linking the Laboratory and the Field in Potato Early Dying Detection: From Spectral Signatures to Vegetation Indices Obtained with Multispectral Cameras Coupled to Drones. Agronomy. 2024; 14(7):1569. https://doi.org/10.3390/agronomy14071569
Chicago/Turabian StyleLeón-Rueda, William A., Sandra Gómez-Caro, Luis A. Mendoza-Vargas, Camilo A. León-Sánchez, and Joaquín G. Ramírez-Gil. 2024. "Linking the Laboratory and the Field in Potato Early Dying Detection: From Spectral Signatures to Vegetation Indices Obtained with Multispectral Cameras Coupled to Drones" Agronomy 14, no. 7: 1569. https://doi.org/10.3390/agronomy14071569
APA StyleLeón-Rueda, W. A., Gómez-Caro, S., Mendoza-Vargas, L. A., León-Sánchez, C. A., & Ramírez-Gil, J. G. (2024). Linking the Laboratory and the Field in Potato Early Dying Detection: From Spectral Signatures to Vegetation Indices Obtained with Multispectral Cameras Coupled to Drones. Agronomy, 14(7), 1569. https://doi.org/10.3390/agronomy14071569







