Molecular Mechanism of Pyrroloquinoline Quinone-Mediated Rice Tolerance to Imidazolinone Herbicide Imazamox
Abstract
:1. Introduction
2. Materials and Methods
2.1. Rice Variety and Chemicals
2.2. Rice Cultivation and Experimental Treatment Design
2.3. Transcriptome Sequencing
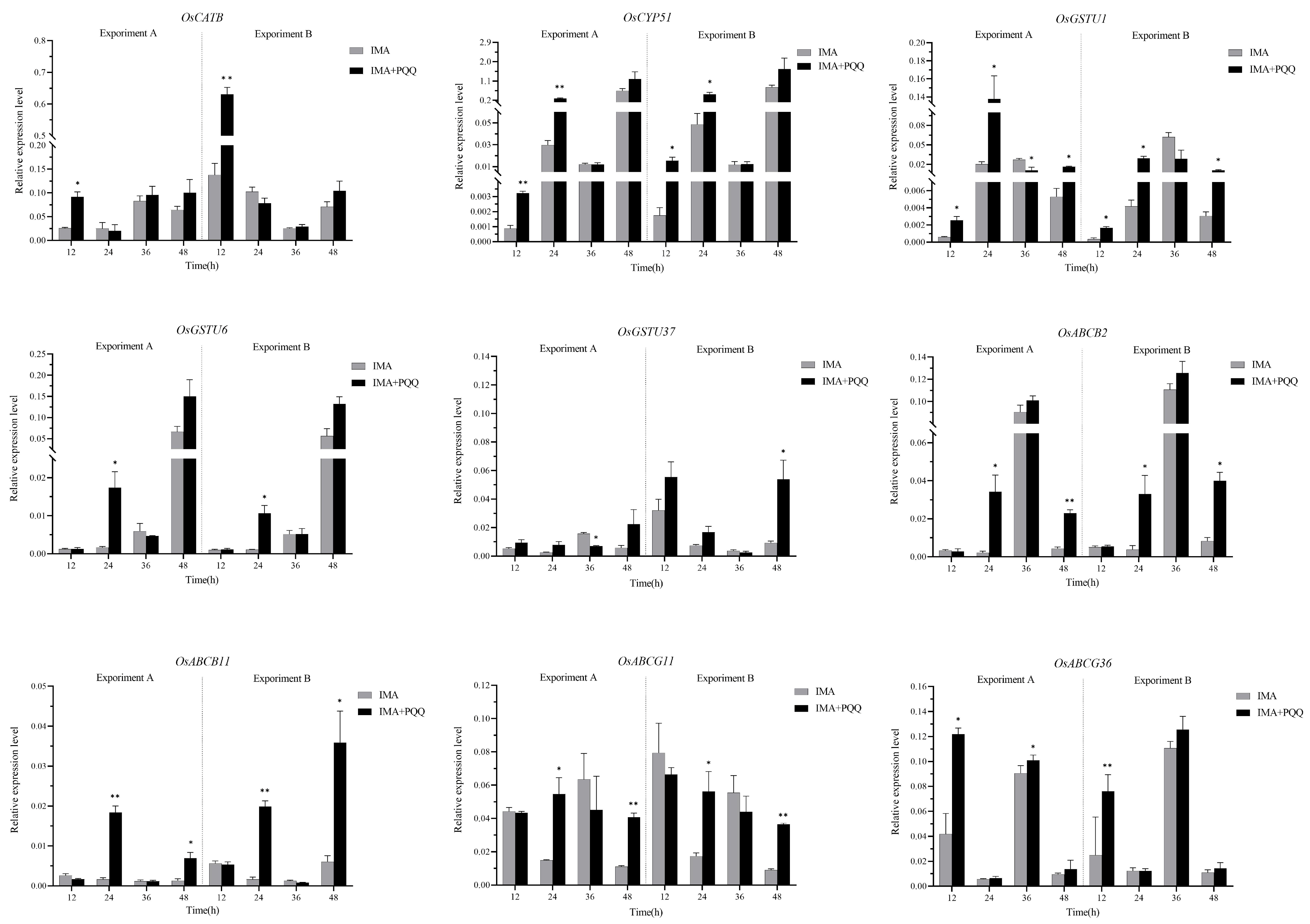
2.4. Validation of the DEGs by RT-qPCR
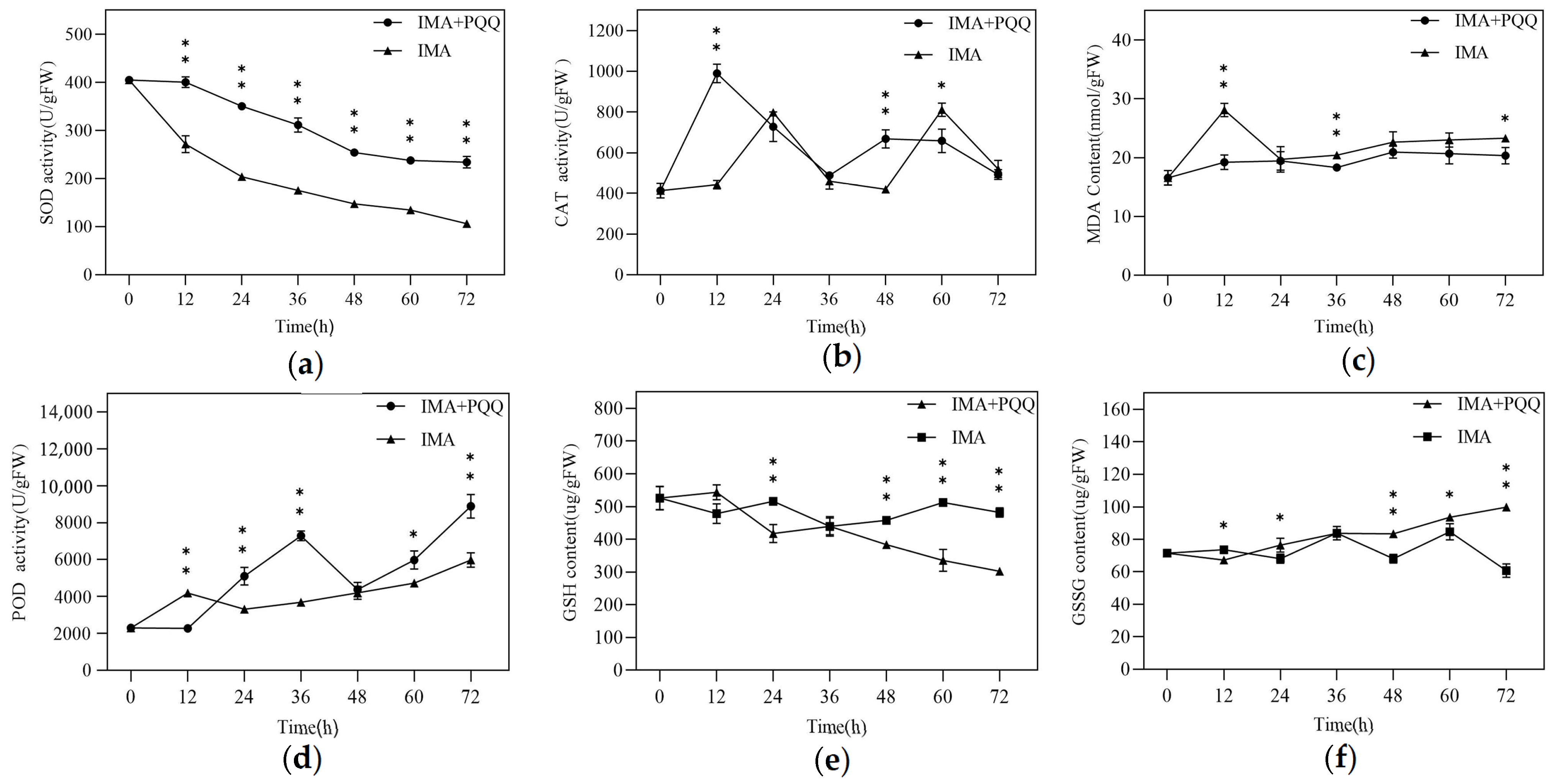
2.5. Determination of Enzymatic Activity Indicators
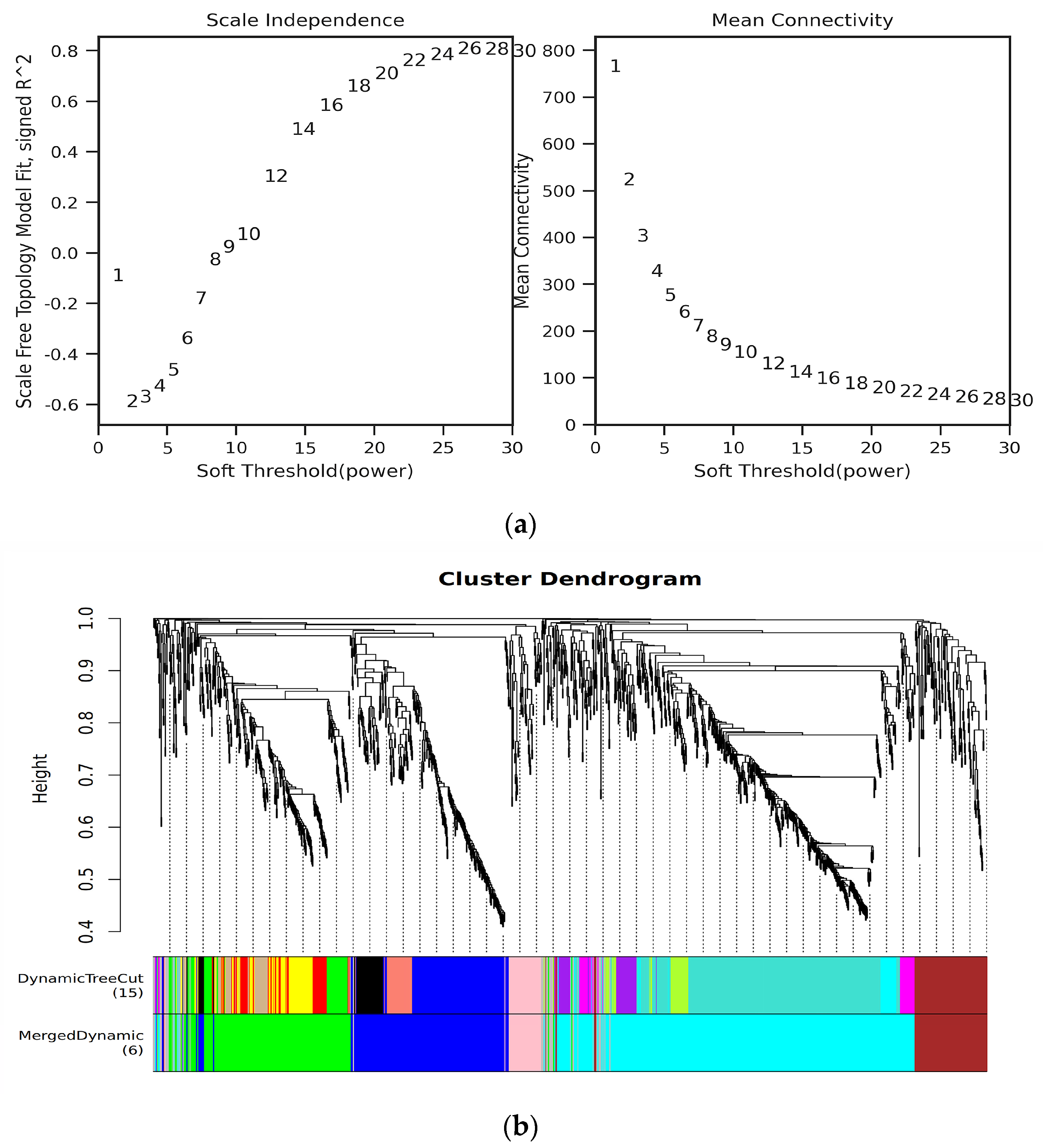
2.6. Data Analyses
3. Results
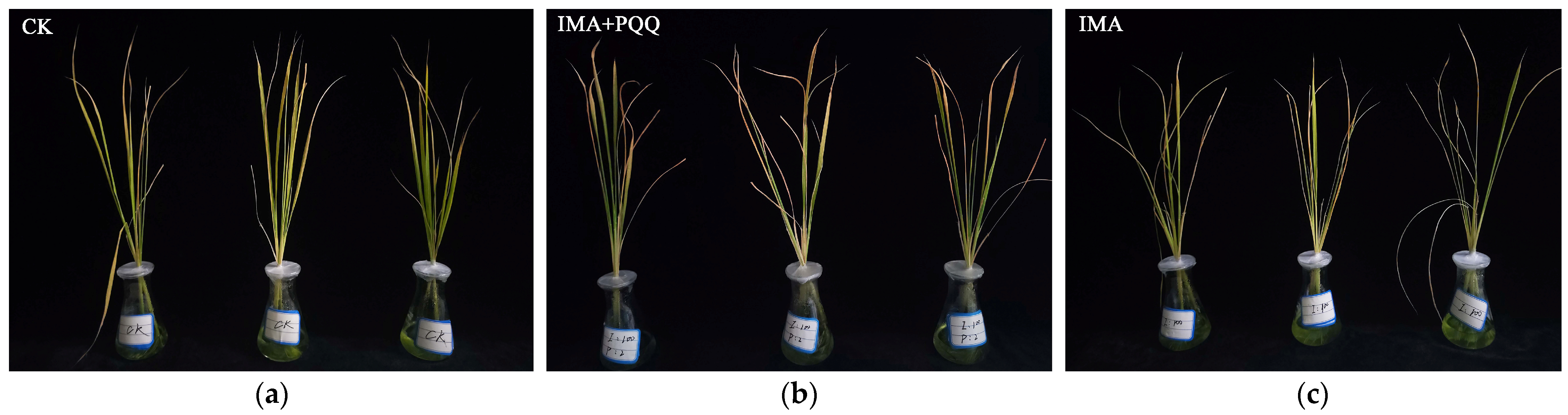
3.1. PQQ-Induced Rice Tolerance to Imazamox
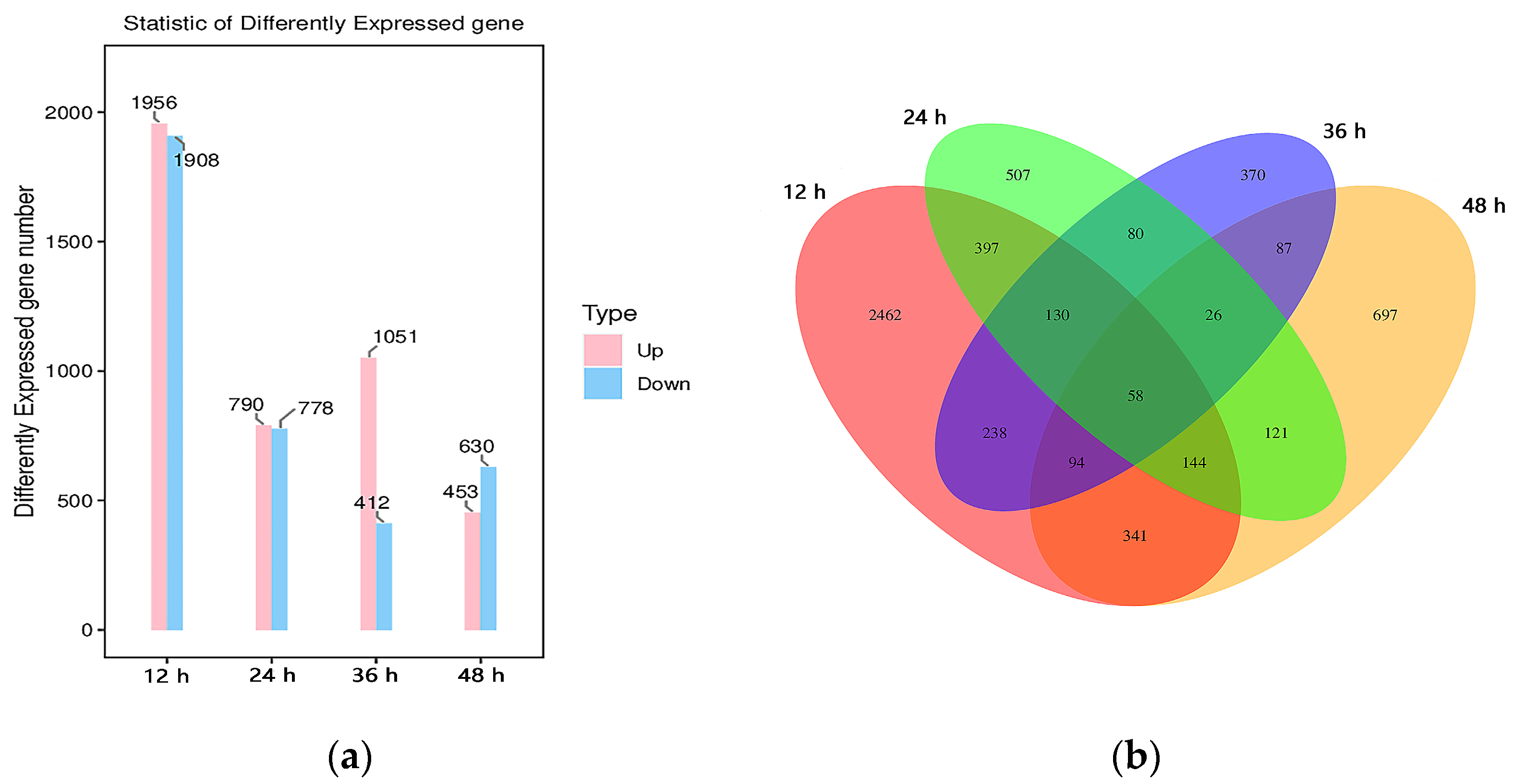
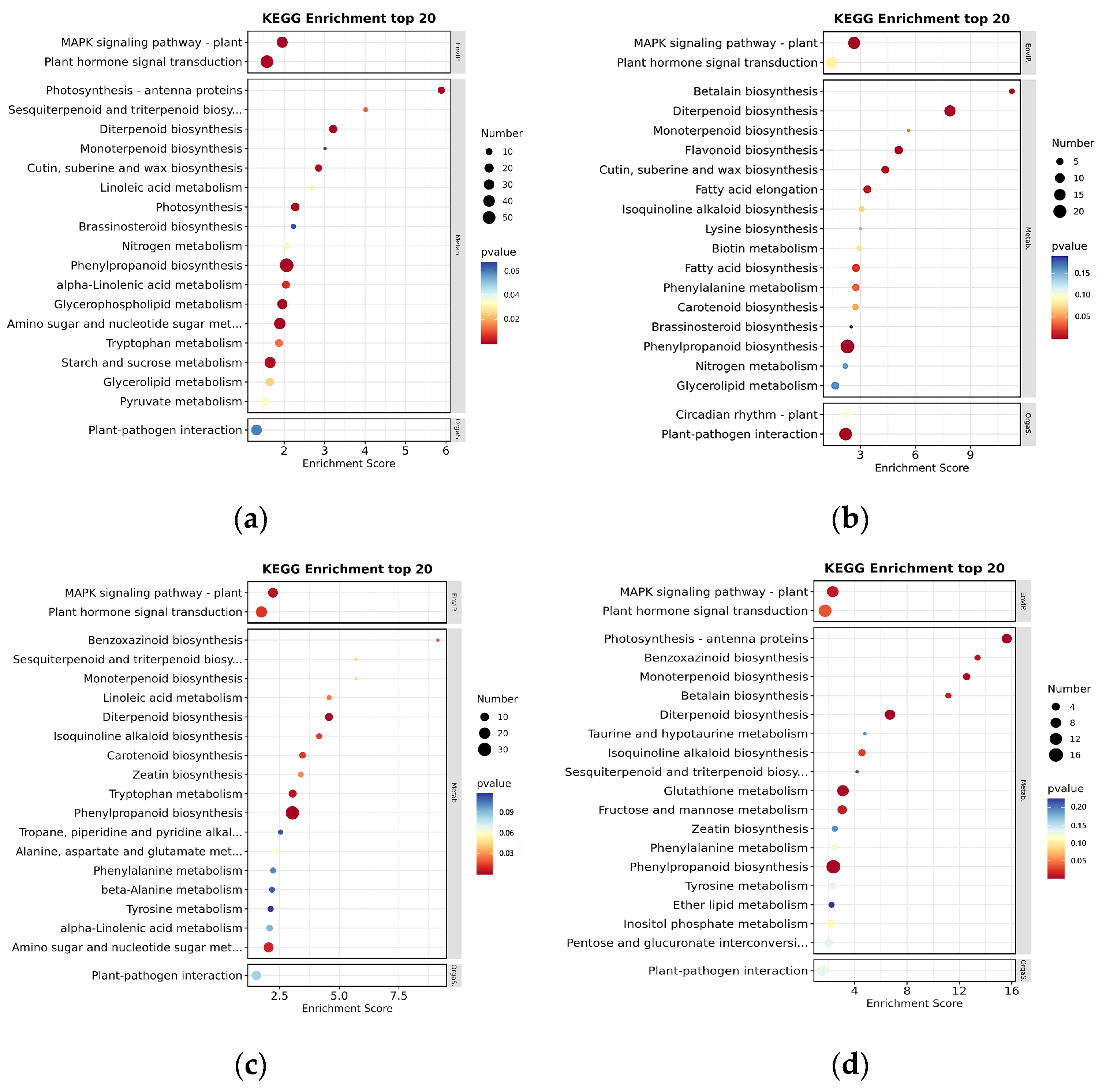
3.2. Analysis of DEGs by RNA-Seq
3.3. Verification of the DEGs by RT-qPCR Assay
3.4. Key Indicators of Enzyme Activity in Redox Processes and Related Detoxification Enzymes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burgos, N.R.; Singh, V.; Tseng, T.M.; Black, H.; Young, N.D.; Huang, Z.; Hyma, K.E.; Caicedo, A.L.; Gealy, D.R. The impact of herbicide-resistant rice technology on phenotypic diversity and population structure of United States weedy rice. Plant Physiol. 2014, 166, 1208–1220. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Ma, X.; Cui, D.; Wang, Y.; Geng, L.; Cao, G.; Zhang, H.; Han, L. Comprehensive evaluation and analysis of the mechanism of cold tolerance based on the transcriptome of weedy rice seedlings. Rice 2020, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H. Weedy rice complexes: Case studies from Malaysia, Vietnam, and Surinam. In Proceedings of the Wild and Weedy Rice in Rice Ecosystems in Asia—A Review (Ho Chi Minh City, Vietnam, 10–11 August 1998); International Rice Research Institute: Laguna, Philippines, 2000; pp. 25–34. [Google Scholar]
- Zhang, Y.; Fang, J.; Wu, X.; Dong, L. Na+/K+ balance and transport regulatory mechanisms in weedy and cultivated rice (Oryza sativa L.) under salt stress. BMC Plant Biol. 2018, 18, 375. [Google Scholar] [CrossRef] [PubMed]
- Baek, J.S.; Chung, N.J. Seed wintering and deterioration characteristics between weedy and cultivated rice. Rice 2012, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Sudianto, E.; Beng-Kah, S.; Ting-Xiang, N.; Saldain, N.E.; Scott, R.C.; Burgos, N.R. Clearfield rice: Its development, success, and key challenges on a global perspective. Crop Prot. 2013, 49, 40–51. [Google Scholar] [CrossRef]
- de Avila, L.A.; Marchesan, E.; Camargo, E.R.; Merotto, A.; da Rosa Ulguim, A.; Noldin, J.A.; Andres, A.; Mariot, C.H.; Agostinetto, D.; Dornelles, S.H.; et al. Eighteen years of Clearfield™ rice in Brazil: What have we learned? Weed Sci. 2021, 69, 585–597. [Google Scholar] [CrossRef]
- Grover, N.; Kumar, A.; Yadav, A.K.; Gopala Krishnan, S.; Ellur, R.K.; Bhowmick, P.K.; Vinod, K.K.; Bollinedi, H.; Nagarajan, M.; Viswanathan, C.; et al. Marker assisted development and characterization of herbicide tolerant near isogenic lines of a Mega Basmati rice variety, “Pusa Basmati 1121”. Rice 2020, 13, 68. [Google Scholar] [CrossRef] [PubMed]
- Shoba, D.; Raveendran, M.; Manonmani, S.; Utharasu, S.; Dhivyapriya, D.; Subhasini, G.; Ramchandar, S.; Valarmathi, R.; Grover, N.; Krishnan, S.G.; et al. Development and genetic characterization of a novel herbicide (imazethapyr) tolerant mutant in rice (Oryza sativa L.). Rice 2017, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Maqbool, I.; Sudharsan, M.; Kanimozhi, G.; Alrashood, S.T.; Khan, H.A.; Prasad, N.R. Crude cell-free extract from Deinococcus radiodurans exhibit anticancer activity by inducing apoptosis in triple-negative breast cancer cells. Front. Cell Dev. Biol. 2020, 8, 707. [Google Scholar] [CrossRef]
- Broderick, J.B.; Duffus, B.R.; Duschene, K.S.; Shepard, E.M. Radical S-adenosylmethionine enzymes. Chem. Rev. 2014, 114, 4229–4317. [Google Scholar] [CrossRef]
- Campillo-Brocal, J.C.; Lucas-Elio, P.; Sanchez-Amat, A. Identification in M arinomonas mediterranea of a novel quinoprotein with glycine oxidase activity. Microbiologyopen 2013, 2, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Richhardt, J.; Luchterhand, B.; Bringer, S.; Büchs, J.; Bott, M. Evidence for a key role of cytochrome bo3 oxidase in respiratory energy metabolism of Gluconobacter oxydans. J. Bacteriol. 2013, 195, 4210–4220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhao, X.; Shen, H.; Zhang, C. Molecular mechanisms of cell death in intervertebral disc degeneration (Review). Int. J. Mol. Med. 2016, 37, 1439–1448. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Yang, M.; Lan, M.; Liu, C.; Zhang, Y.; Huang, B.; Liu, H.; Zhou, Y. ROS: Crucial intermediators in the pathogenesis of intervertebral disc degeneration. Oxid. Med. Cell. Longev. 2017, 1, 5601593. [Google Scholar] [CrossRef] [PubMed]
- Medina, M.; Rizo, A.; Dinh, D.; Chau, B.; Omidvar, M.; Juarez, A.; Ngo, J.; Johnson, H.A. MopA, the Mn oxidizing protein from Erythrobacter sp. SD-21, requires Heme and NAD+ for Mn(II) oxidation. Front. Microbiol. 2018, 9, 2671. [Google Scholar] [CrossRef] [PubMed]
- Rajpurohit, Y.S.; Gopalakrishnan, R.; Misra, H.S. Involvement of a protein kinase activity inducer in DNA double strand break repair and radioresistance of Deinococcus radiodurans. J. Bacteriol. 2008, 190, 3948–3954. [Google Scholar] [CrossRef] [PubMed]
- Choi, O.; Kim, J.; Kim, J.G.; Jeong, Y.; Moon, J.S.; Park, C.S.; Hwang, I. Pyrroloquinoline quinone is a plant growth promotion factor produced by Pseudomonas fluorescens B16. Plant Physiol. 2008, 146, 657. [Google Scholar] [CrossRef]
- Kim, J.G.; Jung, S. Differential antioxidant mechanisms of rice plants in response to oxyfluorfen and paraquat. Weed Turfgrass Sci. 2013, 2, 254–259. [Google Scholar] [CrossRef]
- Vercampt, H.; Koleva, L.; Vassilev, A.; Vangronsveld, J.; Cuypers, A. Short-term phytotoxicity in Brassica napus (L.) in response to pre-emergently applied metazachlor: A microcosm study. Environ. Toxicol. Chem. 2017, 6, 59–70. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, H.; Bei, F.; Wu, C.; Zhang, L.; Jia, S.; Wang, J.; Liu, W. Investigating the mechanism of metabolic resistance to tribenuron-methyl in Capsella bursa-pastoris (L.) Medik. by full-length transcriptome assembly combined with RNA-Seq. J. Agric. Food Chem. 2021, 69, 3692–3701. [Google Scholar] [CrossRef]
- Wang, J.; Cao, W.; Guo, Q.; Yang, Y.; Bai, L.; Pan, L. Resistance to mesosulfuron-methyl in Beckmannia syzigachne may involve ROS burst and non-target-site resistance mechanisms. Ecotoxicol. Environ. Saf. 2022, 229, 113072. [Google Scholar] [CrossRef] [PubMed]
- Busi, R.; Vila-Aiub, M.M.; Powles, S.B. Genetic control of a cytochrome P450 metabolism-based herbicide resistance mechanism in Lolium rigidum. Heredity 2011, 106, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Iwakami, S.; Endo, M.; Saika, H.; Okuno, J.; Nakamura, N.; Yokoyama, M.; Watanabe, H.; Toki, S.; Uchino, A.; Inamura, T. Cytochrome P450 CYP81A12 and CYP81A21 are associated with resistance to two acetolactate synthase inhibitors in Echinochloa phyllopogon. Plant Physiol. 2014, 165, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Guo, Q.; Wang, J.; Shi, L.; Yang, X.; Zhou, Y.; Yu, Q.; Bai, L. CYP81A68 confers metabolic resistance to ALS and ACCase-inhibiting herbicides and its epigenetic regulation in Echinochloa crus-galli. J. Hazard. Mater. 2022, 428, 128225. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.; Buono, D.D.; Fordham, M.; Skipsey, M.; Brazier, M.; Dixon, D.P.; Cummins, I. Differential induction of glutathione transferases and glucosyltransferases in wheat, maize and Arabidopsis thaliana by herbicide safeners. Z. Naturforsch. C 2005, 60, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.F., Jr.; O’Brien, S.R.; Ma, R.; Hager, A.G.; Riggins, C.W.; Lambert, K.N.; Riechers, D.E. Biochemical characterization of metabolism-based atrazine resistance in Amaranthus tuberculatus and identification of an expressed GST associated with resistance. Plant Biotechnol. J. 2017, 15, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Dücker, R.; Zöllner, P.; Lümmen, P.; Ries, S.; Collavo, A.; Beffa, R. Glutathione transferase plays a major role in flufenacet resistance of ryegrass (Lolium spp.) field populations. Pest Manag. Sci. 2019, 75, 3084–3092. [Google Scholar] [CrossRef] [PubMed]
- Licciardello, C.; D’Agostino, N.; Traini, A.; Recupero, G.R.; Frusciante, L.; Chiusano, M.L. Characterization of the glutathione S-transferase gene family through ESTs and expression analyses within common and pigmented cultivars of Citrus sinensis (L.) Osbeck. BMC Plant Biol. 2014, 14, 39. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Yan, Y.; Luo, Y.; Zou, N.; Liu, W.; Wan, J. Unravelling mesosulfuron-methyl phytotoxicity and metabolism-based herbicide resistance in Alopecurus aequalis: Insight into regulatory mechanisms using proteomics. Sci. Total Environ. 2019, 670, 486–497. [Google Scholar] [CrossRef]
- Jugulam, M.; Shyam, C. Non-target-site resistance to herbicides: Recent developments. Plants 2019, 8, 417. [Google Scholar] [CrossRef]
- Moretti, M.L.; Alárcon-Reverte, R.; Pearce, S.; Morran, S.; Hanson, B.D. Transcription of putative tonoplast transporters in response to glyphosate and paraquat stress in Conyza bonariensis and Conyza canadensis and selection of reference genes for qRT-PCR. PLoS ONE 2017, 12, e0180794. [Google Scholar] [CrossRef]
- Zhu, L.; Xu, J.; Zhang, D.B. Molecular evolution, expression and functional network prediction analysis of ABC transporter gene family in Arabidopsis thaliana. Plant Physiol. J. 2012, 48, 1151–1166. [Google Scholar] [CrossRef]
- Thomas, C.; Tampé, R. Structural and mechanistic principles of ABC transporters. Ann. Rev. Biochem. 2020, 89, 605–636. [Google Scholar] [CrossRef] [PubMed]
- Wilkens, S. Structure and mechanism of ABC transporters. F1000prime Rep. 2015, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Biemans-Oldehinkel, E.; Doeven, M.K.; Poolman, B. ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 2006, 580, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.; Annilo, T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genom. Hum. Genet. 2005, 6, 123–142. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Guo, Y.; Liu, X.; Gao, Y.; Sun, Y.; Dong, L.; Li, J. Potential roles of three ABCB genes in quinclorac resistance identified in Echinochloa crus-galli var. zelayensis. Agronomy 2022, 12, 1961. [Google Scholar] [CrossRef]
- Liu, G.; Sánchez-Fernández, R.; Li, Z.S.; Rea, P.A. Enhanced multispecificity of Arabidopsis vacuolar multidrug resistance-associated protein-type ATP-binding cassette transporter, AtMRP2. J. Biol. Chem. 2001, 276, 8648–8656. [Google Scholar] [CrossRef] [PubMed]
- Bartholomew, D.M.; Van Dyk, D.E.; Lau, S.M.; O’Keefe, D.P.; Rea, P.A.; Viitanen, P.V. Alternate energy-dependent pathways for the vacuolar uptake of glucose and glutathione conjugates. Plant Physiol. 2002, 130, 1562–1572. [Google Scholar] [CrossRef]
- Lu, Y.P.; Li, Z.S.; Rea, P.A. AtMRP1 gene of Arabidopsis encodes a glutathione S-conjugate pump: Isolation and functional definition of a plant ATP-binding cassette transporter gene. Proc. Nat. Acad. Sci. USA 1997, 94, 8243–8248. [Google Scholar] [CrossRef]
- Lu, Y.P.; Li, Z.S.; Drozdowicz, Y.M.; Hortensteiner, S.; Martinoia, E.; Rea, P.A. AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: Functional comparisons with AtMRP1. Plant Cell. 1998, 10, 267–282. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Yu, Q.; Wang, J.; Han, H.; Mao, L.; Nyporko, A.; Maguza, A.; Fan, L.; Bai, L.; Powles, S. An ABCC-type transporter endowing glyphosate resistance in plants. Proc. Nat. Acad. Sci. USA 2021, 118, e2100136118. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Hu, S.; Luo, K.; Tang, T.; Ma, G.; Liu, D.; Peng, Y.; Liu, Y.; Zhang, Y.; Bai, L. Molecular Mechanism of Pyrroloquinoline Quinone-Mediated Rice Tolerance to Imidazolinone Herbicide Imazamox. Agronomy 2024, 14, 1572. https://doi.org/10.3390/agronomy14071572
Li S, Hu S, Luo K, Tang T, Ma G, Liu D, Peng Y, Liu Y, Zhang Y, Bai L. Molecular Mechanism of Pyrroloquinoline Quinone-Mediated Rice Tolerance to Imidazolinone Herbicide Imazamox. Agronomy. 2024; 14(7):1572. https://doi.org/10.3390/agronomy14071572
Chicago/Turabian StyleLi, Sifu, Shiyuan Hu, Kai Luo, Tao Tang, Guolan Ma, Ducai Liu, Yajun Peng, Yang Liu, Yuzhu Zhang, and Lianyang Bai. 2024. "Molecular Mechanism of Pyrroloquinoline Quinone-Mediated Rice Tolerance to Imidazolinone Herbicide Imazamox" Agronomy 14, no. 7: 1572. https://doi.org/10.3390/agronomy14071572
APA StyleLi, S., Hu, S., Luo, K., Tang, T., Ma, G., Liu, D., Peng, Y., Liu, Y., Zhang, Y., & Bai, L. (2024). Molecular Mechanism of Pyrroloquinoline Quinone-Mediated Rice Tolerance to Imidazolinone Herbicide Imazamox. Agronomy, 14(7), 1572. https://doi.org/10.3390/agronomy14071572





