Alterations in Soil Bacterial Community and Its Assembly Process within Paddy Field Induced by Integrated Rice–Giant River Prawn (Macrobrachium rosenbergii) Farming
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Experiment and Sample Collection
2.2. Paddy Soil Properties Determination
2.3. DNA Extraction
2.4. Sequencing and Data Processing
2.5. Statistical Analysis
3. Results
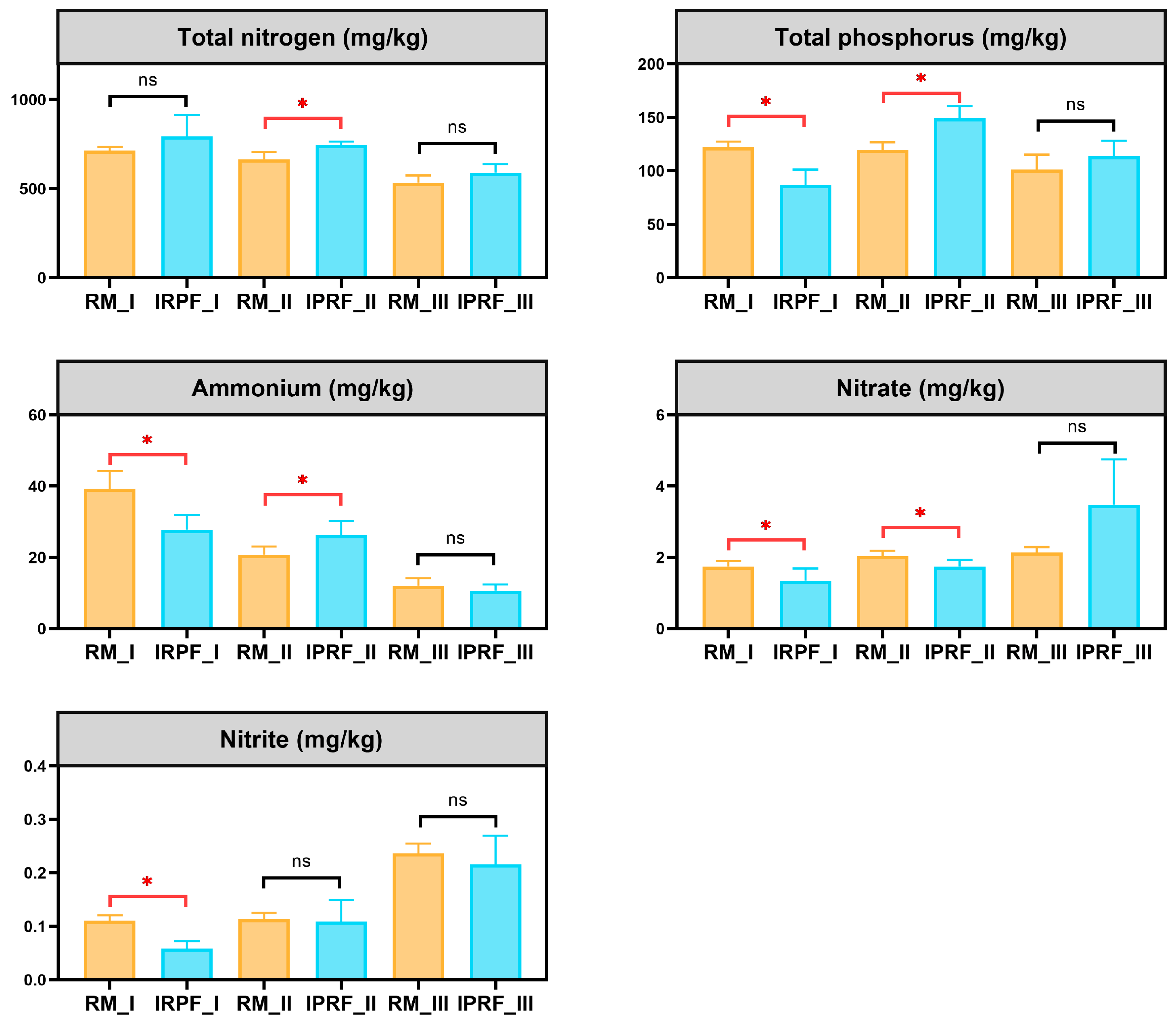
3.1. Environmental Factors
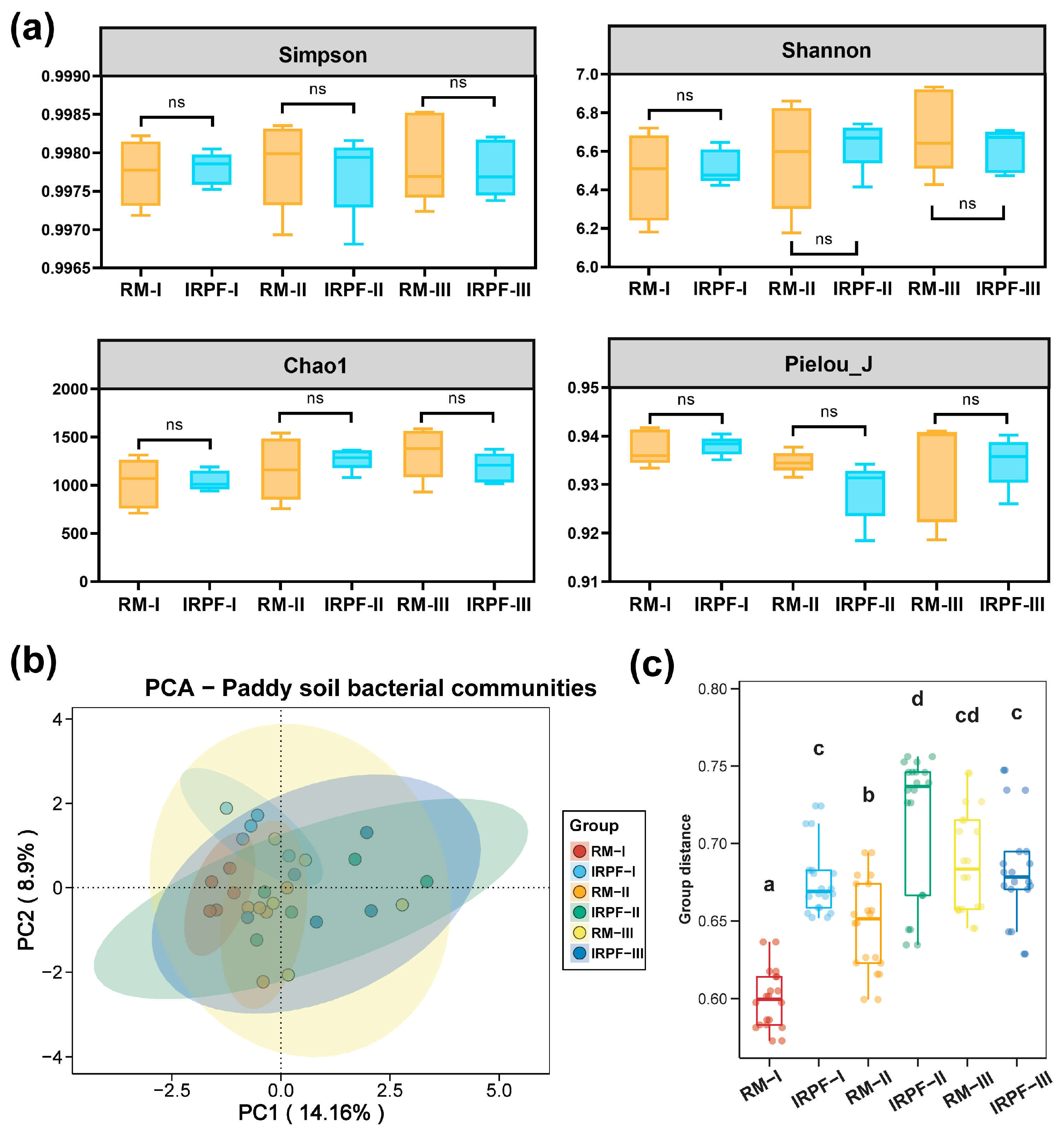
3.2. Soil Bacterial Community Diversities
3.3. Soil Bacterial Community Compositions
3.4. Function Prediction for Soil Bacterial Community
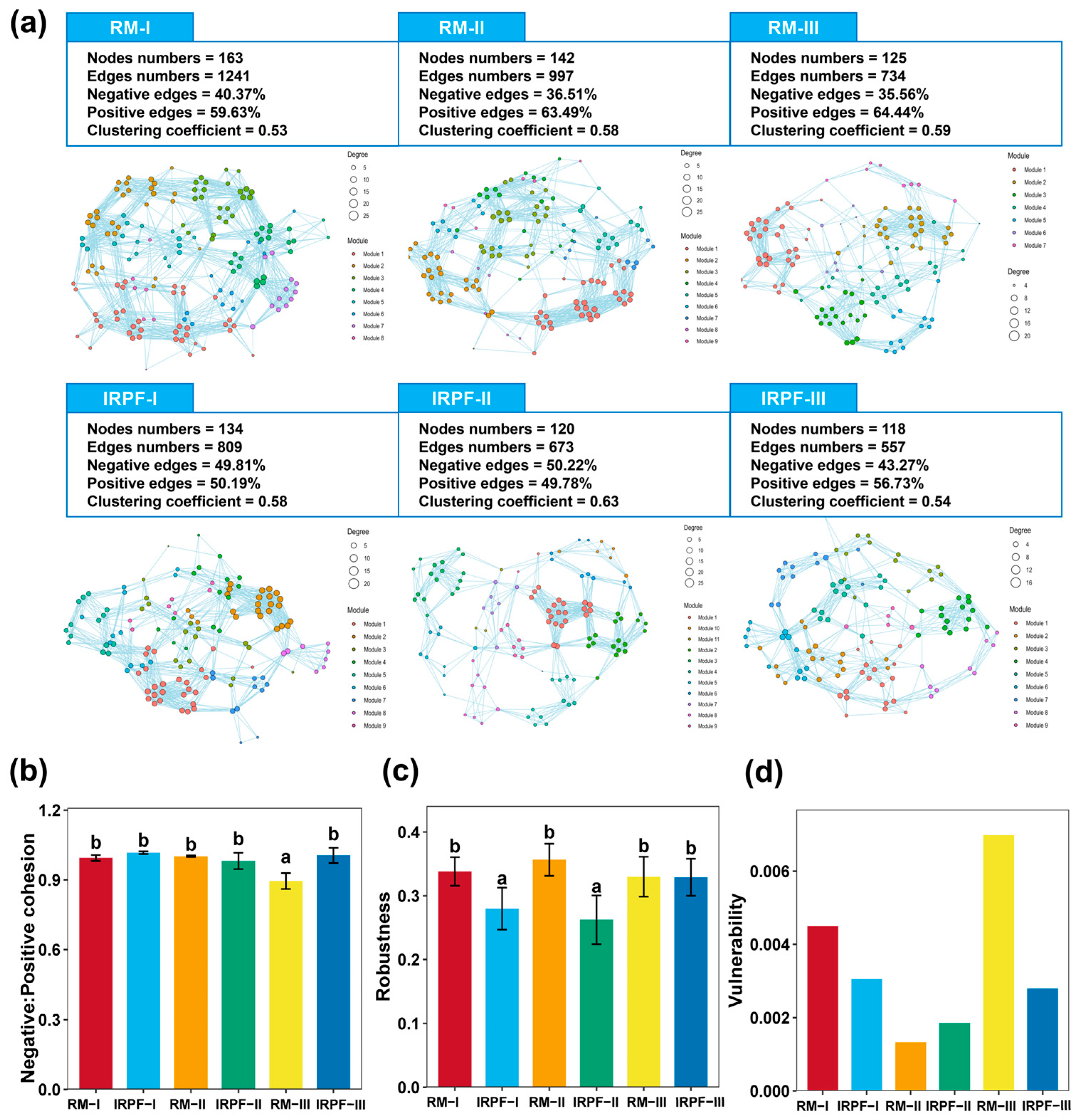
3.5. Co-Occurrence Network for Soil Bacterial Community
3.6. Assembly Processes Shaping the Soil Bacterial Community
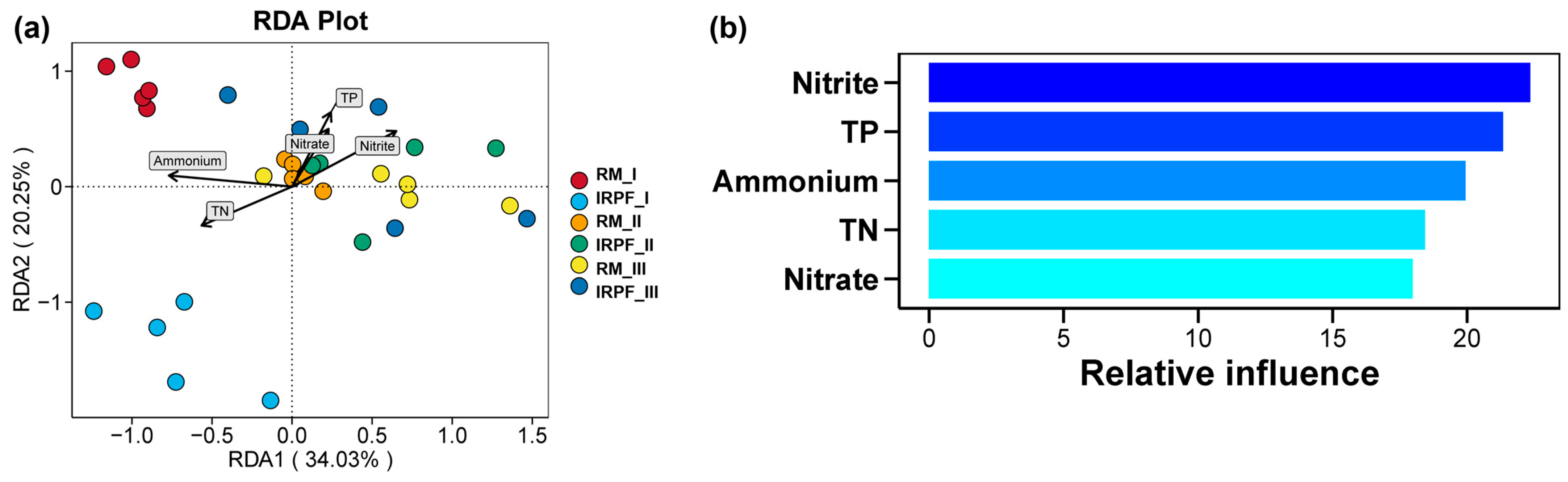
3.7. Correlations of Environmental Factors with Soil Bacterial Communities
4. Discussion
4.1. Variations in the Soil Bacterial Community Composition, Diversity, and Function
4.2. Variations in the Soil Bacterial Co-Occurrence Network Pattern and Community Assembly
4.3. Assiciations of Environmental Factors with Bacterial Communities in Paddy Soil
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ullah, H.; Datta, A.; Samim, N.A.; Din, S.U. Growth and yield of lowland rice as affected by integrated nutrient management and cultivation method under alternate wetting and drying water regime. J. Plant Nutr. 2019, 42, 580–594. [Google Scholar] [CrossRef]
- Jin, T.; Ge, C.D.; Gao, H.; Zhang, H.C.; Sun, X.L. Evaluation and Screening of Co-Culture Farming Models in Rice Field Based on Food Productivity. Sustainability 2020, 12, 2173. [Google Scholar] [CrossRef]
- Hu, L.L.; Zhang, J.; Ren, W.Z.; Guo, L.; Cheng, Y.X.; Li, J.Y.; Li, K.X.; Zhu, Z.W.; Zhang, J.E.; Luo, S.M.; et al. Can the co-cultivation of rice and fish help sustain rice production? Sci. Rep. 2016, 6, 28728. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.B.; Li, X. Review of rice-fish-farming systems in China—One of the Globally Important Ingenious Agricultural Heritage Systems (GIAHS). Aquaculture 2006, 260, 106–113. [Google Scholar] [CrossRef]
- Xie, J.; Hu, L.L.; Tang, J.J.; Wu, X.; Li, N.N.; Yuan, Y.G.; Yang, H.S.; Zhang, J.E.; Luo, S.M.; Chen, X. Ecological mechanisms underlying the sustainability of the agricultural heritage rice-fish coculture system. Proc. Natl. Acad. Sci. USA 2011, 108, E1381–E1387. [Google Scholar] [CrossRef] [PubMed]
- Kangmin, L. Rice-fish culture in China: A review. Aquaculture 1988, 71, 173–186. [Google Scholar] [CrossRef]
- Yu, X.; Hao, X.; Dang, Z.; Yang, L. Industrial development report on integrated rice-fish farming in China (2023). China Fish. News 2023, 3, 1–12. [Google Scholar] [CrossRef]
- Ahmed, N.; Hornbuckle, J.; Turchini, G.M. Blue–green water utilization in rice–fish cultivation towards sustainable food production. Ambio 2022, 51, 1933–1948. [Google Scholar] [CrossRef]
- Halwart, M.; Gupta, M.V. Culture of Fish in Rice Fields; FAO: Rome, Italy, 2004. [Google Scholar]
- National Fisheries Technology Extension Center. “Thirteenth Five-Year Plan” Development Report on China’s Integrated Rice-Fish Farming Industry. China Fish. 2022, 1, 43–52. [Google Scholar]
- Lan, L.M.; Micha, J.-C.; Long, D.N.; Yen, P.T. Effect of Densities and Culture Systems on Growth, Survival, Yield, and Economic Return of Freshwater Prawn, Macrobrachium rosenbergiiFarming in the Rice Field in the Mekong Delta, Vietnam. J. Appl. Aquac. 2006, 18, 43–62. [Google Scholar] [CrossRef]
- Liu, M.; Ma, Q.L.; He, B.; Ni, M.; Zhou, D.; Zhou, S.B.; Yuan, J.L. Assessing nutrient budgets and N2O emission of newly constructed rice-giant freshwater prawn co-culture on reclaimed land. Agric. Ecosyst. Environ. 2023, 357, 108686. [Google Scholar] [CrossRef]
- Kimura, M. Populations, community composition and biomass of aquatic organisms in the floodwater of rice fields and effects of field management. Soil Sci. Plant Nutr. 2005, 51, 159–181. [Google Scholar] [CrossRef]
- Hou, Y.; Jia, R.; Sun, W.; Li, B.; Zhu, J. Influences of the Integrated Rice-Crayfish Farming System with Different Stocking Densities on the Paddy Soil Microbiomes. Int. J. Mol. Sci. 2024, 25, 3786. [Google Scholar] [CrossRef]
- Hou, Y.; Yu, Z.; Jia, R.; Li, B.; Zhu, J. Integrated rice-yellow catfish farming resulting in variations in the agricultural environment, rice growth performance, and soil bacterial communities. Environ. Sci. Pollut. Res. 2024, 31, 28967–28981. [Google Scholar] [CrossRef] [PubMed]
- Tyc, O.; Kulkarni, P.; Ossowicki, A.; Tracanna, V.; Medema, M.H.; Baarlen, P.v.; IJcken, W.F.J.v.; Verhoeven, K.J.F.; Garbeva, P. Exploring the Interspecific Interactions and the Metabolome of the Soil Isolate Hylemonella gracilis. mSystems 2023, 8, e0057422. [Google Scholar] [CrossRef] [PubMed]
- Curtis, T.P.; Sloan, W.T.; Scannell, J.W. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 2002, 99, 10494–10499. [Google Scholar] [CrossRef] [PubMed]
- Torsvik, V.; Ovreås, L. Microbial diversity and function in soil: From genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Buée, M.; Murat, C.; Frey-Klett, P.; Martin, F. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ. Microbiol. Rep. 2010, 2, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Cornforth, D.M.; Foster, K.R. Competition sensing: The social side of bacterial stress responses. Nat. Rev. Microbiol. 2013, 11, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef]
- Stubbendieck, R.M.; Vargas-Bautista, C.; Straight, P.D. Bacterial Communities: Interactions to Scale. Front. Microbiol. 2016, 7, 1234. [Google Scholar] [CrossRef] [PubMed]
- Day, J.; Diener, C.; Otwell, A.; Tams, K.; Bebout, B.; Detweiler, A.; Lee, M.; Scott, M.; Ta, W.; Ha, M.; et al. Lettuce (Lactuca sativa) productivity influenced by microbial inocula under nitrogen-limited conditions in aquaponics. PLoS ONE 2021, 16, e0247534. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef]
- Pineda, A.; Dicke, M.; Pieterse, C.M.J.; Pozo, M.J. Beneficial microbes in a changing environment: Are they always helping plants to deal with insects? Funct. Ecol. 2013, 27, 574–586. [Google Scholar] [CrossRef]
- Herlambang, A.; Murwantoko, M.; Istiqomah, I. Dynamic change in bacterial communities in the integrated rice–fish farming system in Sleman, Yogyakarta, Indonesia. Aquac. Res. 2021, 52, 5566–5578. [Google Scholar] [CrossRef]
- Moriarty, D. The role of microorganisms in aquaculture ponds. Aquaculture 1997, 151, 333–349. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, Q.; Li, Z.; Cheng, W.; Sun, J.; Guo, Z.; Li, Y.; Zhou, J.; Meng, D.; Li, H.; et al. Environmental factors shaping the diversity of bacterial communities that promote rice production. Bmc Microbiol. 2018, 18, 51. [Google Scholar] [CrossRef]
- Zhang, J.; Tang, H.; Zhu, J.; Lin, X.; Feng, Y. Effects of elevated ground-level ozone on paddy soil bacterial community and assembly mechanisms across four years. Sci. Total Environ. 2019, 654, 505–513. [Google Scholar] [CrossRef]
- Zhao, Z.; Jiang, J.; Pan, Y.; Dong, Y.; Chen, Z.; Zhang, G.; Gao, S.; Sun, H.; Guan, X.; Wang, B.; et al. Temporal dynamics of bacterial communities in the water and sediments of sea cucumber (Apostichopus japonicus) culture ponds. Aquaculture 2020, 528, 735498. [Google Scholar] [CrossRef]
- Hou, Y.; Li, B.; Xu, G.; Li, D.; Zhang, C.; Jia, R.; Li, Q.; Zhu, J. Dynamic and Assembly of Benthic Bacterial Community in an Industrial-Scale In-Pond Raceway Recirculating Culture System. Front. Microbiol. 2021, 12, 797817. [Google Scholar] [CrossRef]
- Prijambada, I.D.; Sitompul, R.A.; Widada, J.; Widianto, D. Impact of Agricultural Intensification Practices on Bacterial Community in Agro-ecosystems of Southern Sumatra, Indonesia. Int. J. Agric. Biol. 2012, 14, 816–820. [Google Scholar]
- Singh, U.; Choudhary, A.K.; Sharma, S. Agricultural practices modulate the bacterial communities, and nitrogen cycling bacterial guild in rhizosphere: Field experiment with soybean. J. Sci. Food Agric. 2021, 101, 2687–2695. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Z.; Cai, H.; Lu, W.; Li, J. Long-term agricultural contamination shaped diversity response of sediment microbiome. J. Environ. Sci. 2021, 99, 90–99. [Google Scholar] [CrossRef]
- Zhang, Z.; Deng, Q.; Wan, L.; Cao, X.; Zhou, Y.; Song, C. Bacterial Communities and Enzymatic Activities in Sediments of Long-Term Fish and Crab Aquaculture Ponds. Microorganisms 2021, 9, 501. [Google Scholar] [CrossRef]
- HJ 632-2011; Soil-Determination of Total Phosphorus by Alkali Fusion–Mo-Sb Anti Spectrophotometric Method. Ministry of Ecology and Environment of the People’s Republic of China: Beijing, China, 2012.
- HJ 717-2014; Soil Quality-Determination of Total Nitrogen-Modified Kjeldahl Method. Ministry of Ecology and Environment of the People’s Republic of China: Beijing, China, 2014.
- HJ 634-2012; Soil-Determination of Ammonium, Nitrite and Nitrate by Extraction with Potassium Chloride Solution-Spectrophotometric Methods. Ministry of Ecology and Environment of the People’s Republic of China: Beijing, China, 2012.
- Wang, J.; Shi, X.; Zheng, C.; Suter, H.; Huang, Z. Different responses of soil bacterial and fungal communities to nitrogen deposition in a subtropical forest. Sci. Total Environ. 2021, 755, 142449. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High resolution sample inference from amplicon data. Nat. Methods 2015, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Tukey’s Honestly Significant Difference (HSD) Test. In Encyclopedia of Research Design; Salkind, N., Ed.; Sage: Thousand Oaks, CA, USA, 2010; Volume 3, pp. 1–5. [Google Scholar]
- Barberán, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef]
- Louca, S.; Polz, M.F.; Mazel, F.; Albright, M.B.N.; Huber, J.A.; O’Connor, M.I.; Ackermann, M.; Hahn, A.S.; Srivastava, D.S.; Crowe, S.A.; et al. Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2018, 2, 936–943. [Google Scholar] [CrossRef]
- Song, W.; Liu, J.; Qin, W.; Huang, J.; Yu, X.; Xu, M.; Stahl, D.; Jiao, N.; Zhou, J.; Tu, Q.; et al. Functional Traits Resolve Mechanisms Governing the Assembly and Distribution of Nitrogen-Cycling Microbial Communities in the Global Ocean. mBio 2022, 13, e03832-21. [Google Scholar] [CrossRef]
- Arunrat, N.; Sansupa, C.; Kongsurakan, P.; Sereenonchai, S.; Hatano, R. Soil Microbial Diversity and Community Composition in Rice-Fish Co-Culture and Rice Monoculture Farming System. Biology 2022, 11, 1242. [Google Scholar] [CrossRef]
- Hou, Y.; Jia, R.; Sun, W.; Ding, H.; Li, B.; Zhu, J. Red Claw Crayfish Cherax quadricarinatus Cultivation Influences the Dynamics and Assembly of Benthic Bacterial Communities in Paddy Fields. Environments 2023, 10, 178. [Google Scholar] [CrossRef]
- Jiang, X.; Ma, H.; Zhao, Q.-l.; Yang, J.; Xin, C.-y.; Chen, B. Bacterial communities in paddy soil and ditch sediment under rice-crab co-culture system. AMB Express 2021, 11, 163. [Google Scholar] [CrossRef]
- Li, P.; Wu, G.; Li, Y.; Hu, C.; Ge, L.; Zheng, X.; Zhang, J.; Chen, J.; Zhang, H.; Bai, N. Long-term rice-crayfish-turtle co-culture maintains high crop yields by improving soil health and increasing soil microbial community stability. Geoderma 2022, 413, 115745. [Google Scholar] [CrossRef]
- Zhao, Z.; Chu, C.B.; Zhou, D.P.; Wang, Q.F.; Wu, S.H.; Zheng, X.Q.; Song, K.; Lv, W.G. Soil bacterial community composition in rice-fish integrated farming systems with different planting years. Sci. Rep. 2021, 11, 10855. [Google Scholar] [CrossRef]
- Yi, X.; Yi, K.; Fang, K.; Gao, H.; Dai, W.; Cao, L. Microbial community structures and important associations between soil nutrients and the responses of specific taxa to rice-frog cultivation. Front. Microbiol. 2019, 10, 1752. [Google Scholar] [CrossRef]
- Zhang, C.; Mi, W.; Xu, Y.; Zhou, W.; Bi, Y. Long-term integrated rice-crayfish culture disrupts the microbial communities in paddy soil. Aquac. Rep. 2023, 29, 101515. [Google Scholar] [CrossRef]
- Ettwig, K.F.; Butler, M.K.; Le Paslier, D.; Pelletier, E.; Mangenot, S.; Kuypers, M.M.; Schreiber, F.; Dutilh, B.E.; Zedelius, J.; de Beer, D. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 2010, 464, 543–548. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, P.; Ye, F.; Jiang, Y.; Song, L.; Op den Camp, H.J.; Zhu, G.; Wu, S. Nitrite-dependent anaerobic methane oxidizing bacteria along the water level fluctuation zone of the Three Gorges Reservoir. Appl. Microbiol. Biotechnol. 2016, 100, 1977–1986. [Google Scholar] [CrossRef]
- He, Z.; Cai, C.; Wang, J.; Xu, X.; Zheng, P.; Jetten, M.S.; Hu, B. A novel denitrifying methanotroph of the NC10 phylum and its microcolony. Sci. Rep. 2016, 6, 32241. [Google Scholar] [CrossRef]
- Raghoebarsing, A.A.; Pol, A.; Van de Pas-Schoonen, K.T.; Smolders, A.J.; Ettwig, K.F.; Rijpstra, W.I.C.; Schouten, S.; Damsté, J.S.S.; Op den Camp, H.J.; Jetten, M.S. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 2006, 440, 918–921. [Google Scholar] [CrossRef]
- Versantvoort, W.; Guerrero-Cruz, S.; Speth, D.R.; Frank, J.; Gambelli, L.; Cremers, G.; Van Alen, T.; Jetten, M.S.; Kartal, B.; Op den Camp, H.J. Comparative genomics of Candidatus Methylomirabilis species and description of Ca. Methylomirabilis Lanthanidiphila 2018, 9, 407382. [Google Scholar]
- Alvarez, A.; Saez, J.M.; Davila Costa, J.S.; Colin, V.L.; Fuentes, M.S.; Cuozzo, S.A.; Benimeli, C.S.; Polti, M.A.; Amoroso, M.J. Actinobacteria: Current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 2017, 166, 41–62. [Google Scholar] [CrossRef]
- Benimeli, C.S.; Amoroso, M.J.; Chaile, A.P.; Castro, G.R. Isolation of four aquatic streptomycetes strains capable of growth on organochlorine pesticides. Bioresour. Technol. 2003, 89, 133–138. [Google Scholar] [CrossRef]
- Polti, M.A.; Atjián, M.C.; Amoroso, M.J.; Abate, C.M. Soil chromium bioremediation: Synergic activity of actinobacteria and plants. Int. Biodeterior. Biodegrad. 2011, 65, 1175–1181. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A.; Charter, K.; Bib, M.J.; Bipp, M.; Keiser, T.; Butner, M.J.J.I.F. Practical Streptomyces Genetics; John Innes Foundation: Norwick, UK, 2000. [Google Scholar]
- Li, A.; Wang, Y.; Wang, Y.; Dong, H.; Wu, Q.; Mehmood, K.; Chang, Z.; Li, Y.; Chang, Y.-F.; Shi, L.; et al. Microbiome analysis reveals soil microbial community alteration with the effect of animal excretion contamination and altitude in Tibetan Plateau of China. Int. Soil Water Conserv. Res. 2021, 9, 639–648. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Y.; Qu, Z.; Mu, W.; Mi, W.; Ma, Y.; Su, L.; Si, L.; Li, J.; You, Q. Microbial communities in paddy soil as influenced by nitrogen fertilization and water regimes. Agron. J. 2022, 114, 379–394. [Google Scholar] [CrossRef]
- Song, X.; Yu, D.; Qiu, Y.; Qiu, C.; Xu, L.; Zhao, J.; Wang, X. Unexpected phosphorous removal in a Candidatus_Competibacter and Defluviicoccus dominated reactor. Bioresour. Technol. 2022, 345, 126540. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Lv, X.; Zhao, K.; Xue, R.; Liu, Y.; Xu, J.; Ma, B.J. Strengthening insights in microbial ecological networks from theory to applications. mSystems 2019, 4. [Google Scholar] [CrossRef]
- Ma, B.; Wang, Y.; Ye, S.; Liu, S.; Stirling, E.; Gilbert, J.A.; Faust, K.; Knight, R.; Jansson, J.K.; Cardona, C. Earth microbial co-occurrence network reveals interconnection pattern across microbiomes. Microbiome 2020, 8, 82. [Google Scholar] [CrossRef]
- Mikhailov, I.S.; Zakharova, Y.R.; Bukin, Y.S.; Galachyants, Y.P.; Petrova, D.P.; Sakirko, M.V.; Likhoshway, Y.V. Co-occurrence networks among bacteria and microbial eukaryotes of Lake Baikal during a spring phytoplankton bloom. Microb. Ecol. 2019, 77, 96–109. [Google Scholar] [CrossRef]
- Hou, Y.R.; Jia, R.; Li, B.; Zhu, J. Apex Predators Enhance Environmental Adaptation but Reduce Community Stability of Bacterioplankton in Crustacean Aquaculture Ponds. Int. J. Mol. Sci. 2022, 23, 10785. [Google Scholar] [CrossRef]
- Qin, M.; Xu, H.; Zhao, D.; Zeng, J.; Wu, Q.L. Aquaculture drives distinct patterns of planktonic and sedimentary bacterial communities: Insights into co-occurrence pattern and assembly processes. Environ. Microbiol. 2022, 24, 4079–4093. [Google Scholar] [CrossRef]
- Chen, J.; Guo, Q.; Liu, D.; Hu, C.; Sun, J.; Wang, X.; Liang, G.; Zhou, W. Composition, predicted functions, and co-occurrence networks of fungal and bacterial communities_ Links to soil organic carbon under long-term fertilization in a rice-wheat cropping system. Eur. J. Soil Biol. 2020, 100, 103226. [Google Scholar] [CrossRef]
- Zhang, K.; Shi, Y.; Lu, H.; He, M.; Huang, W.; Siemann, E. Soil bacterial communities and co-occurrence changes associated with multi-nutrient cycling under rice-wheat rotation reclamation in coastal wetland. Ecol. Indic. 2022, 144, 109485. [Google Scholar] [CrossRef]
- Hunt, D.E.; Ward, C.S. A network-based approach to disturbance transmission through microbial interactions. Front. Microbiol. 2015, 6, 1182. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.T.-W.; Liu, A.-C.; Weng, C.-Y.; Chen, Y.-C.; Chen, C.-Y.; Weng, F.C.-H.; Wang, D.; Chou, C.-Y. A network-based approach to deciphering a dynamic microbiome’s response to a subtle perturbation. Sci. Rep. 2020, 10, 19530. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Huang, J.; Zhao, Z.; Feng, W.; Zheng, X.; Du, P. Impact of clomazone on bacterial communities in two soils. Front. Microbiol. 2023, 14, 1198808. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-H.; Chen, S.-Y.; Chen, J.-W.; Xue, K.; Chen, S.-L.; Wang, X.-M.; Chen, T.; Kang, S.-C.; Rui, J.-P.; Thies, J.E.; et al. Reduced microbial stability in the active layer is associated with carbon loss under alpine permafrost degradation. Proc. Natl. Acad. Sci. USA 2021, 118, e2025321118. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, K.; Chen, X.; Wei, K.; Wu, R.; Wang, G. Stochastic and deterministic assembly processes of bacterial communities in different soil aggregates. Appl. Soil Ecol. 2024, 193, 105153. [Google Scholar] [CrossRef]
- Sloan, W.T.; Lunn, M.; Woodcock, S.; Head, I.M.; Nee, S.; Curtis, T.P. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 2006, 8, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Burns, A.R.; Stephens, W.Z.; Stagaman, K.; Wong, S.; Rawls, J.F.; Guillemin, K.; Bohannan, B.J.M. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 2015, 10, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.; Chen, H.; Liu, X.; Wong, M.H.; Xu, F.; Yang, X.; Xu, W.; Zeng, Q.; Wang, W.; Li, S. Characteristics of spatial and seasonal bacterial community structures in a river under anthropogenic disturbances. Environ. Pollut. 2020, 264, 114818. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Pan, Y.; Jiang, J.; Gao, S.; Sun, H.; Dong, Y.; Sun, P.; Guan, X.; Zhou, Z. Unrevealing variation of microbial communities and correlation with environmental variables in a full culture-cycle of Undaria pinnatifida. Mar. Environ. Res. 2018, 139, 46–56. [Google Scholar] [CrossRef]
- Nicholaus, R.; Lukwambe, B.; Zhao, L.; Yang, W.; Zhu, J.; Zheng, Z. Bioturbation of blood clam Tegillarca granosa on benthic nutrient fluxes and microbial community in an aquaculture wastewater treatment system. Int. Biodeterior. Biodegrad. 2019, 142, 73–82. [Google Scholar] [CrossRef]
- Nicholaus, R.; Zheng, Z. The effects of bioturbation by the Venus clam Cyclina sinensis on the fluxes of nutrients across the sediment–water interface in aquaculture ponds. Aquac. Int. 2014, 22, 913–924. [Google Scholar] [CrossRef]
- Qin, Y.; Hou, J.; Deng, M.; Liu, Q.; Wu, C.; Ji, Y.; He, X. Bacterial abundance and diversity in pond water supplied with different feeds. Sci. Rep. 2016, 6, 35232. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Wang, Y.; Tang, J.; Dai, Y. Bacterial communities in Chinese grass carp (Ctenopharyngodon idellus) farming ponds. Aquac. Res. 2013, 45, 138–149. [Google Scholar] [CrossRef]
- Islas-Lima, S.; Thalasso, F.; Gómez-Hernandez, J. Evidence of anoxic methane oxidation coupled to denitrification. Water Res. 2004, 38, 13–16. [Google Scholar] [CrossRef]
- Zhao, W.; Chen, Q.; Ma, C. Nitrite-dependent anaerobic methane oxidation and microbial characteristics: A review. Microbiol. China 2021, 48, 3847–3859. [Google Scholar]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Hou, Y.; Jia, R.; Li, B.; Zhu, J.; Ge, X. Alterations in Soil Bacterial Community and Its Assembly Process within Paddy Field Induced by Integrated Rice–Giant River Prawn (Macrobrachium rosenbergii) Farming. Agronomy 2024, 14, 1600. https://doi.org/10.3390/agronomy14081600
Zhang Y, Hou Y, Jia R, Li B, Zhu J, Ge X. Alterations in Soil Bacterial Community and Its Assembly Process within Paddy Field Induced by Integrated Rice–Giant River Prawn (Macrobrachium rosenbergii) Farming. Agronomy. 2024; 14(8):1600. https://doi.org/10.3390/agronomy14081600
Chicago/Turabian StyleZhang, Yiyun, Yiran Hou, Rui Jia, Bing Li, Jian Zhu, and Xianping Ge. 2024. "Alterations in Soil Bacterial Community and Its Assembly Process within Paddy Field Induced by Integrated Rice–Giant River Prawn (Macrobrachium rosenbergii) Farming" Agronomy 14, no. 8: 1600. https://doi.org/10.3390/agronomy14081600
APA StyleZhang, Y., Hou, Y., Jia, R., Li, B., Zhu, J., & Ge, X. (2024). Alterations in Soil Bacterial Community and Its Assembly Process within Paddy Field Induced by Integrated Rice–Giant River Prawn (Macrobrachium rosenbergii) Farming. Agronomy, 14(8), 1600. https://doi.org/10.3390/agronomy14081600






