Identification and Functional Analysis of the EPF/EPFL Gene Family in Maize (Zea mays L.): Implications for Drought Stress Response
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Evolutionary Analysis of EPF/EPFL Genes
2.2. Analysis of ZmEPF/EPFL Gene Structures and Their Encoded Proteins
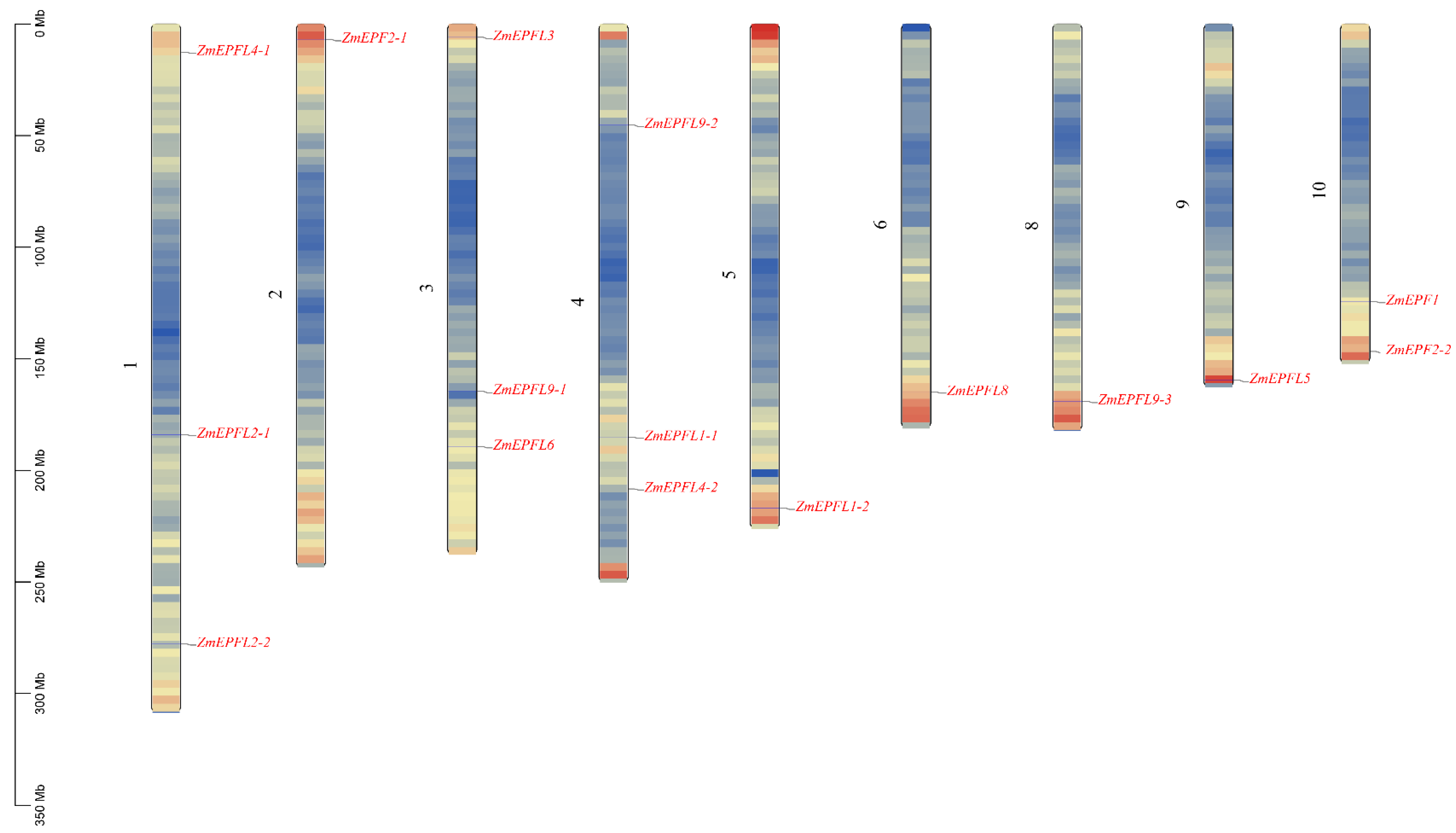
2.3. Chromosomal Localization and Basic Information of ZmEPF/EPFL
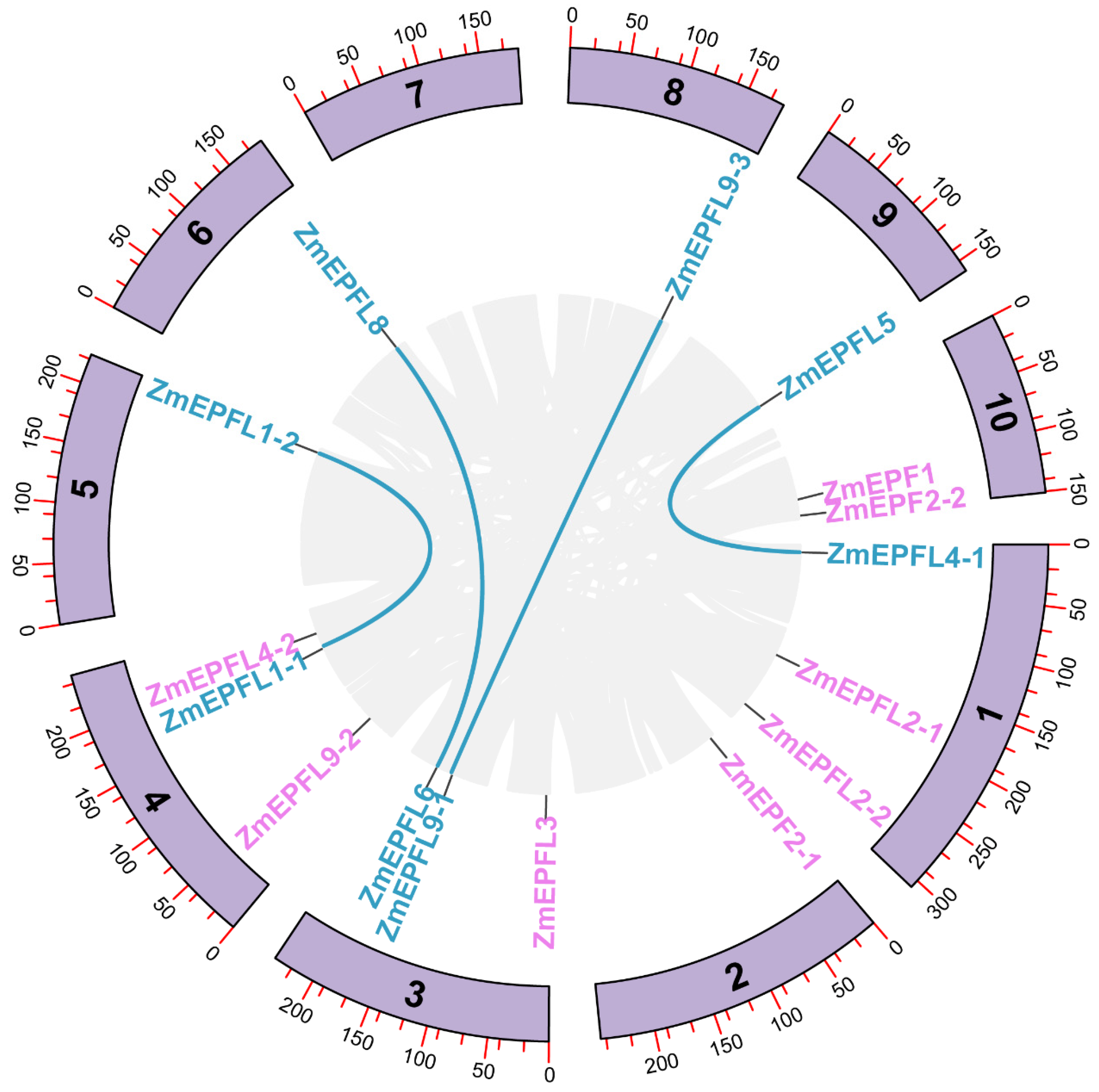
2.4. Collinearity Analysis of EPF/EPFL Genes across Genomes
2.5. ZmEPF/EPFL Protein-Protein Interaction
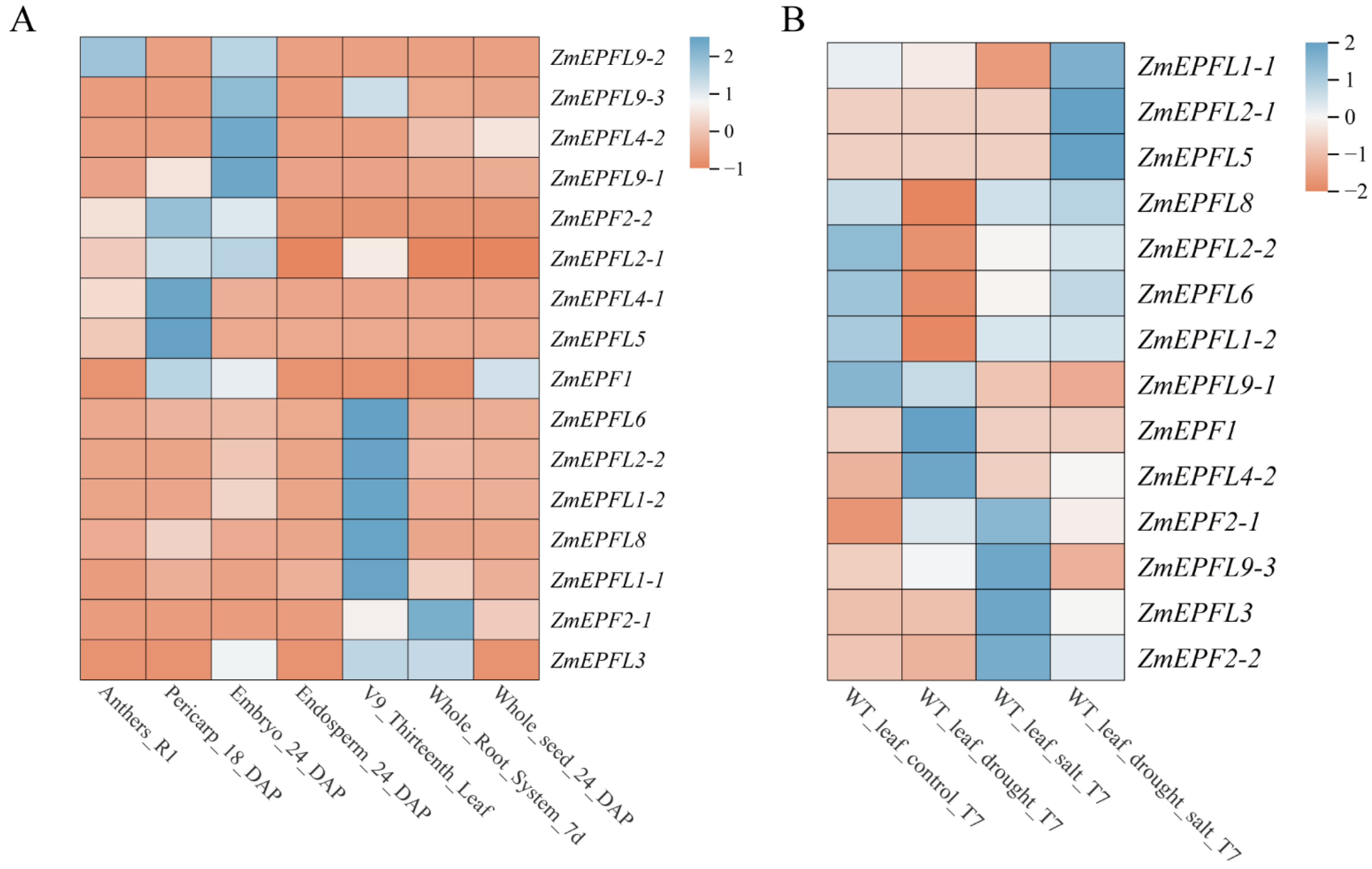
2.6. Expression Profiling of the ZmEPF/EPFL Gene Family
2.7. Drought-Induced Expression Patterns of ZmEPF/EPFL Genes
2.8. Drought Stress in Maize and qRT-PCR Analysis
3. Results
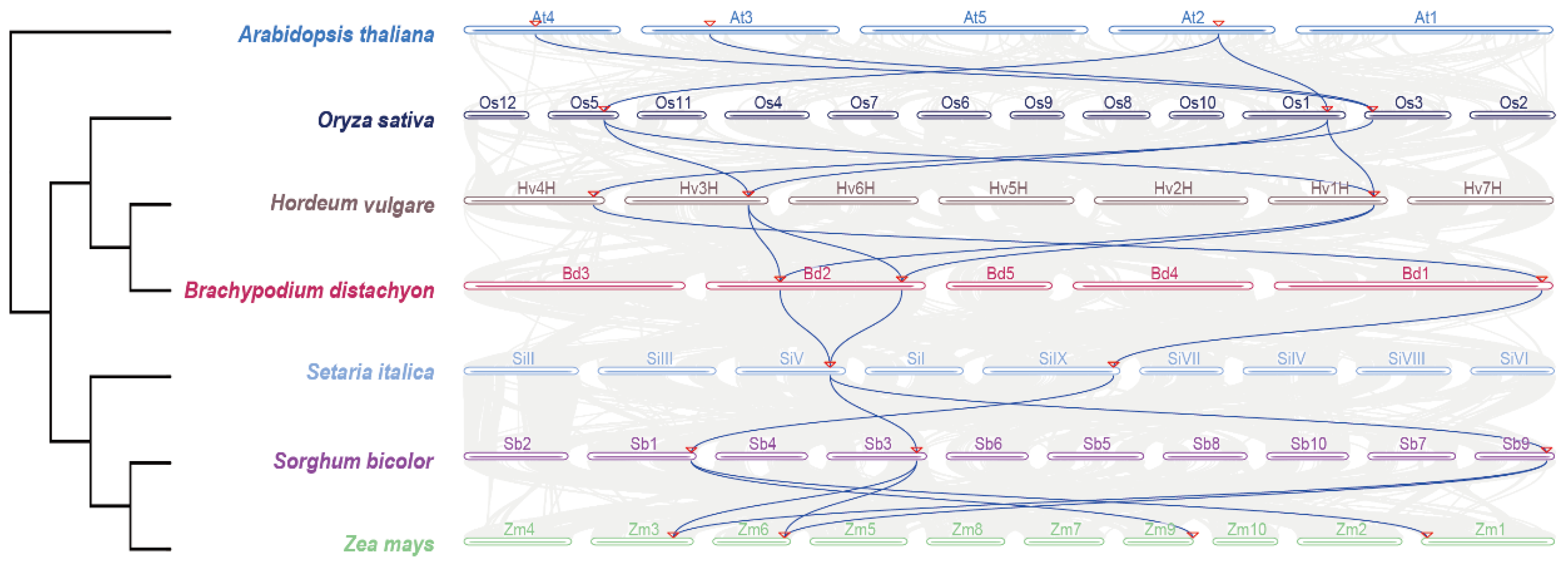
3.1. Comprehensive Identification and Phylogenetic Delineation of EPF/EPFL Gene Family in Seven Poaceae Species
3.2. The Gene Structure, Encoded Protein Motifs, and Conserved Domains of the EPF/EPFL Gene Family in Maize
3.3. Chromosome Localization and Synteny Relationship of ZmEPF/EPFL Genes
3.4. Analysis of Covariance within the ZmEPF/EPFL Family and Evolutionary Relationships among Species
3.5. Expansion and Contraction Analysis of Poaceae Plants and Their EPF/EPFL Gene Families
3.6. Expression Analysis of ZmEPF/EPFL Genes
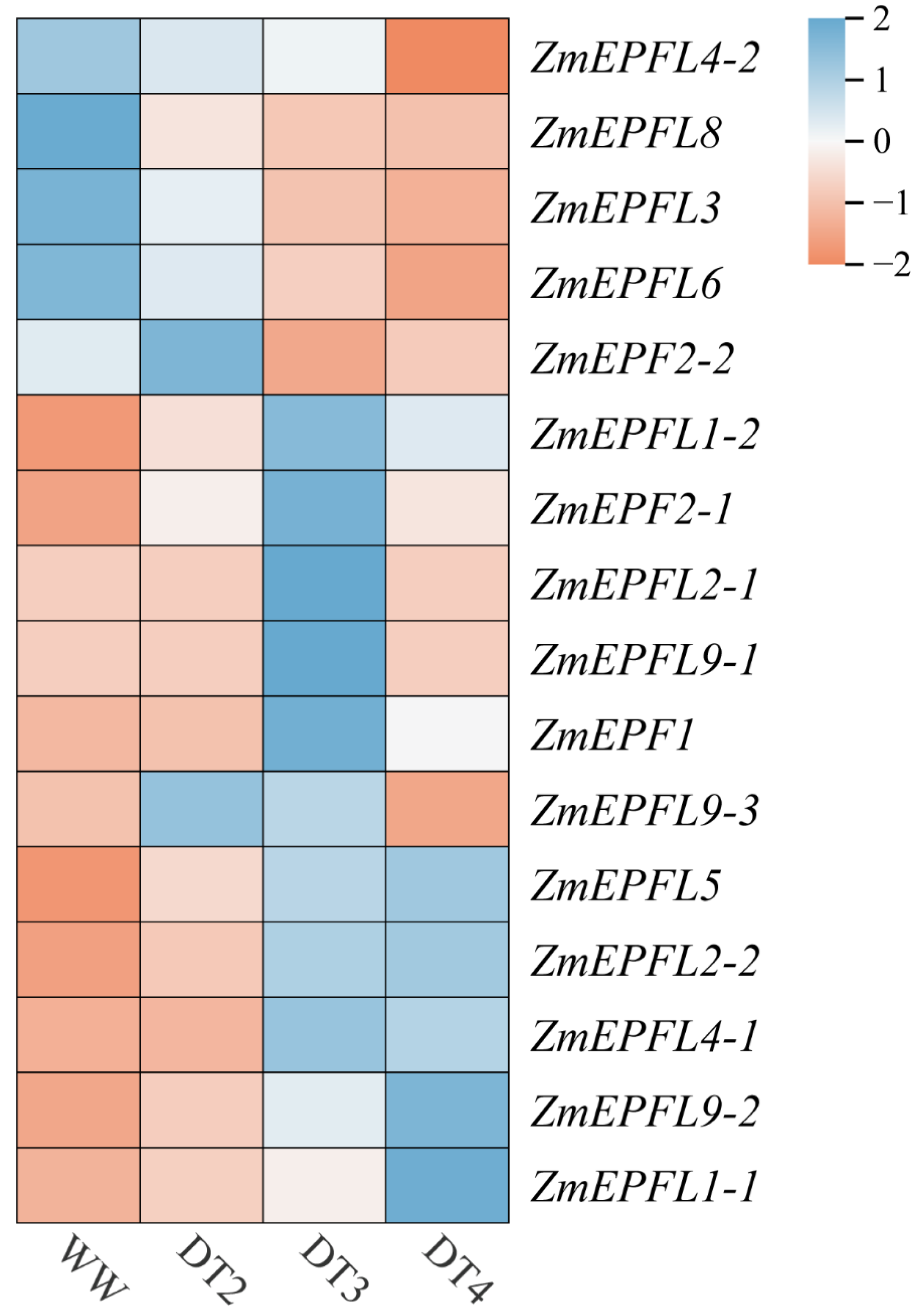
3.7. Expression Pattern Analysis of ZmEPF/EPFL Genes under Varied Drought Conditions and qRT-PCR Validation
3.8. Validation of Drought Stress on Maize EPF/EPFL Gene Family
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.M. Osmoregulation and water stress in higher plants. Annu. Rev. Plant Biol. 1984, 35, 299–319. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Blumwald, E. The roles of ROS and ABA in systemic acquired acclimation. Plant Cell. 2015, 27, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Shekhawat, J.; Upadhyay, S.K. DPY1 as an osmosensor for drought signaling. Trends Plant Sci. 2024, 29, 616–619. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Cui, J.; Liu, S.; Wang, S.; Lian, Y.; Bai, Y.; Zhu, T.; Wu, H.; Wang, Y.; Yang, S.; et al. Natural variations of ZmSRO1d modulate the trade-off between drought resistance and yield by affecting ZmRBOHC-mediated stomatal ROS production in maize. Mol. Plant 2022, 15, 1558–1574. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Dai, X.R.; Yue, X.; Gao, X.Q.; Zhang, X.S. Identification of small secreted peptides (SSPs) in maize and expression analysis of partial SSP genes in reproductive tissues. Planta 2014, 240, 713–728. [Google Scholar] [CrossRef]
- Torii, K.U. Mix-and-match: Ligand-receptor pairs in stomatal development and beyond. Trends Plant Sci. 2012, 17, 711–719. [Google Scholar] [CrossRef]
- Kim, J.S.; Jeon, B.W.; Kim, J. Signaling Peptides Regulating Abiotic Stress Responses in Plants. Front. Plant Sci. 2021, 12, 704490. [Google Scholar] [CrossRef]
- Falquetto-Gomes, P.; Silva, W.J.; Siqueira, J.A.; Araújo, W.L.; Nunes-Nesi, A. From epidermal cells to functional pores: Understanding stomatal development. J. Plant Physiol. 2024, 292, 154163. [Google Scholar] [CrossRef]
- Jangra, R.; Brunetti, S.C.; Wang, X.; Kaushik, P.; Gulick, P.J.; Foroud, N.A.; Wang, S.; Lee, J.S. Duplicated antagonistic EPF peptides optimize grass stomatal initiation. Development 2021, 148, dev199780. [Google Scholar] [CrossRef]
- Jiao, Z.; Wang, J.; Shi, Y.; Wang, Z.; Zhang, J.; Du, Q.; Liu, B.; Jia, X.; Niu, J.; Gu, C.; et al. Genome-Wide Identification and Analysis of the EPF Gene Family in Sorghum bicolor (L.) Moench. Plants 2023, 12, 3912. [Google Scholar] [CrossRef]
- Lu, J.; He, J.; Zhou, X.; Zhong, J.; Li, J.; Liang, Y.K. Homologous genes of epidermal patterning factor regulate stomatal development in rice. J. Plant Physiol. 2019, 234–235, 18–27. [Google Scholar] [CrossRef]
- Harrison, E.L.; Arce Cubas, L.; Gray, J.E.; Hepworth, C. The influence of stomatal morphology and distribution on photosynthetic gas exchange. Plant J. 2020, 101, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Bertolino, L.T.; Caine, R.S.; Gray, J.E. Impact of Stomatal Density and Morphology on Water-Use Efficiency in a Changing World. Front. Plant Sci. 2019, 10, 225. [Google Scholar] [CrossRef] [PubMed]
- Caine, R.S.; Chater, C.C.; Kamisugi, Y.; Cuming, A.C.; Beerling, D.J.; Gray, J.E.; Fleming, A.J. An ancestral stomatal patterning module revealed in the non-vascular land plant Physcomitrella patens. Development 2016, 143, 3306–3314. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.; Hepworth, C.; Dutton, C.; Dunn, J.A.; Hunt, L.; Stephens, J.; Waugh, R.; Cameron, D.D.; Gray, J.E. Reducing Stomatal Density in Barley Improves Drought Tolerance without Impacting on Yield. Plant Physiol. 2017, 174, 776–787. [Google Scholar] [CrossRef]
- Dunn, J.; Hunt, L.; Afsharinafar, M.; Meselmani, M.A.; Mitchell, A.; Howells, R.; Wallington, E.; Fleming, A.J.; Gray, J.E. Reduced stomatal density in bread wheat leads to increased water-use efficiency. J. Exp. Bot. 2019, 70, 4737–4748. [Google Scholar] [CrossRef]
- Yin, X.; Biswal, A.K.; Dionora, J.; Perdigon, K.M.; Balahadia, C.P.; Mazumdar, S.; Chater, C.; Lin, H.C.; Coe, R.A.; Kretzschmar, T.; et al. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 2017, 36, 745–757. [Google Scholar] [CrossRef]
- Caine, R.S.; Yin, X.; Sloan, J.; Harrison, E.L.; Mohammed, U.; Fulton, T.; Biswal, A.K.; Dionora, J.; Chater, C.C.; Coe, R.A.; et al. Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol. 2019, 221, 371–384. [Google Scholar] [CrossRef]
- Meng, X.; Liu, S.; Zhang, C.; He, J.; Ma, D.; Wang, X.; Dong, T.; Guo, F.; Cai, J.; Long, T.; et al. The unique sweet potato NAC transcription factor IbNAC3 modulates combined salt and drought stresses. Plant Physiol. 2023, 191, 747–771. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Jia, F.; Jiao, Z.; Wang, J.; Xia, X.; Yin, W. Ectopic expression of secretory peptide PdEPF3 in Arabidopsis confers drought tolerance with reduced stomatal density. Acta Soc. Bot. Pol. 2019, 88, 3627. [Google Scholar] [CrossRef]
- Hara, K.; Yokoo, T.; Kajita, R.; Onishi, T.; Yahata, S.; Peterson, K.M.; Torii, K.U.; Kakimoto, T. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 2009, 50, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Hnilova, M.; Maes, M.; Lin, Y.C.; Putarjunan, A.; Han, S.K.; Avila, J.; Torii, K.U. Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 2015, 522, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Uchida, N.; Lee, J.S.; Horst, R.J.; Lai, H.H.; Kajita, R.; Kakimoto, T.; Tasaka, M.; Torii, K.U. Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc. Natl. Acad. Sci. USA 2012, 109, 6337–6342. [Google Scholar] [CrossRef] [PubMed]
- Abrash, E.B.; Davies, K.A.; Bergmann, D.C. Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell. 2011, 23, 2864–2879. [Google Scholar] [CrossRef] [PubMed]
- Takata, N.; Yokota, K.; Ohki, S.; Mori, M.; Taniguchi, T.; Kurita, M. Evolutionary relationship and structural characterization of the EPF/EPFL gene family. PLoS ONE 2013, 8, e65183. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Stelpflug, S.C.; Sekhon, R.S.; Vaillancourt, B.; Hirsch, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. An Expanded Maize Gene Expression Atlas based on RNA Sequencing and its Use to Explore Root Development. Plant Genome 2016, 9, plantgenome2015-04. [Google Scholar] [CrossRef] [PubMed]
- Forestan, C.; Aiese Cigliano, R.; Farinati, S.; Lunardon, A.; Sanseverino, W.; Varotto, S. Stress-induced and epigenetic-mediated maize transcriptome regulation study by means of transcriptome reannotation and differential expression analysis. Sci. Rep. 2016, 6, 30446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Fan, Y.; Sun, X.; Chen, L.; Terzaghi, W.; Bucher, E.; Li, L.; Dai, M. A large-scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis. Plant J. 2019, 98, 697–713. [Google Scholar] [CrossRef]
- Zhiling, L.; Wenhua, D.; Fangyuan, Z. Genome-wide identification and phylogenetic and expression pattern analyses of EPF/EPFL family genes in the Rye (Secale cereale L.). BMC Genom. 2024, 25, 532. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cai, K.; Pei, X.; Li, Y.; Hu, Y.; Meng, F.; Song, X.; Tigabu, M.; Ding, C.; Zhao, X. Genome-Wide Identification of NAC Transcription Factor Family in Juglans mandshurica and Their Expression Analysis during the Fruit Development and Ripening. Int. J. Mol. Sci. 2021, 22, 12414. [Google Scholar] [CrossRef]
- Weng, X.; Zhu, L.; Yu, S.; Liu, Y.; Ru, Y.; Zhang, Z.; He, Z.; Zhou, L.; Chen, X. Carbon monoxide promotes stomatal initiation by regulating the expression of two EPF genes in Arabidopsis cotyledons. Front. Plant Sci. 2022, 13, 1029703. [Google Scholar] [CrossRef]
- Chater, C.C.C.; Caine, R.S.; Fleming, A.J.; Gray, J.E. Origins and Evolution of Stomatal Development. Plant Physiol. 2017, 174, 624–638. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Huang, Y.; Liu, Z.; Li, C.; Yu, H.; Shahid, M.Q.; Lin, Y.; Qiao, X.; Xiao, J.; Gray, J.E.; et al. Small EPIDERMAL PATTERNING FACTOR-LIKE2 peptides regulate awn development in rice. Plant Physiol. 2022, 190, 516–531. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.K.; Dubeaux, G.; Takahashi, Y.; Schroeder, J.I. Signaling mechanisms in abscisic acid mediated stomatal closure. Plant J. 2021, 105, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.K. Calcium Channels, OST1 and Stomatal Defence: Current Status and Beyond. Cells 2023, 12, 127. [Google Scholar] [CrossRef] [PubMed]
- Nerva, L.; Chitarra, W.; Fila, G.; Lovat, L.; Gaiotti, F. Variability in Stomatal Adaptation to Drought among Grapevine Cultivars: Genotype-Dependent Responses. Agriculture 2023, 13, 2186. [Google Scholar] [CrossRef]
- Liu, R.; Xu, K.; Li, Y.; Zhao, W.; Ji, H.; Lei, X.; Ma, T.; Ye, J.; Zhang, J.; Du, H.; et al. Investigation on the Potential Functions of ZmEPF/EPFL Family Members in Response to Abiotic Stress in Maize. Int. J. Mol. Sci. 2024, 25, 7196. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, H.; Wang, Q.; Chen, Z.; Sun, X.; Zhao, F.; Zhang, D.; Fei, J.; Zhao, R.; Yin, Y. Identification and Functional Analysis of the EPF/EPFL Gene Family in Maize (Zea mays L.): Implications for Drought Stress Response. Agronomy 2024, 14, 1734. https://doi.org/10.3390/agronomy14081734
Xia H, Wang Q, Chen Z, Sun X, Zhao F, Zhang D, Fei J, Zhao R, Yin Y. Identification and Functional Analysis of the EPF/EPFL Gene Family in Maize (Zea mays L.): Implications for Drought Stress Response. Agronomy. 2024; 14(8):1734. https://doi.org/10.3390/agronomy14081734
Chicago/Turabian StyleXia, Hanchao, Qi Wang, Ziqi Chen, Xiaopeng Sun, Fangfang Zhao, Di Zhang, Jianbo Fei, Rengui Zhao, and Yuejia Yin. 2024. "Identification and Functional Analysis of the EPF/EPFL Gene Family in Maize (Zea mays L.): Implications for Drought Stress Response" Agronomy 14, no. 8: 1734. https://doi.org/10.3390/agronomy14081734
APA StyleXia, H., Wang, Q., Chen, Z., Sun, X., Zhao, F., Zhang, D., Fei, J., Zhao, R., & Yin, Y. (2024). Identification and Functional Analysis of the EPF/EPFL Gene Family in Maize (Zea mays L.): Implications for Drought Stress Response. Agronomy, 14(8), 1734. https://doi.org/10.3390/agronomy14081734






