Application of Loop-Mediated Isothermal Amplification Assay Combined with Lateral Flow Dipstick (LAMP-LFD) for Specific and Sensitive Detection of Acidovorax citrulli (Schaad et al.) Causing Bacterial Fruit Blotch in Cucurbit Plants
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Strains
2.2. Reagents and Seeds
2.3. DNA Extraction
2.4. Primer and Probe Design
2.5. Optimization of LAMP-LFD Reaction Conditions
2.6. Specificity Assay
2.7. Sensitivity Assay
2.8. Evaluation of LAMP-LFD Assay for Detection of A. citrulli in Seeds
2.9. Detection of A. citrulli in Leaf Tissues
3. Results
3.1. Optimization of A. citrulli LAMP-LFD Conditions and Establishment of Detection Method
3.2. LAMP-LFD Specificity
3.3. LAMP-LFD Sensitivity
3.4. Artificially and Commercially Available Seeds Assay
3.5. LAMP-LFD Validation with Leaf Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crall, J.M.; Schenck, N.C. Bacterial fruit rot of watermelon in Florida. Plant Dis. Rep. 1969, 53, 74–75. [Google Scholar]
- Walcott, R.R.; Gitaitis, R.D. Detection of Acidovorax avenae subsp. citrulli in watermelon seed using immunomagnetic separation and the polymerase chain reaction. Plant Dis. 2000, 84, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.L.; Xu, J.; Zhao, Y.Q.; Li, X.H.; Hu, B.S.; Liu, F.Q. Specific detection of Acidovorax avenae subsp. citrulli by PCR. Jiangsu J. Agric. Sci. 2010, 26, 512–516. [Google Scholar]
- Wang, J.; Bi, Y.; Zhu, Y.; Han, S.Y.; Zhu, X.; Sheng, W.J.; Li, M. Nested-PCR rapidly detect Acidovorax avenae subsp. citrulli from watermelon seeds. Sci. Agric. Sin. 2014, 47, 284–291. [Google Scholar]
- Kan, Y.M.; Yun, X.M.; Li, Y.W.; Li, J.Q.; Lou, L.X. Screening of specific primers for detection of Acidovorax citrulli from cucurbits seed by Bio-PCR. Acta Phytopath. Sin. 2018, 48, 263–270. [Google Scholar]
- Husni, A.A.A.; Ismail, S.I.; Jaafar, N.; Zulperi, D. Etiology, diagnostic approaches and management strategies of Acidovorax citrulli, a bacterial fruit blotch pathogen of cucurbits. Plant Prot. Sci. 2021, 57, 75–94. [Google Scholar] [CrossRef]
- Takashi, S. Studies on control of seed borne bacterial vegetable disease: Studies on epidemiology and control of cucurbits bacterial fruit blotch. J. Gen. Plant Pathol. 2021, 87, 408–412. [Google Scholar]
- Cameron Somodi, C.; Jones, J.B.; Hopkins, D.L.; Stall, R.E.; Kucharer, T.A.; Hodge, N.C.; Watterson, J.C. Occurrence of a bacterial watermelon fruit blotch in Florida. Plant Dis. 1991, 75, 1053–1056. [Google Scholar] [CrossRef]
- Latin, R.X.; Hopkins, D.L. Bacterial fruit blotch of watermelon. Plant Dis. 1995, 79, 761–765. [Google Scholar] [CrossRef]
- Cai, X.Q.; Huang, Y.Y.; Yang, J.Z.; Chen, J.; Cai, G.L.; Hu, F.P. Pathogen identification of bacterial fruit blotch of watermelon in Fujian. J. Fujian Agric. For. Univ. 2005, 34, 434–437. [Google Scholar]
- Burdman, S.; Walcott, R. Acidovorax citrulli: Generating basic and applied knowledge to tackle a global threat to the cucurbit industry. Mol. Plant Pathol. 2012, 13, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Toshio, M.; Nakayama, E.; Nanaka, N.; Tabuchi, M. Host-specific activation of a pathogen effector Aave_4606 from Acidovorax citrulli, the causal agent for bacterial fruit blotch. Biochem. Biophys. Res. Commun. 2022, 616, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, D.L.; Thompson, C.M. Seed transmission of Acidovarax avenae subsp. citrulli in cucurbits. HortScience 2002, 37, 924–926. [Google Scholar] [CrossRef]
- Burdman, S.; Kots, N.; Kritzman, G.; Kopelowitz, J. Molecular, physiological, and host-range characterization of Acidovorax avenae subsp. citrulli isolates from watermelon and melon in Israel. Plant Dis. 2005, 89, 1339–1347. [Google Scholar] [CrossRef]
- Wu, P.Y.; Ho, L.C.; Chang, J.J.; Tzeng, K.C.; Deng, W.L.; Lin, Y.H. Development of a TaqMan probe-based insulated isothermal PCR (TiiPCR) for the detection of Acidovorax citrulli, the bacterial pathogen of watermelon fruit blotch. Eur. J. Plant Pathol. 2017, 147, 869–875. [Google Scholar] [CrossRef]
- Feng, J.J.; Li, J.Q.; Wacott, R.R.; Zhang, G.M.; Luo, L.X.; Kang, L.; Zheng, Y.; Schaad, N.W. Advances in detection of Acidovorax citrulli, the causal agent of bacterial fruit blotch of cucurbits. Seed Sci. Technol. 2013, 41, 1–15. [Google Scholar] [CrossRef]
- Tian, Q.; Feng, J.J.; Hu, J.; Zhao, W.J. Selective detection of viable seed-borne Acidovorax citrulli by real-time PCR with propidium monoazide. Sci. Rep. 2016, 6, 35457. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Hossain, R.H.; Kim, H.T.; Jesse, D.M.I.; Abuyusuf, M.; Jung, H.J.; Park, J.I.; Nou, I.S. Development of Molecular Markers for Detection of Acidovorax citrulli Strains Causing Bacterial Fruit Blotch Disease in Melon. Int. J. Mol. Sci. 2019, 20, 2715. [Google Scholar] [CrossRef]
- Fei, N.Y.; Chen, H.M.; Yang, Y.W.; Guan, W.; Liu, B.Y.; Zhao, T.C. Advances of cucurbit bacterial fruit blotch abroad. China Cucurb. Veget. 2022, 35, 1–5. [Google Scholar]
- Wang, Z.L.; Cheng, W.H.; Dong, Z.Y.; Yao, X.M.; Deng, X.; Ou, C. A CCRSPR/LbCas12a-based method for detection of bacterial fruit blotch pathogens in watermelon. Microbiol. Spectr. 2024, 12, 1–11. [Google Scholar]
- Schaad, N.W.; Postnikova, E.; Sechler, A.; Claflin, L.E.; Vidaver, A.K.; Jones, J.B.; Agarkova, I.; Ignatov, A.; Dickstein, E.; Ramundo, B.A. Reclassification of subspecies of Acidovorax avenae as A. avenae (Manns 1905) emend., A. cattleyae (Pavarino 1911) comb. nov., A. citrulli (Schaad et al., 1978) comb. nov., and proposal of A. oryzae sp. nov. Syst. Appl. Microbiol. 2008, 31, 434–446. [Google Scholar] [CrossRef]
- Zhao, T.; Feng, J.J.; Sechler, A. An Improved Assay for detection of Acidovorax avenae subsp. citrulli in watermelon and melon seed. Seed Sci. Technol. 2009, 37, 337–349. [Google Scholar] [CrossRef]
- Xiong, L.B.; Liu, Q.; Wang, T.C.; Gao, L.P.; Wang, J.; Liu, S.T.; Song, R.; Shi, Y.B.; Wang, J.P.; Wen, Z.H. An improved DAS Dot ELISA method for detection of Acidovorax avenae subsp. citrulli. Microbiol. China 2010, 37, 1551–1556. [Google Scholar]
- Himananto, O.; Thummabenjapone, P.; Luxananil, P. Novel and highly specific monoclonal antibody to Acidovorax citrulli and development of ELISA-based detection in cucurbit leaves and seeds. Plant Dis. 2011, 95, 1172–1178. [Google Scholar] [CrossRef][Green Version]
- Charlermroj, R.; Himananto, O.; Seepiban, C. Multiplex detection of plant pathogens using a microsphere immunoassay technology. PLoS ONE 2013, 8, e62344. [Google Scholar] [CrossRef]
- Charlermroj, R.; Makornwattana, M.; Himananto, O.; Seepidan, C.; Phuengwas, S.; Warin, N.; Gajanandana, O.; Karoonuthaisiri, K. An accurate, specific, sensitive, high-throughput method based on a microsphere immunoassay for multiplex detection of three viruses and bacterial fruit blotch bacterium in cucurbits. J. Virol. Methods 2017, 247, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hu, J. Preliminary research on serological method for detecting fruit blotch bacterium in hemi melon seed. J. Inner Mong. Agric. Univ. 2005, 26, 20–23. [Google Scholar]
- Puttharugsa, C.; Wangkam, T.; Huangkamhang, N.; Gajanandana, O.; Himananto, O.; Sutapun, B.; Amarit, R.; Somboonkaew, A.; Srikhirin, T. Development of surface plasmon resonance imaging for detection of Acidovorax avenae subsp. citrulli (Aac) using specific monoclonal antibody. Biosens. Bioelectron. 2011, 26, 2341–2346. [Google Scholar] [CrossRef]
- Kuo, S.Y.; Lin, Y.C.; Lai, Y.C. Production of fluorescent antibody-labeling proteins in plants using a viral vector and the application in the detection of Acidovorax citrulli and Bamboo mosaic virus. Pub. Libr. Sci. 2018, 13, e0192455. [Google Scholar] [CrossRef]
- Saisin, L.; Amarit, R.; Somboonkaew, A. Significant sensitivity improvement for camera-based lateral flow immunoassay readers. Sensors 2019, 18, 4026. [Google Scholar] [CrossRef]
- Venette, J.R.; Lamppa, R.S.; Albaugh, D.A.; Nayes, J.B. Presumptive procedure (dome test) for detection of seedborne bacterial pathogens in dry beans. Plant Dis. 1987, 71, 984–990. [Google Scholar] [CrossRef]
- Zhao, T.C.; Sun, F.Z.; Wang, B.W.; Hui, W.G. Pathogen identification of hami melon bacterial fruit blotch. Acta Phytopathol. Sin. 2001, 31, 357–364. [Google Scholar]
- Yan, L.C.; Zhao, Y.Q.; Zhou, J.J.; Chen, S.; Bai, S.; Tian, Y.L.; Gong, W.R.; Hu, B.S. Rapid and sensitive detection of Acidovorax citrulli in cucurbit seeds by visual loop-mediated isothermal amplification assay. J. Phytopath. 2019, 167, 10–18. [Google Scholar] [CrossRef]
- Zhao, Z.J.; Lu, Y.; Tian, W.; Wang, Y.X.; Wen, C.L.; Li, J.Q.; Xu, X.L.; Luo, L.X. Detection of Acidovorax citrulli and Pseudoonas syringae pv. lachrymans from cucurbit seeds by multiplex droplet digital PCR. Plant Prot. 2021, 47, 156–163. [Google Scholar]
- Bahar, O.; Efrat, M.; Hadar, M. New subspecies-specific polymerase chain reaction-based assay for the detection of Acidovorax avenae subsp. citrulli. Plant Pathol. 2008, 57, 754–763. [Google Scholar] [CrossRef]
- Oya, H.; Nakagawa, H.; Saito, N.; Uematsu, H.; Ohara, T. Detection of Acidovorax avenae subsp. citrulli from seed using LAMP method. Jpn. J. Phythol. 2008, 74, 304–310. [Google Scholar]
- Yang, G.; Erdman, D.; Tondella, M.L.; Fields, B.S. Evaluation tetramethylrhodamine and black hole quencher 1 labeled probes and five commercial amplification mixes in TaqMan® real-time RT-PCR assays for respiratory pathogens. J. Virol. Methods 2009, 162, 288–290. [Google Scholar] [CrossRef]
- Cho, M.S.; Park, D.H.; Ahn, T.Y.; Park, D.S. Rapid and specific detection of Acidovorax avenae subsp. citrulli using SYBR green-based real-time PCR amplification of the YD-repeat protein gene. J. Microbiol. Biotechnol. 2015, 25, 1401–1409. [Google Scholar] [CrossRef] [PubMed]
- Slovareva, O.Y.; Starikova, E.V. A novel qPCR-based test system for Acidovorax citrulli based on the PAS domain S-Box protein gene. Mol. Genet. Microbiol. Virol. 2021, 36, 100–103. [Google Scholar] [CrossRef]
- Tian, Y.L.; Zhao, Y.Q.; Bai, S.; Walcott, R.R.; Hu, B.S.; Liu, F.Q. Reliable and sensitive detection of Acidovorax cirulli in cucurbit seed using padlock-probe-based assay. Plant Dis. 2013, 97, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, L.; Xu, F.S.; Zhao, L.H.; Xie, G.L. Immuno-capture PCR method for detecting Acidovorax avenae subsp. citrulli from watermelon. Chin. J. Agric. Biotechnol. 2007, 4, 173–179. [Google Scholar]
- Zhang, J.; Tian, Q.; Zhu, S.F.; Zhao, W.J.; Liu, F.Q. Rapid on-sit detection of Acidovorax citrulli by cross-priming amplification. Mol. Cell. Probes 2012, 26, 175–176. [Google Scholar] [CrossRef] [PubMed]
- Lan, C.Z.; Gan, L.; Dai, Y.L.; Liu, X.F.; Yang, X.J. Development of loop-mediated isothermal amplification (LAMP) assay for specific and sensitive detection of Mycocentrospora acerina (Hart.) causing round leaf spot disease in Sanqi (Panax notogenseng). Horticulturae 2022, 18, 1060. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Mori, Y.; Tomita, N.; Kanda, H. Loop-mediated isothermal amplification (LAMP): Principle, features, and future prospects. J. Microbiol. 2015, 53, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Rigano, L.A.; Malamud, F.; Orce, I.G.; Filippone, M.P.; Marano, M.R.; Amaral, A.D.; Castagnaro, A.P.; Vojnov, A.A. Rapid and sensitive detection of Candidatus Liberibacter asiaticus by loop mediated isothermal amplification combined with a lateral flow dipstick. BMC Microbiol. 2014, 14, 86. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Wang, R.N.; Zhou, Q.J.; Chen, J.R. Rapid Detection of Vibrio vulnificus by Loop-mediated Isothermal Amplification Combined with Lateral Flow Dipstick Assay. Biotechnol. Bull. 2014, 6, 81–87. [Google Scholar]
- Sridapan, T.; Tangkawsakul, W.; Janvilisri, T.; Kiatpathomchai, W.; Dangtip, S.; Ngamwongsatit, N.; Nacapricha, D.; Ounjai, P.; Chankhamhaengdecha, S. Rapid detection of Clostridium perfringens in food by loop-mediated isothermal amplification combined with a lateral flow biosensor. PLoS ONE 2021, 16, e0245144. [Google Scholar] [CrossRef]
- Liu, W.W.; Lu, G.; Wang, Y.; Cheng, Z.H.; Gao, Y.Y.; Yin, Z.P.; Wu, Y.; Lv, X.Q.; Guo, P.B.; Zhao, Y.H. A novel loop-mediated isothermal amplification-lateral flow dipstick method for Helicobacter pylori detection. Front. Microbiol. 2023, 14, 1094600. [Google Scholar] [CrossRef]
- Peng, Q.D.; Ning, J.C.; Xu, Q.Y.; Yang, T.; Wang, Y.R.; Zheng, T.R.; Zhuang, Q.G.; Xi, D.H. Development and application of a reverse transcription loop-mediated isothermal amplification combined with lateral flow dipstick for rapid and visual detection of Citrus leaf blotch virus in kiwifruit. Crop Prot. 2021, 143, 105555. [Google Scholar] [CrossRef]
- Kim, N.K.; Lee, H.J.; Kim, S.M.; Jeong, R.D. Rapid and visual detection of barley yellow dwarf virus by reverse transcription recombinase polymerase amplification with lateral flow strips. Plant Pathol. J. 2022, 38, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.S.; Brennan, M.S.; Khan, A.; Ali, G.S. Implementation of loop-mediated isothermal amplification methods in lateral flow devices for the detection of Rhizoctonia solani. Can. J. Plant Pathol. 2015, 37, 118–129. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, H.L.; Li, X.J.; Shan, C.L.; Li, X.S.; Chen, Y.; Shao, W.D.; Zhu, P. Establishment of Leptosphaeria maculans detection by loop-mediated isothermal amplification combined with a lateral flow dipstick. Plant Quar. 2016, 30, 32–37. [Google Scholar]
- Jiang, L.; Gu, R.; Li, X.; Mu, D. Simple and rapid detection Aspergillus fumigatus by loop-mediated isothermal amplification coupled with lateral flow biosensor assay. J. Appl. Mcriobiol. 2021, 131, 2352–2360. [Google Scholar] [CrossRef]
- Cai, Y.; Zhou, Q.J.; Gu, J.F.; Chen, X.F.; Chen, J. Rapid and Sensitive Detection of Meloidogyne camelliae by LAMP-LFD. J. Agric. Biotechnol. 2016, 24, 770–780. [Google Scholar]
- Zhou, Q.J.; Cai, Y.; Gu, J.F.; Wang, X.; Chen, J. Rapid and sensitive detection of Meloidogyne mali by loop-mediated isothermal amplification combined with a lateral flow dipstick. Eur. J. Plant Pathol. 2017, 148, 755–769. [Google Scholar] [CrossRef]
- Sun, P.P.; Shi, J.L.; Peng, Z.; Wang, S.; Wang, Y.; Wu, X.Y.; Xu, S.J.; Li, J. Rapid and sensitive diagnosis of Mycoplasma hyopneumoniae by loop-mediated isothermal amplification combined with a lateral flow dipstic. Acta Vet. Zootech. Sin. 2020, 51, 1419–1428. [Google Scholar]
- Han, X.T.; Zhao, T.; Yan, T.; Yu, R.C. Rapid and sensitive detection of Karenia mikimotoi by loop-mediated isothermal amplification combined with lateral flow dipstick. Environ. Sci. Pollut. Res. 2022, 29, 24696–24703. [Google Scholar] [CrossRef]
- Hui, W.G.; Zhao, T.C.; Schaad, N.W.; Sun, F.Z.; Wang, J.R. Establishment of rapid detection method for the pathogen of hami melon fruit blotch. Sci. Agric. Sin. 2007, 10, 2495–2501. [Google Scholar]
- Walcott, R.R.; Castro, A.C.; Fessehaie, A.; Ling, K. Progress towards a commercial PCR-based seed assay for Acidovorax avenae subsp. citrulli. Seed Sci. Technol. 2006, 34, 101–116. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, H.J.; Zhao, Z.J.; Wen, C.L.; Wu, P.; Song, S.H.; Yu, S.C.; Lou, L.X.; Xu, X.L. Application of droplet digital PCR in detection of seed-transmitted pathogen Acidovorax citrulli. J. Integr. Agric. 2020, 19, 561–569. [Google Scholar]
- Walcott, R.R.; Langston, D.B.; Sanders, J.F.H.; Gitatitis, R.D. Investigating intraspecific variation of Acidovorax avenae subsp. citrulli using DNA fingerprinting and whole cell fatty acid analysis. Phytopathology 2000, 90, 191–196. [Google Scholar] [CrossRef]
- Ren, Y.Z.; Li, H.; Li, G.Y.; Wang, X.D.; Wan, G.; Fang, L. Detection of Acidovorax avenae subsp. citrulli in melon seed using the polymerase chain reaction. Xinjiang Agric. Sci. 2004, 41, 329–332. [Google Scholar]
- Zhang, X.L.; Wu, Y.M.; Wang, C.; Li, B.J. Sequence analysis of 16S rDNA and specified primers design of Acidovorax avenae subsp. citrulli. Acta Phytopathol. Sin. 2007, 37, 225–231. [Google Scholar]
- Eckshtain-Levi, N.; Shkedy, D.; Gershovits, M.; Da Silva, G.M.; Tamir-Ariel, D.; Walcott, R.; Pupko, T.; Burdman, S. Insights from the genome sequence of Acidovorax citrulli M6, a group I strain of the causal agent of bacterial fruit blotch of cucurbits. Front. Microbiol. 2016, 7, 430. [Google Scholar]
- Song, J.Y.; Oo, M.M.; Park, S.Y.; Seo, M.W.; Lee, S.C.; Jeon, N.B.; Nam, M.H.; Lee, Y.S.; Kim, H.G.; Oh, S.K. Analysis of intraspecific genetic diversity in Acidovorax citrulli causing bacterial fruit blotch on cucurbits in Korea. Korean J. Agric. Sci. 2018, 45, 575–582. [Google Scholar]
- Jackson, A.P.; Thomas, G.H.; Parkhill, J.; Thomson, N.R. Evolutionary diversification of an ancient gene family (rhs) through C-terminal displacement. BMC Genom. 2009, 10, 584. [Google Scholar] [CrossRef]
- Innis, M.A.; Gelfand, D.H. Optimization of PCR’s. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 3–12. [Google Scholar]
- Infantino, A.; Pucci, N. A PCR-based assay for the detection and identification of Pyrenochaeta lycopersici. Eur. J. Plant Pathol. 2005, 112, 337–347. [Google Scholar] [CrossRef]
- Zeng, H.J.; Zhang, D.Q.; Zhai, X.Z.; Wang, S.J.; Liu, Q. Enhancing the immunofluorescent sensitivity for detection of Acidovorax citrulli using fluorescein isothiocyanate labeled antigen and antibody. Anal. Bioanal. Chem. 2018, 410, 71–77. [Google Scholar]
- Wang, R.B.; Chen, S.Z.; Zhao, Y.M.; Li, B.J.; Liu, P.Q.; Chen, Q.H. Development of a recombinase polymerase amplification-lateral flow dipstick assay for rapid detection of the taro leaf blight pathogen Phytophthora colocasiae. J. Plant Prot. 2022, 49, 1654–1662. [Google Scholar]
- Song, Q.L.R. Rapid detection of seed borne Acidovorax avenae subsp. citrulli by “pathogen immune-concentration” PCR. J. Plant Pathol. 2009, 90, 600. [Google Scholar]
- Nurhan, Ö.; Hüseyi, B. A real-time PCR assay using locked nucleic acid probe for detection of Acrdovarax citrulli. J. Plant Dis. Prot. 2022, 129, 395–409. [Google Scholar]
- Ha, Y.; Fessehaie, A.; Ling, K.S. Simultaneous detection of Acidovarax avenae subsp. citrulli and Didymella bryoniae in cucurbit seedlots using magnetic capture hybridization and real-time polymerase chain reaction. Phytopathology 2009, 99, 666–678. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kirshner, B.; Kritzman, G. Sweat boxes and selective media for the detection of Acidovorax avenae subsp. citrulli in melon and watermelon seeds. Phytoparasitica 2008, 36, 134. [Google Scholar]
- DeBoer, S.H.; Ward, L.J.; Li, X.; Chittaranjan, S. Attenuation of PCR inhibition in the presence of plant compounds by addition of Blotto. Nucleic Acids Res. 1995, 23, 2567–2568. [Google Scholar] [CrossRef] [PubMed]
- Kontanis, E.J.; Reed, F.A. Evaluation of real-time PCR amplification efficiencies to detect PCR inhibitors. J. Forensic Sci. 2006, 51, 795–804. [Google Scholar] [CrossRef]
- Ozakman, M.; Schaad, N.W. A real-time BIO-PCR assay for detection of Ralstonia solanacearum race 3, biovar 2, in asymptomatic potato tubers. Can. J. Plant Pathol. 2003, 25, 232–239. [Google Scholar] [CrossRef]



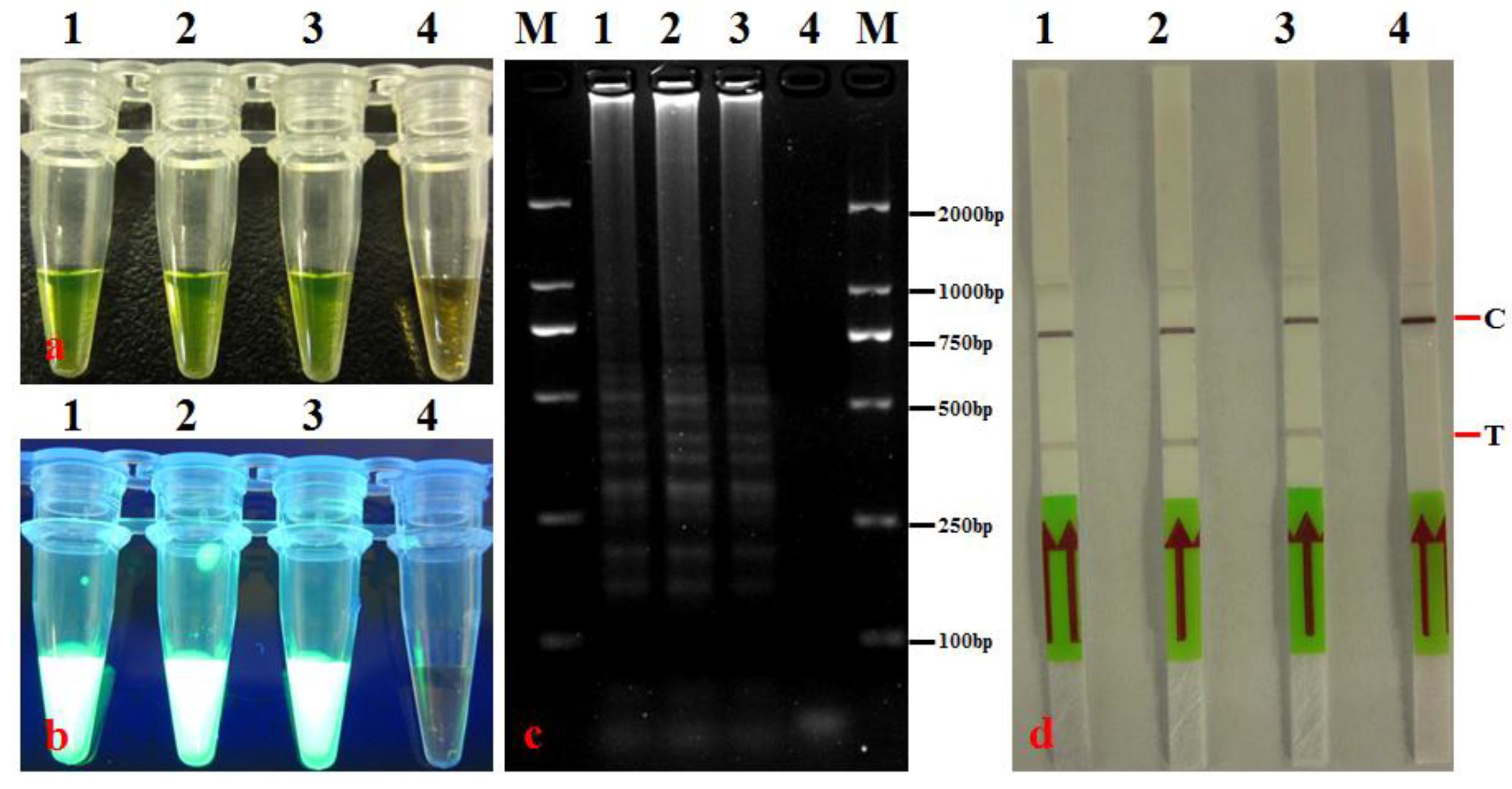


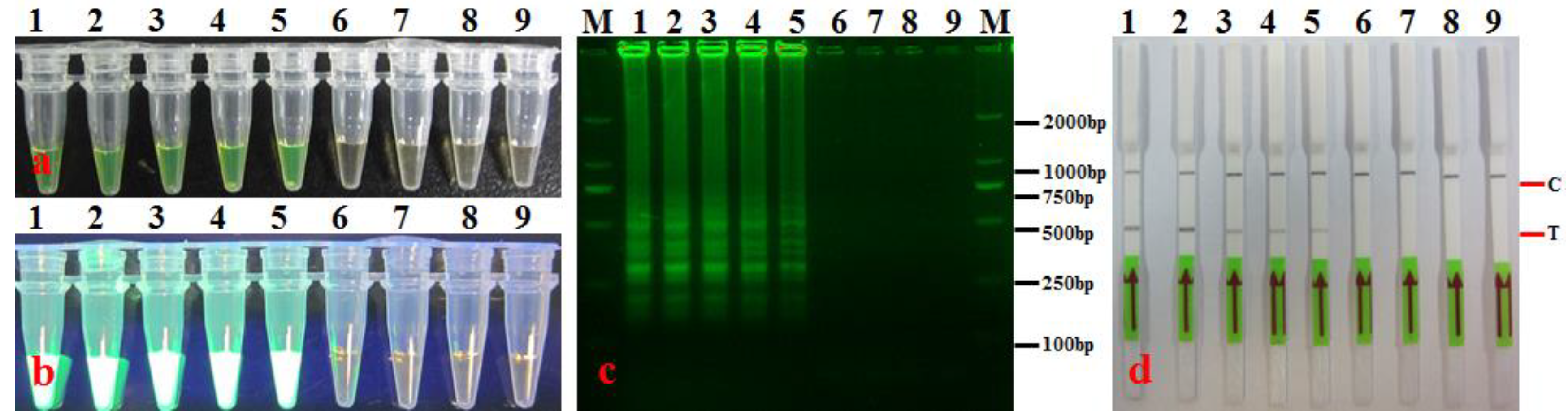
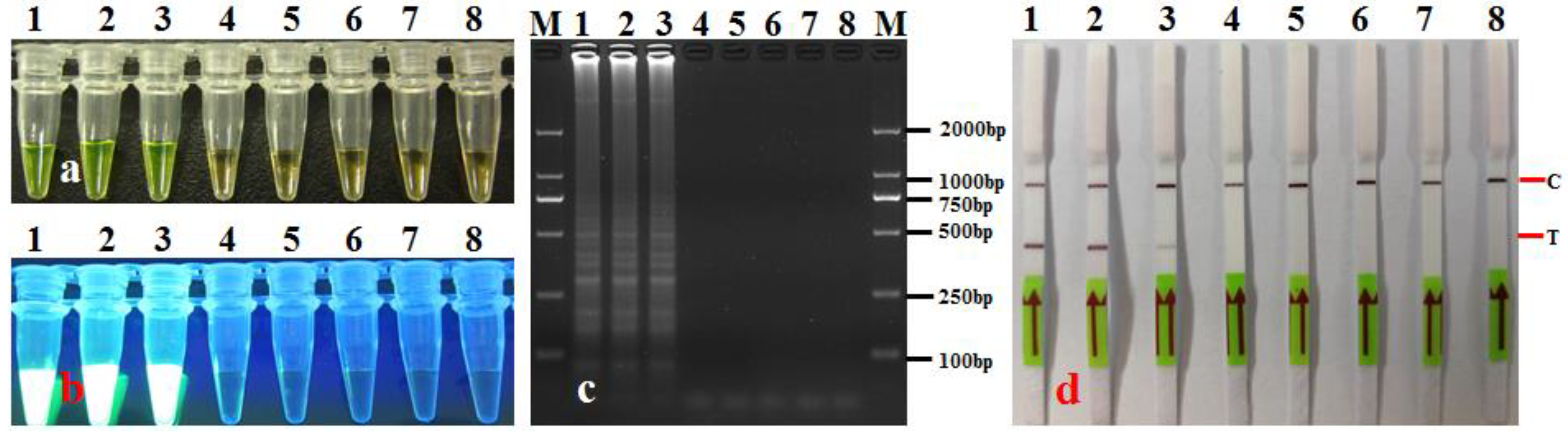
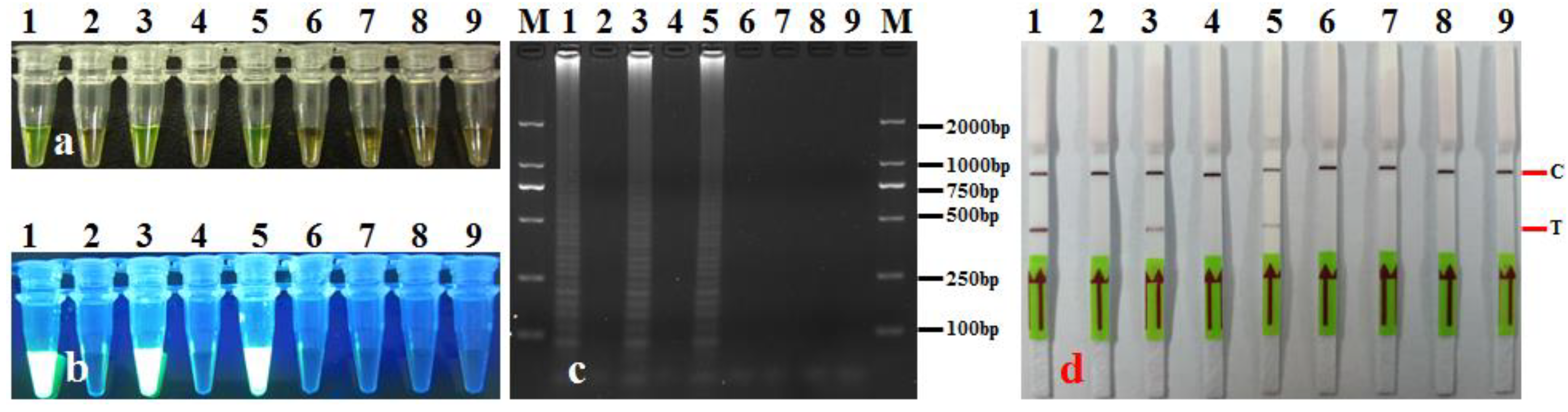
| No. | Species/Strains | Source/Origin | Host | LAMP-LFD |
|---|---|---|---|---|
| 1 | Acidovorax citrulli Ac-1 | IPPFAAS | Citrullus lanatus | + |
| 2 | A. citrulli Ac-2 | IPPFAAS | Cucumis melo | + |
| 3 | A. citrulli Ac-3 | FAFU | Citrullus lanatus | + |
| 4 | A. citrulli Ac-4 | FAFU | Cucumis melo | + |
| 5 | A. citrulli Ac-5 | PPSX | Cucumis melo | + |
| 6 | A. citrulli Ac-6 | PPSX | Cucumis melo | + |
| 7 | A. oryzae OS-1 | CGMCC | Oryza sativa | − |
| 8 | A. delafieldii UK-1 | CGMCC | Unknown | − |
| 9 | A. avenae subsp. avenae AS-1 | CGMCC | Avena sativa | − |
| 10 | A. caeni UK-1 | CGMCC | Unknown | − |
| 11 | A. avenae AS-1 | CGMCC | Avena sativa | − |
| 12 | Bacillus amyloliquefaciens UK-1 | IPPFAAS | Unknown | − |
| 13 | B. atrophaeus UK-1 | IPPFAAS | Unknown | − |
| 14 | B. megatherium CS-1 | IPPFAAS | Cucumis sativus | − |
| 15 | B. mojavensis UK-1 | IPPFAAS | Unknown | − |
| 16 | B. licheniformis CA-1 | IPPFAAS | Capsicum annuum | − |
| 17 | B. pumilus CA-1 | IPPFAAS | Capsicum annuum | − |
| 18 | B. velezensis CS-1 | IPPFAAS | Cucumis sativus | − |
| 19 | Burkholderia vietnamiensis UK-1 | IPPFAAS | Unknown | − |
| 20 | Escherichia coli CS-1 | IPPFAAS | Cucumis sativus | − |
| 21 | Lysinibacillus fusiformis CR-1 | IPPFAAS | Citrus reticulata | − |
| 22 | Paenibacillus polymyxa UK-1 | IPPFAAS | Unknown | − |
| 23 | Pseudomonas syringae pv. Actinidiae AC-1 | SICAU | Actinidia chinensis | − |
| 24 | P. viridiflava UK-1 | IPPFAAS | Unknown | − |
| 25 | P. cichorii UK-1 | IPPFAAS | Unknown | − |
| 26 | P. xiamenensis UK-1 | IPPFAAS | Unknown | − |
| 27 | P. syringae pv. tomato SL-1 | IPPFAAS | Solanum lycopersicon | − |
| 28 | Ralstonia solanacearum NT-1 | FAFU | Nicotiana tabacum | − |
| 29 | R. solanacearum DE-1 | IPPFAAS | Dioscorea esculenta | − |
| 30 | Xanthomonas oryzae pv. oryzae OS-1 | SICAU | Oryza sativa | − |
| 31 | X. oryzae pv. oryzicola OS-1 | IPPFAAS | Oryza sativa | − |
| 32 | X. axonopodis pv. citri CR-1 | IPPFAAS | Citrus reticulata | − |
| No. | Variety | Manufacturing Enterprise/Origin | LAMP-LFD Results of Five Replicates | ||||
|---|---|---|---|---|---|---|---|
| 1 | Watermelon cv. ‘Xiaodelei’ | Changchun Jixiangdi Seed Industry Co., Ltd., Changchun, China | + | + | + | + | + |
| 2 | Watermelon cv. ‘Lanhanteida’ | Hebei Qingfeng Agricultural Technology Co., Ltd., Qingxian, China | + | + | − | + | + |
| 3 | Watermelon cv. ‘Zhaojia 84-24’ | Qingxian Chunfeng Seed Industry Co., Ltd., Qingxian, China | − | − | − | − | − |
| 4 | Watermelon cv. ‘Hongxiaoyu’ | Shenyang Bosite Agricultural Technology Co., Ltd., Shenyang, China | + | − | + | − | + |
| 5 | Watermelon cv. ‘Lvlongbawang’ | Qingxian Aisen Vegetable Variety Promotion Center, Qiangxian, China | + | + | + | − | − |
| 6 | Watermelon cv. ‘Jingmihuanshuai’ | Qingxian Chunfeng Seed Industry Co., Ltd., Qingxian, China | − | − | − | − | − |
| 7 | Watermelon cv. ‘Zhimacui’ | Hebei Maohua Seed Industry Co., Ltd., Cangzhou, China | − | − | − | − | − |
| 8 | Watermelon cv. ‘Lanhanxigua’ | Henan Fangzhen Seed Co., Ltd., Xingxian, China | − | − | − | − | − |
| 9 | Watermelon cv. ‘Gailianjingxin’ | Qingxian Chunfeng Seed Industry Co., Ltd., Qingxian, China | − | − | − | − | − |
| 10 | Watermelon cv. ‘Lanhangxiwang’ | Henan Fangzhen Seed Co., Ltd., Xingxian, China | − | − | − | − | − |
| 11 | Watermelon cv. ‘Lanhanxinong 8’ | Henan Fangzhen Seed Co., Ltd., Xingxian, China | − | − | − | − | − |
| 12 | Watermelon cv. ‘Bintanxiaoxigua’ | Henan Fangzhen Seed Co., Ltd., Xingxian, China | − | − | − | − | − |
| 13 | Watermelon cv. ‘Bintanxiaoxigua 6’ | Henan Fangzhen Seed Co., Ltd., Xingxian, China | − | − | − | − | − |
| 14 | Watermelon cv. ‘Fangzheng 209’ | Henan Fangzhen Seed Co., Ltd., Xingxian, China | − | − | − | − | − |
| 15 | Watermelon cv. ‘Fangzhengheilong’ | Henan Fangzhen Seed Co., Ltd., Xingxian, China | − | − | − | − | − |
| 16 | Watermelon cv. ‘Bintan 8424’ | Henan Fangzhen Seed Co., Ltd., Xingxian, China | − | − | − | − | − |
| 17 | Watermelon cv. ‘Fangzheng 208’ | Henan Fangzhen Seed Co., Ltd., Xingxian, China | − | − | − | − | − |
| 18 | Watermelon cv. ‘Fangzhengmuzhi’ | Henan Fangzhen Seed Co., Ltd., Xingxian, China | − | − | − | − | − |
| 19 | Muskmelon cv. ‘Balixian’ | Henan Fangzhen Seed Co., Ltd., Xingxian, China | − | − | − | − | − |
| 20 | Muskmelon cv. ‘Ribentiaobao’ | Shouguang Meinong Information Technology Co., Ltd., Shouguang, China | − | − | − | − | − |
| 21 | Muskmelon cv. ‘Fangzhengyanjiaomi’ | Henan Fangzhen Seed Co., Ltd., Xingxian, China | − | − | − | − | − |
| 22 | Muskmelon cv. ‘Huancuili F1’ | Hebei Maohua Seed Industry Co., Ltd., Cangzhou, China | − | − | − | − | − |
| 23 | Muskmelon cv. ‘Xiantianxuemi 208’ | Hebei Maohua Seed Industry Co., Ltd., Cangzhou, China | − | − | − | − | − |
| 24 | Muskmelon cv. ‘Jinhongrui’ | Hebei Maohua Seed Industry Co., Ltd., Cangzhou, China | − | − | − | − | − |
| 25 | Muskmelon cv. ‘Tianla 008’ | Shouguang Meinong Information Technology Co., Ltd., Shouguang, China | − | + | + | + | + |
| 26 | Muskmelon cv. ‘Jinhuabao F1’ | Hebei Maohua Seed Industry Co., Ltd., Cangzhou, China | − | − | − | − | − |
| 27 | Muskmelon cv. ‘Jinmi No.1 F1’ | Hebei Maohua Seed Industry Co., Ltd., Cangzhou, China | − | − | − | − | − |
| 28 | Muskmelon cv. ‘Xianmangyuan F1’ | Hebei Maohua Seed Industry Co., Ltd., Cangzhou, China | − | − | − | − | − |
| 29 | Muskmelon cv. ‘Teixuanyanjiaocui’ | Hebei Maohua Seed Industry Co., Ltd., Cangzhou, China | − | − | − | − | − |
| 30 | Muskmelon cv. ‘Cuixiansu’ | Hebei Maohua Seed Industry Co., Ltd., Cangzhou, China | − | − | − | − | − |
| Primer/ Probe | Length (bp) | Sequence (5′–3′) | Purpose |
|---|---|---|---|
| F3 | 17 | CGGAAGGCGAACAGCAG | LAMP |
| B3 | 19 | GGTATCACTGCAGGGTCGA | LAMP |
| FIP | 36 | Biotin-GATGCGCTCGTAGCGCAGCCCGCGTCGTAGGCATCA | LAMP |
| BIP | 36 | 6-FAM-CCGGGACAGCACAGACAAGTTGCAGTCCACGCCCAC | LAMP |
| HP | 18 | GGATGCCGGGACAGCACA | LAMP-LFD |
| Seed Carrying Different Bacterial Cells | Seed with Different Infestation Rates | |||
|---|---|---|---|---|
| Bacterial Cells (CFU·g−1) | Results | Infestation Rate (%) | Frequency (%) (Positive/All Replicates) | Statistical Significance # |
| 104 | + | 0.1 | 100(10/10) | a |
| 103 | + | 0.09 | 100(10/10) | a |
| 102 | + | 0.08 | 90(9/10) | b |
| 101 | + | 0.07 | 80(8/10) | c |
| 5 | + | 0.06 | 50(5/10) | d |
| 2 | − | 0.05 | 20(2/10) | e |
| 1 | − | 0.04 | 0(0/10) | f |
| 0.5 | − | 0.03 | 0(0/10) | f |
| 0.2 | − | 0.02 | 0(0/10) | f |
| 0.1 | − | 0.01 | 0(0/10) | f |
| 0 | − | 0 | 0 | / |
| Sample Code | Location | Collection Time | Varieties | Number of Samples | Frequency of Positives (%) |
|---|---|---|---|---|---|
| XP1~XP10 | Ningde Xiapu, China | 2023-7-14 | Watermelon cv. ‘Xinhuanghou’ | 10 | 20.00 |
| SM1~SM12 | Sanming Sanyuan, China | 2023-8-9 | Watermelon cv. ‘Heimeireng’ | 12 | 0 |
| NP1~NP8 | Nanping Jianyang, China | 2023-8-22 | Muskmelon cv. ‘Balixian’ | 8 | 12.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lan, C.; Luo, M.; Gan, L.; Hu, M.; Ruan, H.; Dai, Y.; Liu, X.; Yang, X. Application of Loop-Mediated Isothermal Amplification Assay Combined with Lateral Flow Dipstick (LAMP-LFD) for Specific and Sensitive Detection of Acidovorax citrulli (Schaad et al.) Causing Bacterial Fruit Blotch in Cucurbit Plants. Agronomy 2024, 14, 1804. https://doi.org/10.3390/agronomy14081804
Lan C, Luo M, Gan L, Hu M, Ruan H, Dai Y, Liu X, Yang X. Application of Loop-Mediated Isothermal Amplification Assay Combined with Lateral Flow Dipstick (LAMP-LFD) for Specific and Sensitive Detection of Acidovorax citrulli (Schaad et al.) Causing Bacterial Fruit Blotch in Cucurbit Plants. Agronomy. 2024; 14(8):1804. https://doi.org/10.3390/agronomy14081804
Chicago/Turabian StyleLan, Chengzhong, Minsang Luo, Lin Gan, Meiling Hu, Hongchun Ruan, Yuli Dai, Xiaofei Liu, and Xiujuan Yang. 2024. "Application of Loop-Mediated Isothermal Amplification Assay Combined with Lateral Flow Dipstick (LAMP-LFD) for Specific and Sensitive Detection of Acidovorax citrulli (Schaad et al.) Causing Bacterial Fruit Blotch in Cucurbit Plants" Agronomy 14, no. 8: 1804. https://doi.org/10.3390/agronomy14081804
APA StyleLan, C., Luo, M., Gan, L., Hu, M., Ruan, H., Dai, Y., Liu, X., & Yang, X. (2024). Application of Loop-Mediated Isothermal Amplification Assay Combined with Lateral Flow Dipstick (LAMP-LFD) for Specific and Sensitive Detection of Acidovorax citrulli (Schaad et al.) Causing Bacterial Fruit Blotch in Cucurbit Plants. Agronomy, 14(8), 1804. https://doi.org/10.3390/agronomy14081804






