Assessing the Capabilities of UV-NIR Spectroscopy for Predicting Macronutrients in Hydroponic Solutions with Single-Task and Multi-Task Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
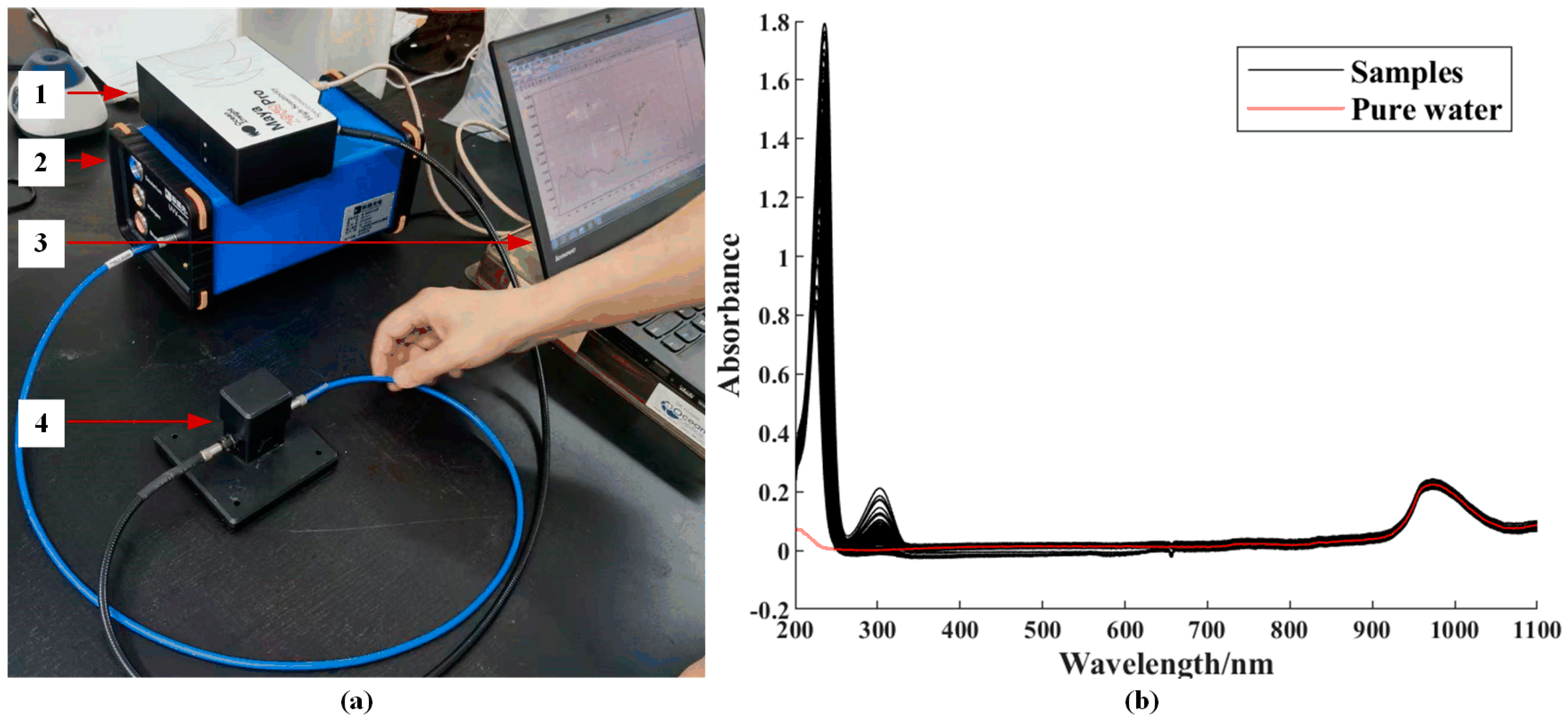
2.2. Spectral Collection and Preprocessing
2.3. Learning Algorithms
2.4. Modelling and Evaluation
3. Results and Discussion
3.1. Characteristics of the Content of Nutrients
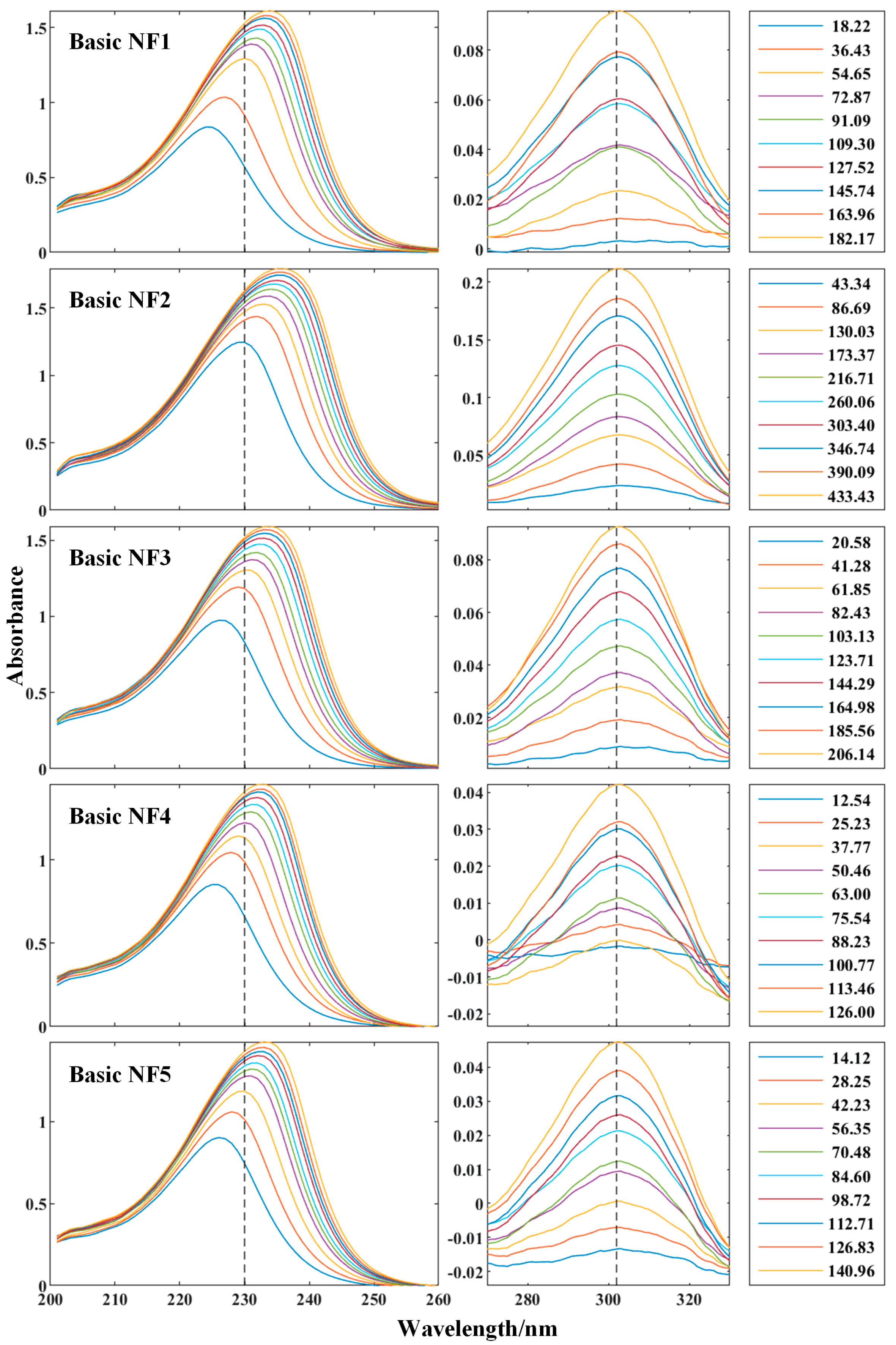
3.2. Spectral Response
3.3. Model Performance
3.3.1. Prediction Results
3.3.2. Comparison of Different Algorithms
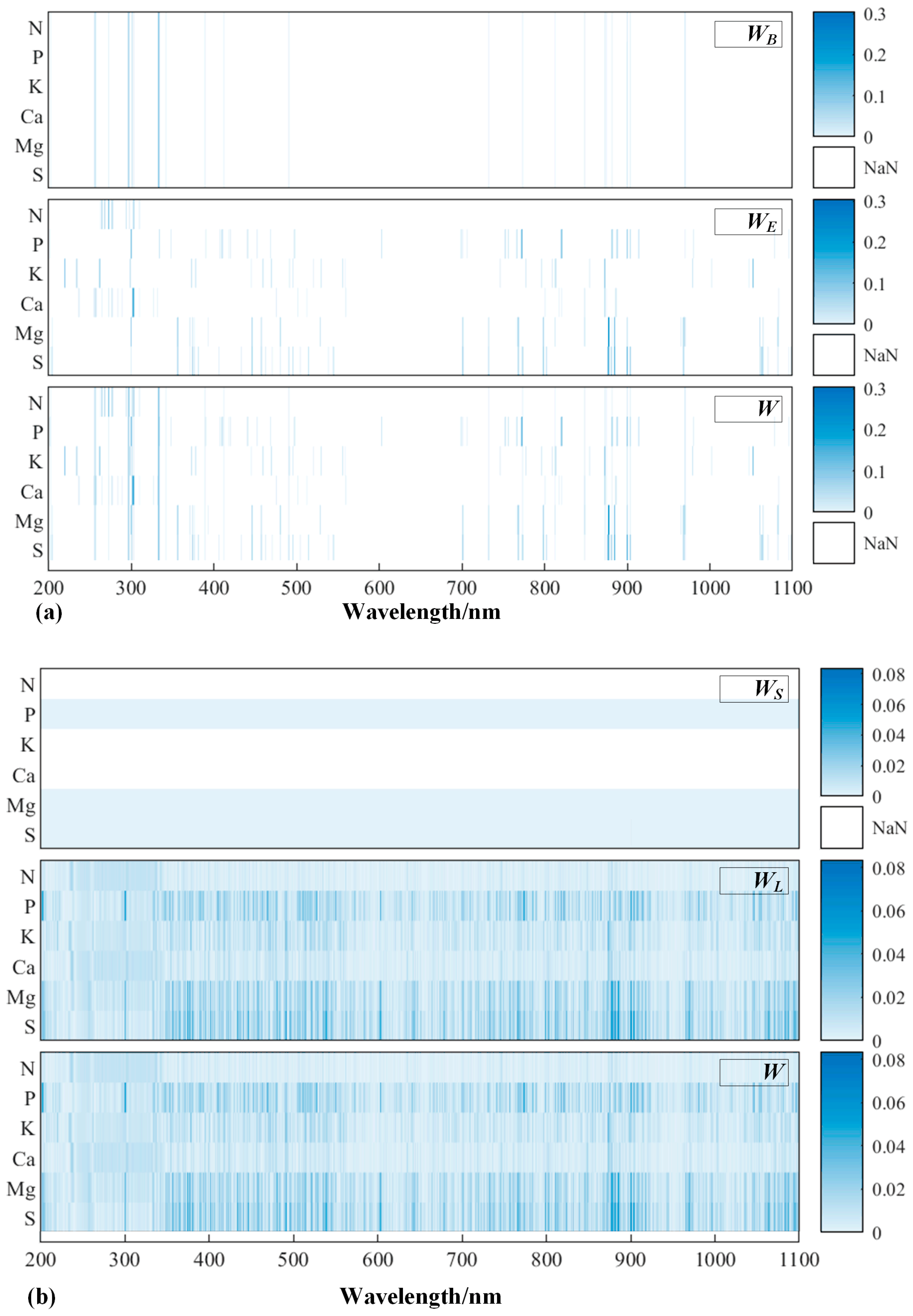
3.3.3. Interpretation of the Prediction Mechanism
3.3.4. Comparison with Previous Studies
4. Conclusions
- N and Ca could be predicted with good accuracy (RPD > 2), K could be predicted with moderate accuracy (1.4 < RPD < 2), and P, Mg, and S could not be successfully predicted (RPD < 1.4);
- Significant spectral absorptions mainly caused by N could be found around 230 nm and 302 nm, and regression features were thereby generated. Other macronutrients did not show any obvious absorption characteristic along UV-NIR, but K and Ca have significant shared features with N;
- Multi-task algorithms usually showed stronger learning ability compared to single-task algorithms, especially with RMTL, which could improve prediction performance for relevant tasks, namely predicting K and Ca and identifying the irrelevant (outlier) tasks—predicting P, Mg, and S.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Trejo-Téllez, L.I.; Gómez-Merino, F.C. Nutrient Solutions for Hydroponic Systems. In Hydroponics—A Standard Methodology for Plant Biological Researches; IntechOpen: London, UK, 2012; ISBN 978-953-51-0386-8. [Google Scholar]
- Sambo, P.; Nicoletto, C.; Giro, A.; Pii, Y.; Valentinuzzi, F.; Mimmo, T.; Lugli, P.; Orzes, G.; Mazzetto, F.; Astolfi, S.; et al. Hydroponic Solutions for Soilless Production Systems: Issues and Opportunities in a Smart Agriculture Perspective. Front. Plant Sci. 2019, 10, 923. [Google Scholar] [CrossRef] [PubMed]
- Bugbee, B. Nutrient Management in Recirculating Hydroponic Culture. In Proceedings of the South Pacific Soilless Culture Conference, Palmerston North, New Zealand, 10–13 February 2003; Acta Hort: Brussels, Belgium, 2004; Volume 648, pp. 99–112. [Google Scholar]
- Son, J.E.; Kim, H.J.; Ahn, T.I. Chapter 20—Hydroponic systems. In Plant Factory, 2nd ed.; Kozai, T., Niu, G., Takagaki, M., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 273–283. ISBN 978-0-12-816691-8. [Google Scholar]
- Surantha, N. Intelligent Monitoring and Controlling System for Hydroponics Precision Agriculture. In Proceedings of the 2019 7th International Conference on Information and Communication Technology (ICoICT), Kuala Lumpur, Malaysia, 24–26 July 2019; pp. 1–6. [Google Scholar]
- Wada, T. Chapter 1.1—Theory and Technology to Control the Nutrient Solution of Hydroponics. In Plant Factory Using Artificial Light; Anpo, M., Fukuda, H., Wada, T., Eds.; Elsevier: Amsterdam, Netherlands, 2019; pp. 5–14. ISBN 978-0-12-813973-8. [Google Scholar]
- Cho, W.-J.; Kim, H.-J.; Jung, D.-H.; Kim, D.-W.; Ahn, T.I.; Son, J.-E. On-Site Ion Monitoring System for Precision Hydroponic Nutrient Management. Comput. Electron. Agric. 2018, 146, 51–58. [Google Scholar] [CrossRef]
- Bamsey, M.; Graham, T.; Thompson, C.; Berinstain, A.; Scott, A.; Dixon, M. Ion-Specific Nutrient Management in Closed Systems: The Necessity for Ion-Selective Sensors in Terrestrial and Space-Based Agriculture and Water Management Systems. Sensors 2012, 12, 13349–13392. [Google Scholar] [CrossRef]
- Asao, T. (Ed.) Hydroponics: A Standard Methodology for Plant Biological Researches; InTech: London, UK, 2012; ISBN 978-953-51-0386-8. [Google Scholar]
- Kim, H.-J.; Kim, D.-W.; Kim, W.K.; Cho, W.-J.; Kang, C.I. PVC Membrane-Based Portable Ion Analyzer for Hydroponic and Water Monitoring. Comput. Electron. Agric. 2017, 140, 374–385. [Google Scholar] [CrossRef]
- Jung, D.-H.; Kim, H.-J.; Cho, W.-J.; Park, S.H.; Yang, S.-H. Validation Testing of an Ion-Specific Sensing and Control System for Precision Hydroponic Macronutrient Management. Comput. Electron. Agric. 2019, 156, 660–668. [Google Scholar] [CrossRef]
- Xu, K.; Kitazumi, Y.; Kano, K.; Shirai, O. Automatic Management of Nutrient Solution for Hydroponics—Construction of Multi-ion Stat-. Anal. Sci. 2020, 36, 1141–1144. [Google Scholar] [CrossRef] [PubMed]
- Ban, B.; Ryu, D.; Lee, M. Machine Learning Approach to Remove Ion Interference Effect in Agricultural Nutrient Solutions. In Proceedings of the 2019 International Conference on Information and Communication Technology Convergence (ICTC), Jeju Island, Korea, 16–18 October 2019; pp. 1156–1161. [Google Scholar]
- Kim, H.-J.; Kim, W.-K.; Roh, M.-Y.; Kang, C.-I.; Park, J.-M.; Sudduth, K.A. Automated Sensing of Hydroponic Macronutrients Using a Computer-Controlled System with an Array of Ion-Selective Electrodes. Comput. Electron. Agric. 2013, 93, 46–54. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, C.; Ye, R.; Duan, Q. Advances on Water Quality Detection by UV-Vis Spectroscopy. Appl. Sci. 2020, 10, 6874. [Google Scholar] [CrossRef]
- Chen, X.; Yin, G.; Zhao, N.; Gan, T.; Yang, R.; Xia, M.; Feng, C.; Chen, Y.; Huang, Y. Simultaneous Determination of Nitrate, Chemical Oxygen Demand and Turbidity in Water Based on UV-Vis Absorption Spectrometry Combined with Interval Analysis. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 244, 118827. [Google Scholar] [CrossRef]
- Causse, J.; Thomas, O.; Jung, A.-V.; Thomas, M.-F. Direct DOC and Nitrate Determination in Water Using Dual Pathlength and Second Derivative UV Spectrophotometry. Water Res. 2017, 108, 312–319. [Google Scholar] [CrossRef]
- Karlsson, M.; Karlberg, B.; Olsson, R.J.O. Determination of Nitrate in Municipal Waste Water by UV Spectroscopy. Anal. Chim. Acta 1995, 312, 107–113. [Google Scholar] [CrossRef]
- Li, J.; Tong, Y.; Guan, L.; Wu, S.; Li, D. Optimization of COD Determination by UV-Vis Spectroscopy Using PLS Chemometrics Algorithms. Optik 2018, 174, 591–599. [Google Scholar] [CrossRef]
- Brito, R.S.; Pinheiro, H.M.; Ferreira, F.; Matos, J.S.; Lourenço, N.D. In Situ UV-Vis Spectroscopy to Estimate COD and TSS in Wastewater Drainage Systems. Urban Water J. 2014, 11, 261–273. [Google Scholar] [CrossRef]
- Shi, W.; Zhuang, W.-E.; Hur, J.; Yang, L. Monitoring Dissolved Organic Matter in Wastewater and Drinking Water Treatments Using Spectroscopic Analysis and Ultra-High Resolution Mass Spectrometry. Water Res. 2021, 188, 116406. [Google Scholar] [CrossRef]
- Cook, S.; Peacock, M.; Evans, C.D.; Page, S.E.; Whelan, M.J.; Gauci, V.; Kho, L.K. Quantifying Tropical Peatland Dissolved Organic Carbon (DOC) Using UV-Visible Spectroscopy. Water Res. 2017, 115, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Monteiro-Silva, F.; Jorge, P.A.S.; Martins, R.C. Optical Sensing of Nitrogen, Phosphorus and Potassium: A Spectrophotometrical Approach toward Smart Nutrient Deployment. Chemosensors 2019, 7, 51. [Google Scholar] [CrossRef]
- Silva, A.F.; Löfkvist, K.; Gilbertsson, M.; Os, E.V.; Franken, G.; Balendonck, J.; Pinho, T.M.; Boaventura-Cunha, J.; Coelho, L.; Jorge, P.; et al. Hydroponics Monitoring through UV-Vis Spectroscopy and Artificial Intelligence: Quantification of Nitrogen, Phosphorous and Potassium. Chem. Proc. 2021, 5, 88. [Google Scholar] [CrossRef]
- Stevens, J.D.; Murray, D.; Diepeveen, D.; Toohey, D. Development and Testing of an IoT Spectroscopic Nutrient Monitoring System for Use in Micro Indoor Smart Hydroponics. Horticulturae 2023, 9, 185. [Google Scholar] [CrossRef]
- Jung, D.-H.; Kim, H.-J.; Kim, H.; Choi, J.; Kim, J.; Park, S. Fusion of Spectroscopy and Cobalt Electrochemistry Data for Estimating Phosphate Concentration in Hydroponic Solution. Sensors 2019, 19, 2596. [Google Scholar] [CrossRef]
- Han, H.; Jung, D.-H.; Kim, H.-J.; Lee, T.S.; Kim, H.S.; Kim, H.-Y.; Park, S.H. Application of a Spectroscopic Analysis-Based Portable Sensor for Phosphate Quantitation in Hydroponic Solutions. J. Sens. 2020, 2020, 9251416. [Google Scholar] [CrossRef]
- Sulaiman, R.; Azeman, N.H.; Mokhtar, M.H.H.; Mobarak, N.N.; Abu Bakar, M.H.; Bakar, A.A.A. Hybrid Ensemble-Based Machine Learning Model for Predicting Phosphorus Concentrations in Hydroponic Solution. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2024, 304, 123327. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Paz-Kagan, T.; Karnieli, A.; Li, S. Linear Multi-Task Learning for Predicting Soil Properties Using Field Spectroscopy. Remote Sens. 2017, 9, 1099. [Google Scholar] [CrossRef]
- Curcio, J.A.; Petty, C.C. The Near Infrared Absorption Spectrum of Liquid Water. JOSA 1951, 41, 302–304. [Google Scholar] [CrossRef]
- Qi, H.; Paz-Kagan, T.; Karnieli, A.; Jin, X.; Li, S. Evaluating Calibration Methods for Predicting Soil Available Nutrients Using Hyperspectral VNIR Data. Soil Tillage Res. 2018, 175, 267–275. [Google Scholar] [CrossRef]
- Rinnan, Å.; Van Den Berg, F.; Engelsen, S.B. Review of the Most Common Pre-Processing Techniques for near-Infrared Spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Helland, I.S.; Næs, T.; Isaksson, T. Related Versions of the Multiplicative Scatter Correction Method for Preprocessing Spectroscopic Data. Chemom. Intell. Lab. Syst. 1995, 29, 233–241. [Google Scholar] [CrossRef]
- Duckworth, J. Mathematical Data Preprocessing. In Near-Infrared Spectroscopy in Agriculture; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2004; pp. 113–132. ISBN 978-0-89118-236-8. [Google Scholar]
- Westad, F.; Martens, H. Variable Selection in near Infrared Spectroscopy Based on Significance Testing in Partial Least Squares Regression. J. Infrared Spectrosc. 2000, 8, 117–124. [Google Scholar] [CrossRef]
- Yu, H.; Chang, G.; Zhang, S.; Zhu, Y.; Yu, Y. Application of Sparse Regularization in Spherical Radial Basis Functions-Based Regional Geoid Modeling in Colorado. Remote Sens. 2023, 15, 4870. [Google Scholar] [CrossRef]
- Pan, T.; Li, M. Feature Extraction From Spectroscopy Using LASSO and Net Analyte Signal. IEEE Sens. J. 2022, 22, 12997–13004. [Google Scholar] [CrossRef]
- Wold, S.; Ruhe, A.; Wold, H.; Dunn, W.J., III. The Collinearity Problem in Linear Regression. The Partial Least Squares (PLS) Approach to Generalized Inverses. SIAM J. Sci. Stat. Comput. 1984, 5, 735–743. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression Shrinkage and Selection Via the Lasso. J. R. Stat. Soc. Ser. B Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Q. An Overview of Multi-Task Learning. Natl. Sci. Rev. 2018, 5, 30–43. [Google Scholar] [CrossRef]
- Thung, K.-H.; Wee, C.-Y. A Brief Review on Multi-Task Learning. Multimed. Tools Appl. 2018, 77, 29705–29725. [Google Scholar] [CrossRef]
- Jalali, A.; Sanghavi, S.; Ruan, C.; Ravikumar, P. A Dirty Model for Multi-task Learning. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 6–9 December 2010; Curran Associates, Inc.: New York, NY, USA, 2010; Volume 23. [Google Scholar]
- Zhou, J.; Chen, J.; Ye, J. MALSAR: Multi-Task Learning via Structural Regularization. Available online: https://jiayuzhou.github.io/MALSAR/ (accessed on 12 February 2024).
- Negahban, S.; Wainwright, M.J. Joint Support Recovery under High-Dimensional Scaling: Benefits and Perils of L1,∞-regularization. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 8–10 December 2008; Curran Associates, Inc.: New York, NY, USA, 2008; Volume 21. [Google Scholar]
- Zhang, C.-H.; Huang, J. The Sparsity and Bias of the Lasso Selection in High-Dimensional Linear Regression. Ann. Stat. 2008, 36, 1567–1594. [Google Scholar] [CrossRef]
- Liu, W. Improved Bounds for Multi-Task Learning with Trace Norm Regularization. In Proceedings of the Thirty Sixth Conference on Learning Theory, Bangalore, India, 12–15 July 2023; PMLR: London, UK, 2023; pp. 700–714. [Google Scholar]
- Boursier, E.; Konobeev, M.; Flammarion, N. Trace Norm Regularization for Multi-Task Learning with Scarce Data. In Proceedings of the Thirty Fifth Conference on Learning Theory, London, UK, 2–5 July 2022; PMLR: London, UK, 2022; pp. 1303–1327. [Google Scholar]
- Chen, J.; Zhou, J.; Ye, J. Integrating Low-Rank and Group-Sparse Structures for Robust Multi-Task Learning. In Proceedings of the 17th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Diego, CA, USA, 21–24 August 2011; Association for Computing Machinery: New York, NY, USA, 2011; pp. 42–50. [Google Scholar]
- Kennard, R.W.; Stone, L.A. Computer Aided Design of Experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Jiao, Y.; Li, Z.; Chen, X.; Fei, S. Preprocessing Methods for Near-Infrared Spectrum Calibration. J. Chemom. 2020, 34, e3306. [Google Scholar] [CrossRef]
- Chang, C.-W.; Laird, D.A.; Mausbach, M.J.; Hurburgh, C.R. Near-Infrared Reflectance Spectroscopy–Principal Components Regression Analyses of Soil Properties. Soil Sci. Soc. Am. J. 2001, 65, 480–490. [Google Scholar] [CrossRef]
- Hastie, T.; Friedman, J.; Tibshirani, R. The Elements of Statistical Learning; Springer Series in Statistics; Springer: New York, NY, USA, 2001; ISBN 978-1-4899-0519-2. [Google Scholar]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A Basic Tool of Chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Malgady, R.G.; Krebs, D.E. Understanding Correlation Coefficients and Regression. Phys. Ther. 1986, 66, 110–120. [Google Scholar] [CrossRef]
- Matcher, S.J.; Cope, M.; Delpy, D.T. Use of the Water Absorption Spectrum to Quantify Tissue Chromophore Concentration Changes in Near-Infrared Spectroscopy. Phys. Med. Biol. 1994, 39, 177. [Google Scholar] [CrossRef] [PubMed]

| Mother Solution | Chemical Reagents | Application Amounts (mg/L) | ||||
|---|---|---|---|---|---|---|
| Basic NF1/ Tomato | Basic NF2/ Sweet Potato | Basic NF3/ Lettuce | Basic NF4/ Grape | Basic NF5/ Watermelon | ||
| A | KNO3 | 500 | 750 | 600 | 404 | 238 |
| MgSO4·7H2O | 300 | 450 | 433 | 246 | 500 | |
| NH4H2PO4 | 50 | 75 | 133 | - | - | |
| KH2PO4 | 150 | 225 | - | 136 | 185 | |
| B | Ca(NO3)2·4H2O | 900 | 2700 | 900 | 590 | 910 |
| Units | Mean | STD | Min | Median | Max | |
|---|---|---|---|---|---|---|
| pH | 6.96 | 0.31 | 6.17 | 6.95 | 7.59 | |
| EC | µS/cm | 288.95 | 149.66 | 72.09 | 257.90 | 864.00 |
| N | mg/L | 111.81 | 80.89 | 12.54 | 91.91 | 433.43 |
| P | mg/L | 24.08 | 15.84 | 0.46 | 22.11 | 71.50 |
| K | mg/L | 122.00 | 74.00 | 2.70 | 115.82 | 354.13 |
| Ca | mg/L | 103.77 | 86.20 | 3.05 | 86.95 | 457.63 |
| Mg | mg/L | 20.86 | 14.03 | 0.17 | 18.13 | 64.75 |
| S | mg/L | 24.19 | 14.47 | 0.23 | 22.85 | 57.55 |
| pH | EC | N | P | K | Ca | Mg | S | |
|---|---|---|---|---|---|---|---|---|
| pH | 1.00 | |||||||
| EC | −0.72 | 1.00 | ||||||
| N | −0.66 | 0.99 | 1.00 | |||||
| P | −0.85 | 0.66 | 0.61 | 1.00 | ||||
| K | −0.67 | 0.88 | 0.86 | 0.65 | 1.00 | |||
| Ca | −0.64 | 0.95 | 0.97 | 0.60 | 0.73 | 1.00 | ||
| Mg | −0.61 | 0.76 | 0.70 | 0.62 | 0.56 | 0.71 | 1.00 | |
| S | −0.52 | 0.57 | 0.48 | 0.45 | 0.40 | 0.47 | 0.90 | 1.00 |
| Algorithm | Macronutrient | Parameter 1 | N 2 | Calibration | Validation | Accuracy Category | |||
|---|---|---|---|---|---|---|---|---|---|
| RPD | SSR/SST | RPD | SSR/SST | ||||||
| PLS | N | 5 | 898 | 11.16 | 0.99 | 9.46 | 0.91 | A | |
| P | 2 | 898 | 1.60 | 0.61 | 1.04 | 0.41 | C | ||
| K | 4 | 898 | 3.16 | 0.90 | 1.70 | 0.87 | B | ||
| Ca | 5 | 898 | 6.94 | 0.98 | 3.93 | 0.86 | A | ||
| Mg | 2 | 898 | 1.46 | 0.52 | 1.08 | 0.38 | C | ||
| S | 2 | 898 | 1.35 | 0.44 | 0.91 | 0.59 | C | ||
| LASSO | N | 0.03 | 12 | 9.62 | 0.93 | 10.12 | 0.90 | A | |
| P | 0.08 | 21 | 2.38 | 0.59 | 1.03 | 0.35 | C | ||
| K | 0.09 | 18 | 2.98 | 0.70 | 1.66 | 0.68 | B | ||
| Ca | 0.03 | 25 | 6.33 | 0.90 | 3.27 | 0.72 | A | ||
| Mg | 0.2 | 10 | 1.57 | 0.33 | 1.32 | 0.19 | C | ||
| S | 0.16 | 13 | 1.58 | 0.35 | 0.96 | 0.30 | C | ||
| DMTL | N | 29 | 7 | 31 | 8.28 | 0.90 | 7.50 | 0.84 | A |
| P | 51 | 2.90 | 0.69 | 1.04 | 0.40 | C | |||
| K | 49 | 4.02 | 0.80 | 1.68 | 0.82 | B | |||
| Ca | 44 | 4.82 | 0.85 | 3.43 | 0.69 | A | |||
| Mg | 46 | 2.84 | 0.67 | 1.15 | 0.45 | C | |||
| S | 59 | 2.80 | 0.64 | 0.95 | 0.55 | C | |||
| RMTL | N | 5 | 21 | 898 | 51.06 | 0.99 | 8.58 | 0.91 | A |
| P | 898 | 40.49 | 0.99 | 1.07 | 0.49 | C | |||
| K | 898 | 37.65 | 0.98 | 1.73 | 0.93 | B | |||
| Ca | 898 | 44.03 | 0.99 | 4.35 | 0.81 | A | |||
| Mg | 898 | 38.76 | 1.00 | 1.28 | 0.44 | C | |||
| S | 898 | 32.09 | 1.03 | 1.09 | 0.58 | C | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, H.; Li, B.; Nie, J.; Luo, Y.; Yuan, Y.; Zhou, X. Assessing the Capabilities of UV-NIR Spectroscopy for Predicting Macronutrients in Hydroponic Solutions with Single-Task and Multi-Task Learning. Agronomy 2024, 14, 1974. https://doi.org/10.3390/agronomy14091974
Qi H, Li B, Nie J, Luo Y, Yuan Y, Zhou X. Assessing the Capabilities of UV-NIR Spectroscopy for Predicting Macronutrients in Hydroponic Solutions with Single-Task and Multi-Task Learning. Agronomy. 2024; 14(9):1974. https://doi.org/10.3390/agronomy14091974
Chicago/Turabian StyleQi, Haijun, Bin Li, Jun Nie, Yizhi Luo, Yu Yuan, and Xingxing Zhou. 2024. "Assessing the Capabilities of UV-NIR Spectroscopy for Predicting Macronutrients in Hydroponic Solutions with Single-Task and Multi-Task Learning" Agronomy 14, no. 9: 1974. https://doi.org/10.3390/agronomy14091974






