Identification and Antagonistic Potential of Bacillus atrophaeus against Wheat Crown Rot Caused by Fusarium pseudograminearum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Isolation of Strains
2.2. Co-Inoculation Assay for the Screening of Biocontrol Strains
2.3. Minimum Inhibitory Dose
2.4. Strain Identification
2.5. Assays for Plant Growth-Promoting Traits
2.6. Whole Genome Sequencing, Annotation, and Analysis
2.7. Biofilm Adhesion Assay
2.8. In Vitro Infection of Wheat Kernels
2.9. Biocontrol Efficiency of SW in Pot Experiment
2.10. Colonization of SW and Plant Growth Promotion Pot Experiment
2.11. Statistical Analysis
3. Results
3.1. Characterization and Identification of SW
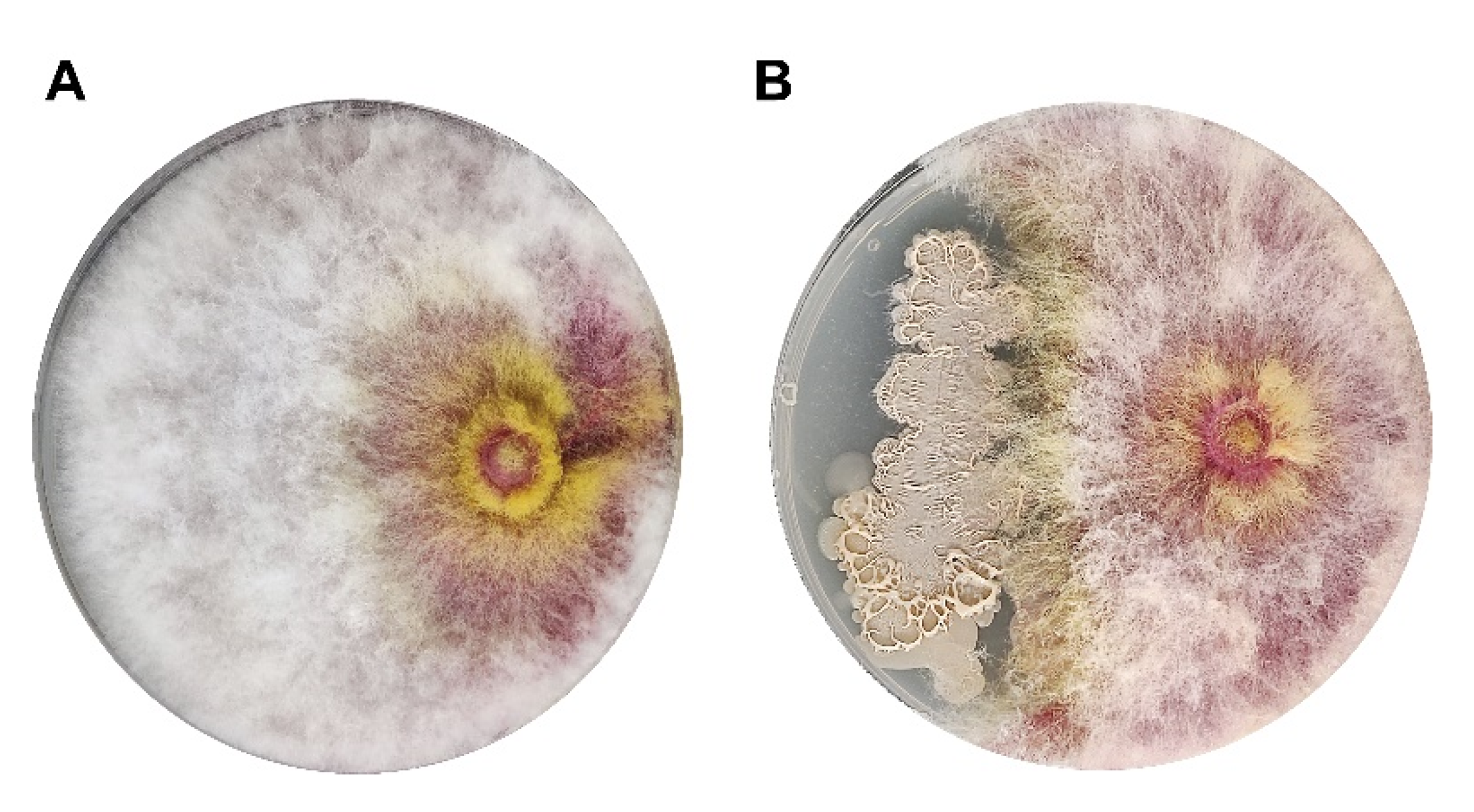
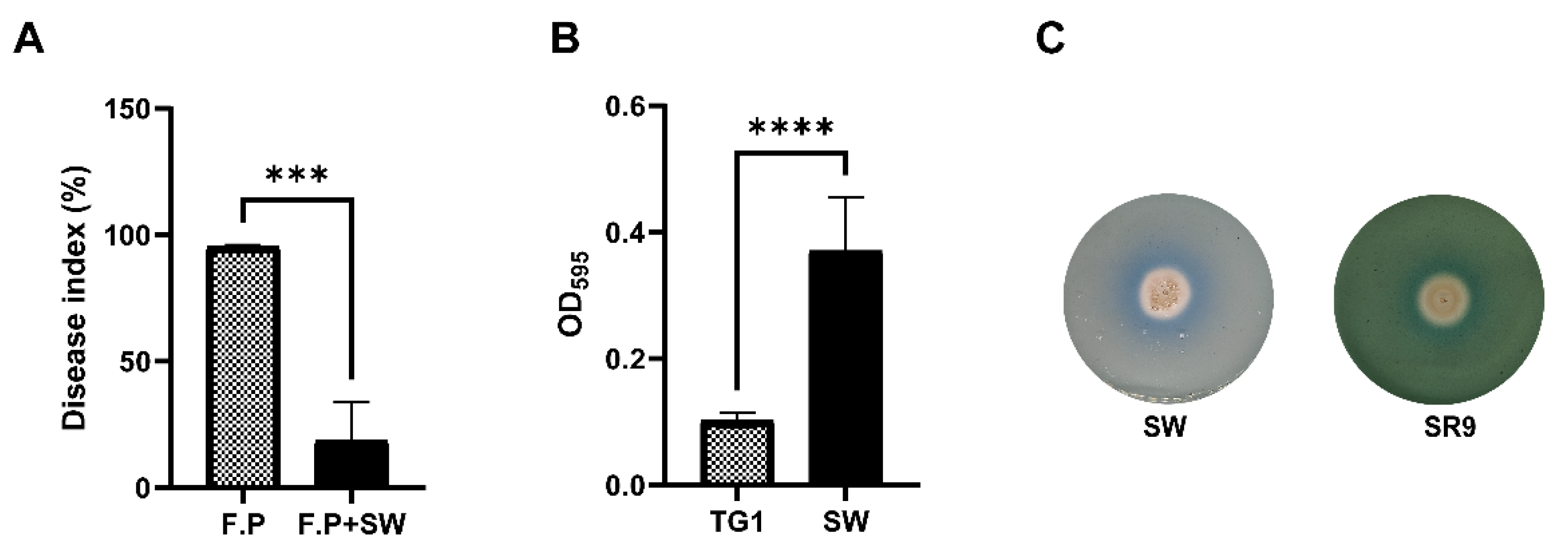
3.2. Biocontrol Efficacy of Wheat Endophytic Strain SW
3.3. Secondary Metabolite Analysis and Plant Beneficial Characters
3.4. Minimum Inhibitory Dose Estimation of SW Strain to Antagonize Fpg
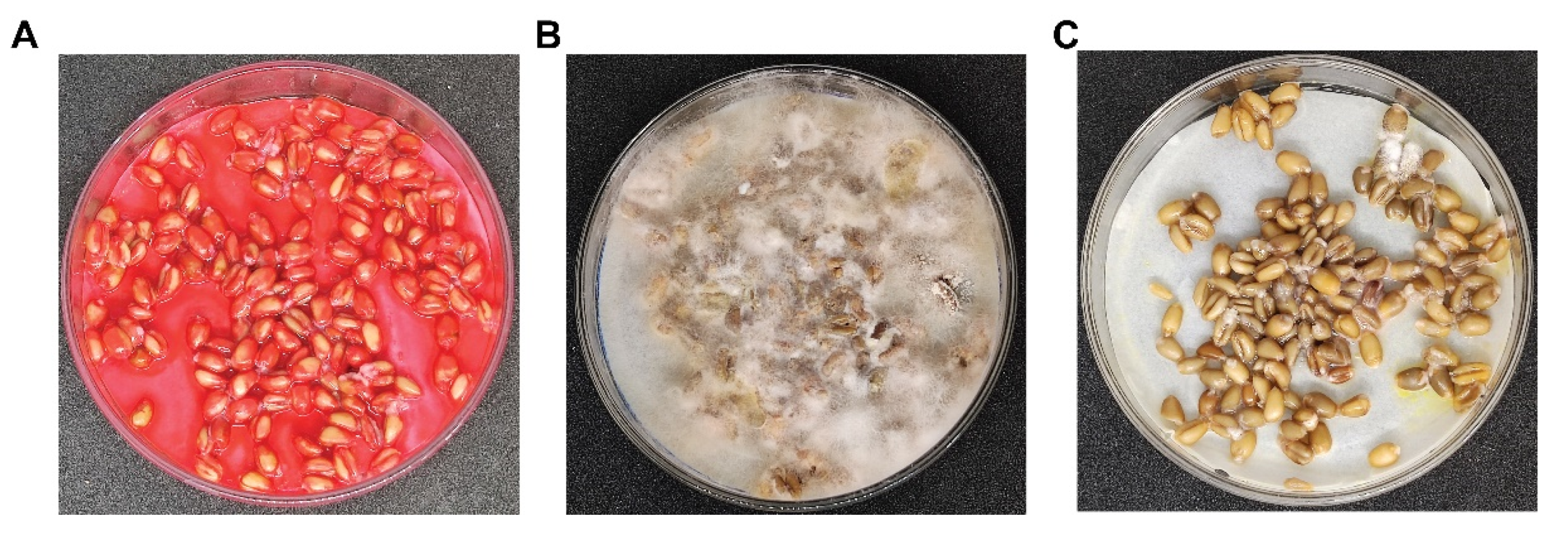
3.5. SW Strain Mimics Fludioxonil Function in Wheat Kernel Experiments
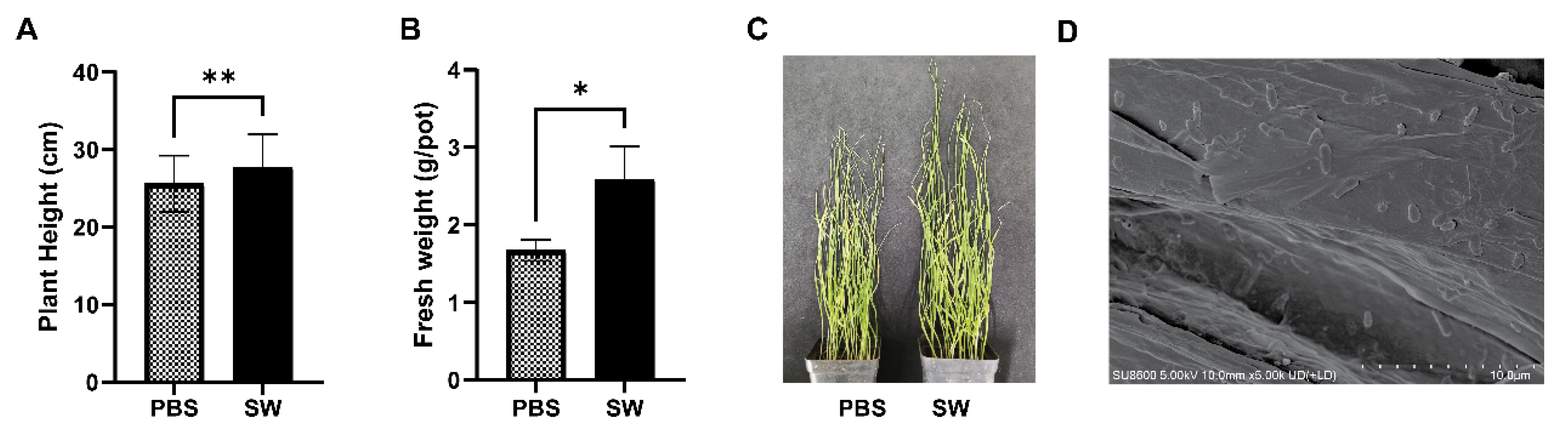
3.6. Growth Promotion and Colonization Effects of the SW Strain in Wheat Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dong, Q.; Liu, Q.; Goodwin, P.H.; Deng, X.; Xu, W.; Xia, M.; Zhang, J.; Sun, R.; Wu, C.; Wang, Q. Isolation and Genome-Based Characterization of Biocontrol Potential of Bacillus Siamensis YB-1631 against Wheat Crown Rot Caused by Fusarium pseudograminearum. J. Fungi 2023, 9, 547. [Google Scholar] [CrossRef] [PubMed]
- Smiley, R.W.; Gourlie, J.A.; Easley, S.A.; Patterson, L.-M.; Whittaker, R.G. Crop Damage Estimates for Crown Rot of Wheat and Barley in the Pacific Northwest. Plant Dis. 2005, 89, 595–604. [Google Scholar] [CrossRef]
- Xu, F.; Yang, G.; Wang, J.; Song, Y.; Liu, L.; Zhao, K.; Li, Y.; Han, Z. Spatial Distribution of Root and Crown Rot Fungi Associated with Winter Wheat in the North China Plain and Its Relationship with Climate Variables. Front. Microbiol. 2018, 9, 1054. [Google Scholar] [CrossRef] [PubMed]
- Backhouse, D.; Abubakar, A.A.; Burgess, L.W.; Dennisc, J.I.; Hollaway, G.J.; Wildermuth, G.B.; Wallwork, H.; Henry, F.J. Survey of Fusarium Species Associated with Crown Rot of Wheat and Barley in Eastern Australia. Australas. Plant Pathol. 2004, 33, 255–261. [Google Scholar] [CrossRef]
- Li, H.L.; Yuan, H.X.; Fu, B.; Xing, X.P.; Sun, B.J.; Tang, W.H. First Report of Fusarium Pseudograminearum Causing Crown Rot of Wheat in Henan, China. Plant Dis. 2012, 96, 1065. [Google Scholar] [CrossRef]
- Xu, F.; Song, Y.L.; Zhou, Y.L.; Zhang, H.; Wang, J.M.; Li, Y.H.; Han, Z.H. Occurrence Dynamics and Characteristics of Fusarium Root and Crown Rot of Wheat in Henan Province during 2013–2016. Plant Prot 2016, 42, 126–132. [Google Scholar]
- Beccari, G.; Prodi, A.; Pisi, A.; Nipoti, P.; Onofri, A.; Nicholson, P.; Pfohl, K.; Karlovsky, P.; Gardiner, D.M.; Covarelli, L. Development of Three Fusarium Crown Rot Causal Agents and Systemic Translocation of Deoxynivalenol Following Stem Base Infection of Soft Wheat. Plant Pathol. 2018, 67, 1055–1065. [Google Scholar] [CrossRef]
- Moretti, A.; Panzarini, G.; Somma, S.; Campagna, C.; Ravaglia, S.; Logrieco, A.F.; Solfrizzo, M. Systemic Growth of F. Graminearum in Wheat Plants and Related Accumulation of Deoxynivalenol. Toxins 2014, 6, 1308–1324. [Google Scholar] [CrossRef] [PubMed]
- Ülger, T.G.; Uçar, A.; Çakıroğlu, F.P.; Yilmaz, S. Genotoxic Effects of Mycotoxins. Toxicon 2020, 185, 104–113. [Google Scholar] [CrossRef]
- Pier, A.C. Major Biological Consequences of Aflatoxicosis in Animal Production. J. Anim. Sci. 1992, 70, 3964–3967. [Google Scholar] [CrossRef]
- Xu, F.; Shi, R.; Liu, L.; Li, S.; Wang, J.; Han, Z.; Liu, W.; Wang, H.; Liu, J.; Fan, J. Fusarium Pseudograminearum Biomass and Toxin Accumulation in Wheat Tissues with and without Fusarium Crown Rot Symptoms. Front. Plant Sci. 2024, 15, 1356723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Xu, Y.; Zhang, Q.; Zhao, L.; Zhu, Y.; Wu, Y.; Li, Z.; Yang, W. Detection of Fungicide Resistance to Fludioxonil and Tebuconazole in Fusarium Pseudograminearum, the Causal Agent of Fusarium Crown Rot in Wheat. PeerJ 2023, 11, e14705. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Yuan, S.; Zhang, Q.; Liu, W.; Zhou, Y.; Yang, W. Screening Fungicides for Controlling Wheat Crown Rot Caused by Fusarium Pseudograminearum across Hebei Province in China. Agriculture 2022, 12, 1643. [Google Scholar] [CrossRef]
- Hou, Y.; Guo, Y.; Wang, L.; He, S.; Zheng, W.; Liu, S.; Xu, J. Impact of Phenamacril on the Growth and Development of Fusarium Pseudograminearum and Control of Crown Rot of Wheat. Plant Dis. 2023, 107, 3843–3850. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Gu, Y.; Zhang, H.; Wang, X.; Liu, X.; Chen, C.; Wang, C.; Zhang, X.; Liu, X.; Shen, X. Synthetic Microbial Community Promotes Bacterial Communities Leading to Soil Multifunctionality in Desertified Land. Microorganisms 2024, 12, 1117. [Google Scholar] [CrossRef]
- Mukhtar, T.; Vagelas, I.; Javaid, A. New Trends in Integrated Plant Disease Management. Front. Agron. 2023, 4, 1104122. [Google Scholar] [CrossRef]
- Chen, C.; Xin, K.; Liu, H.; Cheng, J.; Shen, X.; Wang, Y.; Zhang, L. Pantoea Alhagi, a Novel Endophytic Bacterium with Ability to Improve Growth and Drought Tolerance in Wheat. Sci. Rep. 2017, 7, 41564. [Google Scholar] [CrossRef]
- Farhat, F.; Tariq, A.; Waseem, M.; Masood, A.; Raja, S.; Ajmal, W.; Iftikhar, I.; Zulfiqar, U.; Maqsood, M.F. Plant Growth Promoting Rhizobacteria (PGPR) Induced Improvements in the Growth, Photosynthesis, Antioxidants, and Nutrient Uptake of Rapeseed (Brassica napus L.). Gesunde Pflanz. 2023, 75, 2075–2088. [Google Scholar] [CrossRef]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological Control of Plant Pathogens by Bacillus Species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Ahn, I.-P.; Lee, S.-W.; Suh, S.-C. Rhizobacteria-Induced Priming in Arabidopsis Is Dependent on Ethylene, Jasmonic Acid, and NPR1. Mol. Plant-Microbe Interact. 2007, 20, 759–768. [Google Scholar] [CrossRef]
- Rahman, A.; Uddin, W.; Wenner, N.G. Induced Systemic Resistance Responses in Perennial Ryegrass against M Agnaporthe Oryzae Elicited by Semi-purified Surfactin Lipopeptides and Live Cells of Bacillus amyloliquefaciens. Mol. Plant Pathol. 2015, 16, 546–558. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Hartmann, A.; Gao, X.; Borriss, R. Biocontrol Mechanism by Root-Associated Bacillus Amyloliquefaciens FZB42–a Review. Front. Microbiol. 2015, 6, 780. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Duffy, B.; Nowak, J.; Clément, C.; Barka, E.A. Use of Plant Growth-Promoting Bacteria for Biocontrol of Plant Diseases: Principles, Mechanisms of Action, and Future Prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, C. Fengycins, Cyclic Lipopeptides from Marine Bacillus Subtilis Strains, Kill the Plant-Pathogenic Fungus Magnaporthe Grisea by Inducing Reactive Oxygen Species Production and Chromatin Condensation. Appl. Environ. Microbiol. 2018, 84, e00445-18. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; He, F.; Ding, M.; Geng, C.; Chen, L.; Zou, S.; Liang, Y.; Yu, J.; Dong, H. Thioredoxin Reductase Is Involved in Development and Pathogenicity in Fusarium graminearum. Front. Microbiol. 2019, 10, 393. [Google Scholar] [CrossRef]
- Farzand, A.; Moosa, A.; Zubair, M.; Khan, A.R.; Massawe, V.C.; Tahir, H.A.S.; Sheikh, T.M.M.; Ayaz, M.; Gao, X. Suppression of Sclerotinia Sclerotiorum by the Induction of Systemic Resistance and Regulation of Antioxidant Pathways in Tomato Using Fengycin Produced by Bacillus Amyloliquefaciens FZB42. Biomolecules 2019, 9, 613. [Google Scholar] [CrossRef]
- Gu, Q.; Yang, Y.; Yuan, Q.; Shi, G.; Wu, L.; Lou, Z.; Huo, R.; Wu, H.; Borriss, R.; Gao, X. Bacillomycin D Produced by Bacillus amyloliquefaciens Is Involved in the Antagonistic Interaction with the Plant-Pathogenic Fungus Fusarium graminearum. Appl. Environ. Microbiol. 2017, 83, e01075-17. [Google Scholar] [CrossRef]
- Zhang, X.; Li, B.; Wang, Y.; Guo, Q.; Lu, X.; Li, S.; Ma, P. Lipopeptides, a Novel Protein, and Volatile Compounds Contribute to the Antifungal Activity of the Biocontrol Agent Bacillus Atrophaeus CAB-1. Appl. Microbiol. Biotechnol. 2013, 97, 9525–9534. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, H.; Zhao, S.; Xiang, B.; Yao, Z. Lipopeptide Production by Bacillus Atrophaeus Strain B44 and Its Biocontrol Efficacy against Cotton Rhizoctoniosis. Biotechnol. Lett. 2021, 43, 1183–1193. [Google Scholar] [CrossRef]
- Mu, Y.; Yue, Y.; Gu, G.; Deng, Y.; Jin, H.; Tao, K. Identification and Characterization of the Bacillus Atrophaeus Strain J-1 as Biological Agent of Apple Ring Rot Disease. J. Plant Dis. Prot. 2020, 127, 367–378. [Google Scholar] [CrossRef]
- Pang, F.; Wang, T.; Zhao, C.; Tao, A.; Yu, Z.; Huang, S.; Yu, G. Novel Bacterial Endophytes Isolated from Winter Wheat Plants as Biocontrol Agent against Stripe Rust of Wheat. BioControl 2016, 61, 207–219. [Google Scholar] [CrossRef]
- Njoloma, J.P.B. Isolation of Endophytic Bacteria from Leaves, Stems, or Roots. In Endophytic Microbes: Isolation, Identification, and Bioactive Potentials; Humana: New York, NY, USA, 2022; pp. 3–6. [Google Scholar]
- Atlas, R.M. Handbook of Microbiological Media; CRC Press: Boca Raton, FL, USA, 2004; ISBN 0429129033. [Google Scholar]
- Chen, J.; Hu, L.; Chen, N.; Jia, R.; Ma, Q.; Wang, Y. The Biocontrol and Plant Growth-Promoting Properties of Streptomyces Alfalfae XN-04 Revealed by Functional and Genomic Analysis. Front. Microbiol. 2021, 12, 745766. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Guo, S.-Z.; Zhu, H.-Z.; Yan, L.; Liu, Z.-P.; Li, D.-F.; Jiang, C.-Y.; Corvini, P.F.-X.; Shen, X.-H.; Liu, S.-J. The Sulfonamide-Resistance Dihydropteroate Synthase Gene Is Crucial for Efficient Biodegradation of Sulfamethoxazole by Paenarthrobacter Species. Appl. Microbiol. Biotechnol. 2023, 107, 5813–5827. [Google Scholar] [CrossRef]
- Guo, S.; Liu, Y.; Yin, Y.; Chen, Y.; Jia, S.; Wu, T.; Liao, J.; Jiang, X.; Kareem, H.A.; Li, X. Unveiling the Multifaceted Potential of Pseudomonas khavaziana Strain SR9: A Promising Biocontrol Agent for Wheat Crown Rot. Microbiol. Spectr. 2024, e00712-24. [Google Scholar] [CrossRef]
- Xiao, C.-L.; Chen, Y.; Xie, S.-Q.; Chen, K.-N.; Wang, Y.; Han, Y.; Luo, F.; Xie, Z. MECAT: Fast Mapping, Error Correction, and de Novo Assembly for Single-Molecule Sequencing Reads. Nat. Methods 2017, 14, 1072–1074. [Google Scholar] [CrossRef]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying Bacterial Genes and Endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and Rapid Annotation of Ribosomal RNA Genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. TRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Bateman, A.; Marshall, M.; Khanna, A.; Eddy, S.R. Rfam: An RNA Family Database. Nucleic Acids Res. 2003, 31, 439–441. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-Fold Faster RNA Homology Searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Fouts, D.E. Phage_Finder: Automated Identification and Classification of Prophage Regions in Complete Bacterial Genome Sequences. Nucleic Acids Res. 2006, 34, 5839–5851. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M. The SEED and the Rapid Annotation of Microbial Genomes Using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes Database (CAZy): An Expert Resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N. AntiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Zuo, G. CVTree: A Parallel Alignment-Free Phylogeny and Taxonomy Tool Based on Composition Vectors of Genomes. Genom. Proteom. Bioinforma 2021, 19, 662–667. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A Database Tandem for Fast and Reliable Genome-Based Classification and Nomenclature of Prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Zhang, L.; Li, S.; Liu, X.; Wang, Z.; Jiang, M.; Wang, R.; Xie, L.; Liu, Q.; Xie, X.; Shang, D. Sensing of Autoinducer-2 by Functionally Distinct Receptors in Prokaryotes. Nat. Commun. 2020, 11, 5371. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, W.; Goodwin, P.H.; Lin, Q.; Xia, M.; Xu, W.; Sun, R.; Liang, J.; Wu, C.; Li, H. Response of Fusarium Pseudograminearum to Biocontrol Agent Bacillus Velezensis YB-185 by Phenotypic and Transcriptome Analysis. J. Fungi 2022, 8, 763. [Google Scholar] [CrossRef]
- Paulitz, T.C.; Steffenson, B.J. Biotic Stress in Barley: Disease Problems and Solutions. Barley 2010, 307–354. [Google Scholar] [CrossRef]
- Yadav, A.N. Beneficial Plant-Microbe Interactions for Agricultural Sustainability. J. Appl. Biol. Biotechnol. 2021, 9, 1–4. [Google Scholar] [CrossRef]
- Liu, H.; Li, J.; Carvalhais, L.C.; Percy, C.D.; Prakash Verma, J.; Schenk, P.M.; Singh, B.K. Evidence for the Plant Recruitment of Beneficial Microbes to Suppress Soil-borne Pathogens. New Phytol. 2021, 229, 2873–2885. [Google Scholar] [CrossRef]
- Ayaz, M.; Li, C.-H.; Ali, Q.; Zhao, W.; Chi, Y.-K.; Shafiq, M.; Ali, F.; Yu, X.-Y.; Yu, Q.; Zhao, J.-T. Bacterial and Fungal Biocontrol Agents for Plant Disease Protection: Journey from Lab to Field, Current Status, Challenges, and Global Perspectives. Molecules 2023, 28, 6735. [Google Scholar] [CrossRef]
- Bonaterra, A.; Badosa, E.; Daranas, N.; Francés, J.; Roselló, G.; Montesinos, E. Bacteria as Biological Control Agents of Plant Diseases. Microorganisms 2022, 10, 1759. [Google Scholar] [CrossRef]
- Ons, L.; Bylemans, D.; Thevissen, K.; Cammue, B.P.A. Combining Biocontrol Agents with Chemical Fungicides for Integrated Plant Fungal Disease Control. Microorganisms 2020, 8, 1930. [Google Scholar] [CrossRef]
- Sun, G.; Yao, T.; Feng, C.; Chen, L.; Li, J.; Wang, L. Identification and Biocontrol Potential of Antagonistic Bacteria Strains against Sclerotinia Sclerotiorum and Their Growth-Promoting Effects on Brassica napus. Biol. Control 2017, 104, 35–43. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, Y.; Li, Y.; Fu, X.; Wang, Q. Screening and Characterization of Endophytic Bacillus for Biocontrol of Grapevine Downy Mildew. Crop Prot. 2017, 96, 173–179. [Google Scholar] [CrossRef]
- Zalila-Kolsi, I.; Mahmoud, A.B.; Ali, H.; Sellami, S.; Nasfi, Z.; Tounsi, S.; Jamoussi, K. Antagonist Effects of Bacillus spp. Strains against Fusarium graminearum for Protection of Durum Wheat (Triticum turgidum L. subsp. durum). Microbiol. Res. 2016, 192, 148–158. [Google Scholar] [CrossRef]
- Kabeerdass, N.; Al Otaibi, A.; Rajendran, M.; Manikandan, A.; Kashmery, H.A.; Rahman, M.M.; Madhu, P.; Khan, A.; Asiri, A.M.; Mathanmohun, M. Bacillus-Mediated Silver Nanoparticle Synthesis and Its Antagonistic Activity against Bacterial and Fungal Pathogens. Antibiotics 2021, 10, 1334. [Google Scholar] [CrossRef] [PubMed]
- Balthazar, C.; Novinscak, A.; Cantin, G.; Joly, D.L.; Filion, M. Biocontrol Activity of Bacillus Spp. and Pseudomonas Spp. against Botrytis Cinerea and Other Cannabis Fungal Pathogens. Phytopathology 2022, 112, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Villa-Rodriguez, E.; Parra-Cota, F.; Castro-Longoria, E.; López-Cervantes, J.; de los Santos-Villalobos, S. Bacillus Subtilis TE3: A Promising Biological Control Agent against Bipolaris Sorokiniana, the Causal Agent of Spot Blotch in Wheat (Triticum turgidum L. subsp. durum). Biol. Control 2019, 132, 135–143. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-Growth-Promoting Rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The Biofilm Life Cycle: Expanding the Conceptual Model of Biofilm Formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Goswami, D.; Thakker, J.N.; Dhandhukia, P.C. Portraying Mechanics of Plant Growth Promoting Rhizobacteria (PGPR): A Review. Cogent Food Agric. 2016, 2, 1127500. [Google Scholar] [CrossRef]
- Khurana, H.; Sharma, M.; Verma, H.; Lopes, B.S.; Lal, R.; Negi, R.K. Genomic Insights into the Phylogeny of Bacillus Strains and Elucidation of Their Secondary Metabolic Potential. Genomics 2020, 112, 3191–3200. [Google Scholar] [CrossRef]
- Zubair, M.; Farzand, A.; Mumtaz, F.; Khan, A.R.; Sheikh, T.M.; Haider, M.S.; Yu, C.; Wang, Y.; Ayaz, M.; Gu, Q.; et al. Novel Genetic Dysregulations and Oxidative Damage in Fusarium graminearum Induced by Plant Defense Eliciting Psychrophilic Bacillus atrophaeus TS1. Int. J. Mol. Sci. 2021, 22, 12094. [Google Scholar] [CrossRef]
- Fan, H.; Ru, J.; Zhang, Y.; Wang, Q.; Li, Y. Fengycin Produced by Bacillus Subtilis 9407 Plays a Major Role in the Biocontrol of Apple Ring Rot Disease. Microbiol. Res. 2017, 199, 89–97. [Google Scholar] [CrossRef]
- Li, Y.; Han, L.-R.; Zhang, Y.; Fu, X.; Chen, X.; Zhang, L.; Mei, R.; Wang, Q. Biological Control of Apple Ring Rot on Fruit by Bacillus Amyloliquefaciens 9001. Plant Pathol. J. 2013, 29, 168. [Google Scholar] [CrossRef]
- Guillén-Navarro, K.; López-Gutiérrez, T.; García-Fajardo, V.; Gómez-Cornelio, S.; Zarza, E.; De la Rosa-García, S.; Chan-Bacab, M. Broad-Spectrum Antifungal, Biosurfactants and Bioemulsifier Activity of Bacillus Subtilis Subsp. Spizizenii—A Potential Biocontrol and Bioremediation Agent in Agriculture. Plants 2023, 12, 1374. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Bie, X.; Lu, Z.; Lv, F.; Tao, Y.; Qu, X. Effects of Fengycin from Bacillus Subtilis FmbJ on Apoptosis and Necrosis in Rhizopus Stolonifer. J. Microbiol. 2014, 52, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Sun, L.; Xu, H.; Gu, Y.; Lei, P. Bacillus Atrophaeus NX-12 Utilizes Exosmotic Glycerol from Fusarium Oxysporum f. Sp. Cucumerinum for Fengycin Production. J. Agric. Food Chem. 2023, 71, 10565–10574. [Google Scholar] [CrossRef]
- Wu, L.; Xie, Y.; Li, J.; Han, M.; Yang, X.; Chang, F. The Effect of Two Siderophore-Producing Bacillus Strains on the Growth Promotion of Perennial Ryegrass under Cadmium Stress. Microorganisms 2024, 12, 1083. [Google Scholar] [CrossRef]
- Nithyapriya, S.; Lalitha, S.; Sayyed, R.Z.; Reddy, M.S.; Dailin, D.J.; El Enshasy, H.A.; Luh Suriani, N.; Herlambang, S. Production, Purification, and Characterization of Bacillibactin Siderophore of Bacillus Subtilis and Its Application for Improvement in Plant Growth and Oil Content in Sesame. Sustainability 2021, 13, 5394. [Google Scholar] [CrossRef]
- Crosa, J.H.; Walsh, C.T. Genetics and Assembly Line Enzymology of Siderophore Biosynthesis in Bacteria. Microbiol. Mol. Biol. Rev. 2002, 66, 223–249. [Google Scholar] [CrossRef]
- Ni, M.; Wu, Q.; Wang, J.; Liu, W.C.; Ren, J.H.; Zhang, D.P.; Zhao, J.; Liu, D.E.W.; Rao, Y.H.; Lu, C.G. Identification and Comprehensive Evaluation of a Novel Biocontrol Agent Bacillus Atrophaeus JZB120050. J. Environ. Sci. Health Part B 2018, 53, 777–785. [Google Scholar] [CrossRef]
- Knights, H.E.; Jorrin, B.; Haskett, T.L.; Poole, P.S. Deciphering Bacterial Mechanisms of Root Colonization. Environ. Microbiol. Rep. 2021, 13, 428–444. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Leveau, J.H.J. Biocontrol of Plant Pathogens: Principles, Promises and Pitfalls. In The Rhizosphere: Biochemistry and Organic Substances at the Soil-Plant Interface; CRC Press, Inc.: Boca Raton, FL, USA, 2007; pp. 267–296. [Google Scholar]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.-L.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant Growth-Promoting Rhizobacteria and Root System Functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef]
- Magno-Pérez-Bryan, M.C.; Martinez-Garcia, P.M.; Hierrezuelo, J.; Rodriguez-Palenzuela, P.; Arrebola, E.; Ramos, C.; De Vicente, A.; Pérez-García, A.; Romero, D. Comparative Genomics within the Bacillus Genus Reveal the Singularities of Two Robust Bacillus Amyloliquefaciens Biocontrol Strains. Mol. Plant-Microbe Interact. 2015, 28, 1102–1116. [Google Scholar] [CrossRef]
- Guo, S.; Li, X.; He, P.; Ho, H.; Wu, Y.; He, Y. Whole-Genome Sequencing of Bacillus Subtilis XF-1 Reveals Mechanisms for Biological Control and Multiple Beneficial Properties in Plants. J. Ind. Microbiol. Biotechnol. 2015, 42, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Malik, M.S.; Rehman, A.; Khan, I.U.; Khan, T.A.; Jamil, M.; Rha, E.S.; Anees, M. Thermo-Neutrophilic Cellulases and Chitinases Characterized from a Novel Putative Antifungal Biocontrol Agent: Bacillus Subtilis TD11. PLoS ONE 2023, 18, e0281102. [Google Scholar] [CrossRef] [PubMed]
- Gharieb, M.M.; Abo-Zaid, G.A.; Bashir, S.I.; Hafez, E.E. Screening and Molecular Identification of Cellulase-Producing Bacillus Spp. from Agricultural Soil: Its Potential in Biological Control. Middle East J. Appl. Sci 2020, 10, 272–278. [Google Scholar]
- EFSA PLH Panel (EFSA Panel on Plangt Health); Bragard, C.; Baptista, P.; Chatzivassiliou, E.; Di Serio, F.; Gonthier, P.; Jaques Miret, J.A.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S. Pest Categorisation of Fusarium Pseudograminearum. EFSA J. 2022, 20, e07399. [Google Scholar]
- Bencheikh, A.; Hicham, M.; Meriem, D.B.; Asma, G.; Khalida, B.; Rouag, N. Efficiency of Durum Wheat Seeds Biopriming by Rhizobacteria in the Biocontrol of Fusarium Culmorum and Fusarium Chlamydosporum Infecting Durum Wheat in Algeria. Arch. Phytopathol. Plant Prot. 2022, 55, 653–675. [Google Scholar] [CrossRef]

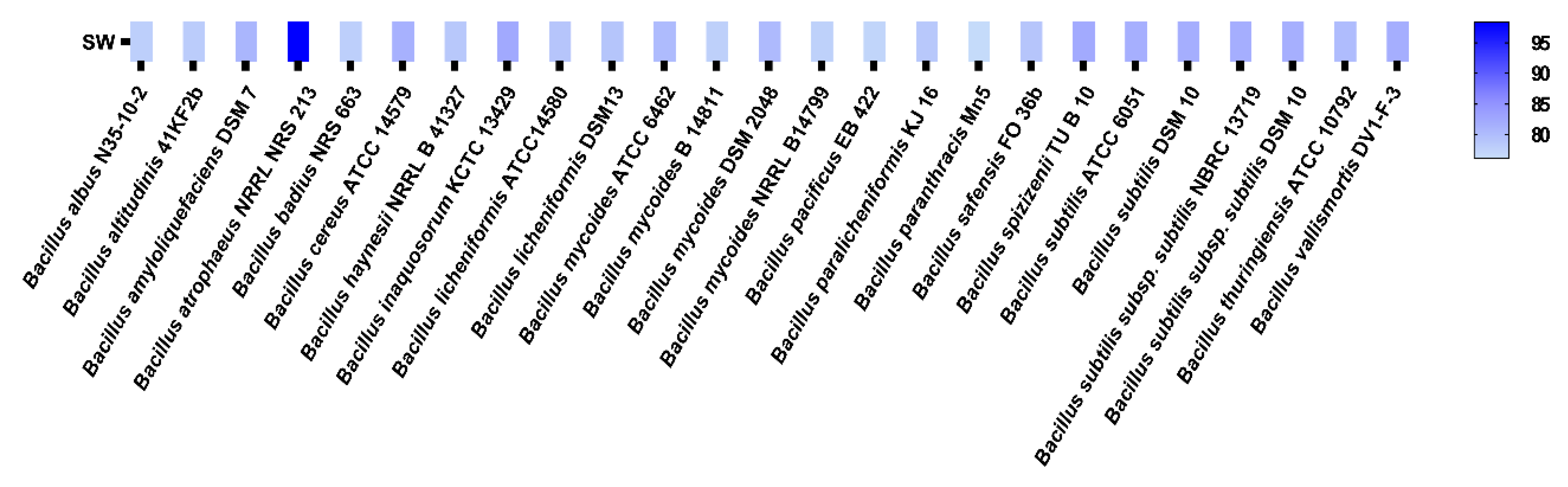

| Cluster | Type | Most Similar Known Cluster | Similarity (%) | Location | Length (bp) |
|---|---|---|---|---|---|
| 1 | NRP a-metallophore, NRPS b | bacillibactin | 100 | 305,340–357,206 | 51,867 |
| 2 | thiopeptide, LAP c | / | / | 370,220–400,322 | 30,103 |
| 3 | NRPS | / | / | 1,514,481–1,561,488 | 47,008 |
| 4 | NRPS | surfactin | 86 | 1,688,924–1,754,337 | 65,414 |
| 5 | NRPS, T1PKS d | zwittermicin A | 18 | 2,034,896–2,116,243 | 81,348 |
| 6 | terpene | / | / | 2,517,449–2,538,291 | 20,843 |
| 7 | TransAT-PKS, PKS-like, T3PKS, NRPS, NRPS-like | bacillaene | 100 | 3,120,507–3,245,201 | 124,685 |
| 8 | NRPS, betalactone, transAT-PKS | fengycin | 100 | 3,349,068–3,491,645 | 142,578 |
| 9 | terpene | / | / | 3,537,638–3,559,527 | 21,890 |
| 10 | NRP-metallophore, NRPS | bacillibactin | 57 | 3,579,460–3,625,986 | 46,527 |
| 11 | T3PKS | 1-carbapen-2-em-3-carboxylic acid | 16 | 3,657,809–3,698,906 | 41,098 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Tariq, A.; Liao, J.; Yang, A.; Jiang, X.; Yin, Y.; Shi, Y.; Li, C.; Pan, J.; Han, D.; et al. Identification and Antagonistic Potential of Bacillus atrophaeus against Wheat Crown Rot Caused by Fusarium pseudograminearum. Agronomy 2024, 14, 2135. https://doi.org/10.3390/agronomy14092135
Guo S, Tariq A, Liao J, Yang A, Jiang X, Yin Y, Shi Y, Li C, Pan J, Han D, et al. Identification and Antagonistic Potential of Bacillus atrophaeus against Wheat Crown Rot Caused by Fusarium pseudograminearum. Agronomy. 2024; 14(9):2135. https://doi.org/10.3390/agronomy14092135
Chicago/Turabian StyleGuo, Shengzhi, Arneeb Tariq, Jun Liao, Aowei Yang, Xinyan Jiang, Yanling Yin, Yuan Shi, Changfu Li, Junfeng Pan, Dejun Han, and et al. 2024. "Identification and Antagonistic Potential of Bacillus atrophaeus against Wheat Crown Rot Caused by Fusarium pseudograminearum" Agronomy 14, no. 9: 2135. https://doi.org/10.3390/agronomy14092135
APA StyleGuo, S., Tariq, A., Liao, J., Yang, A., Jiang, X., Yin, Y., Shi, Y., Li, C., Pan, J., Han, D., & Shen, X. (2024). Identification and Antagonistic Potential of Bacillus atrophaeus against Wheat Crown Rot Caused by Fusarium pseudograminearum. Agronomy, 14(9), 2135. https://doi.org/10.3390/agronomy14092135







