The Role of Plant DNA Methylation in Development, Stress Response, and Crop Breeding
Abstract
:1. Introduction
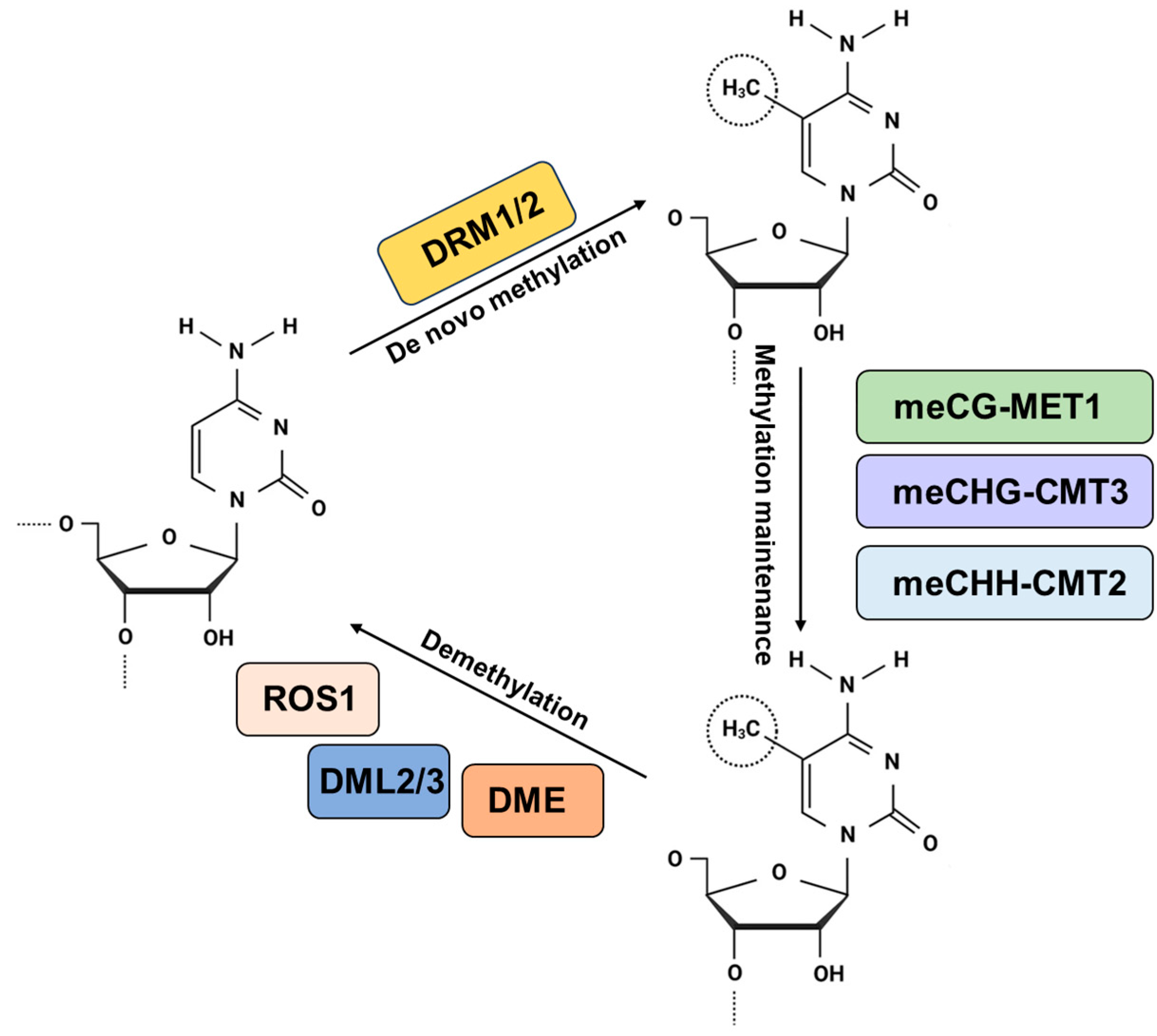
2. DNA Methylation Kinetics
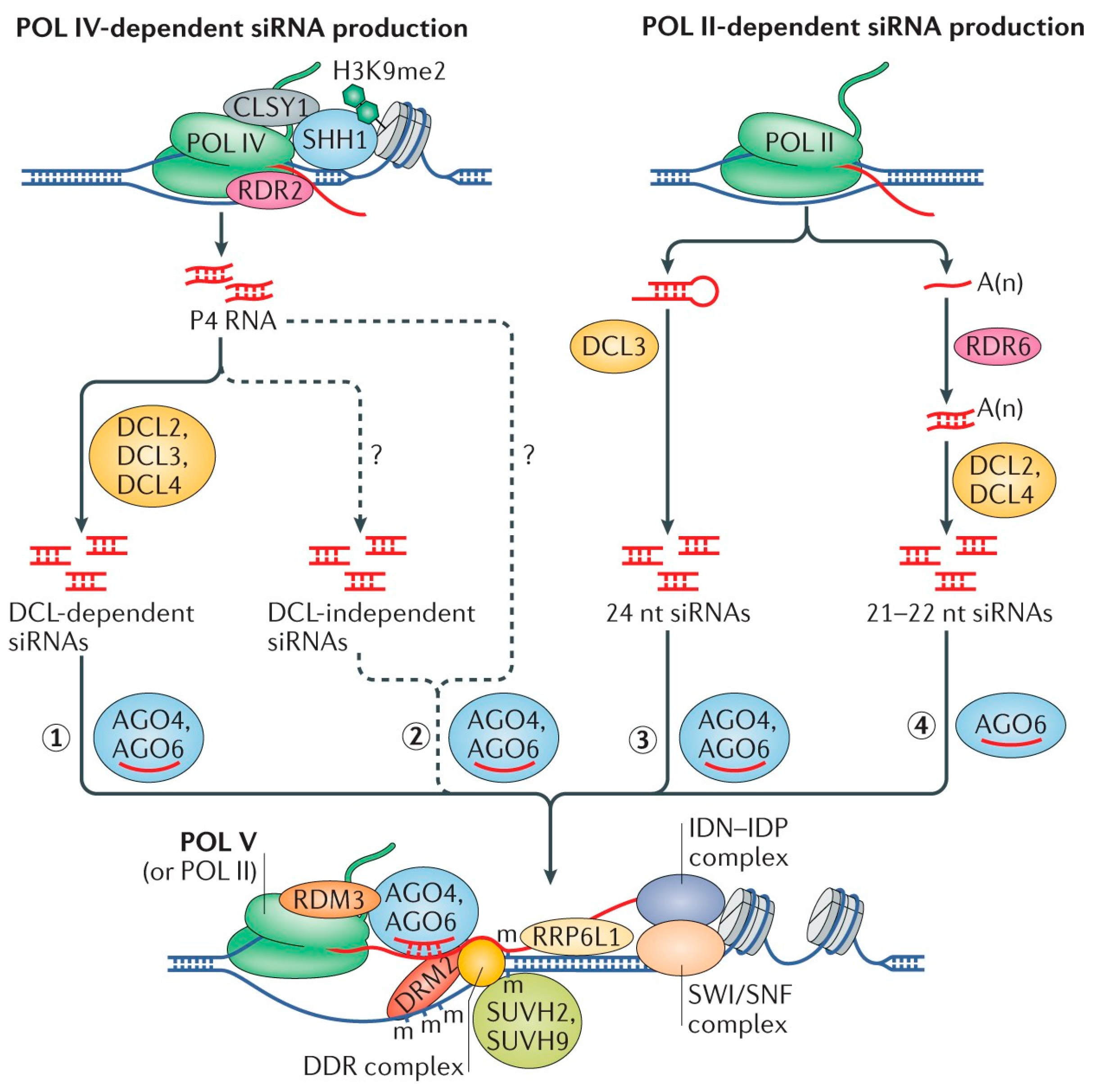
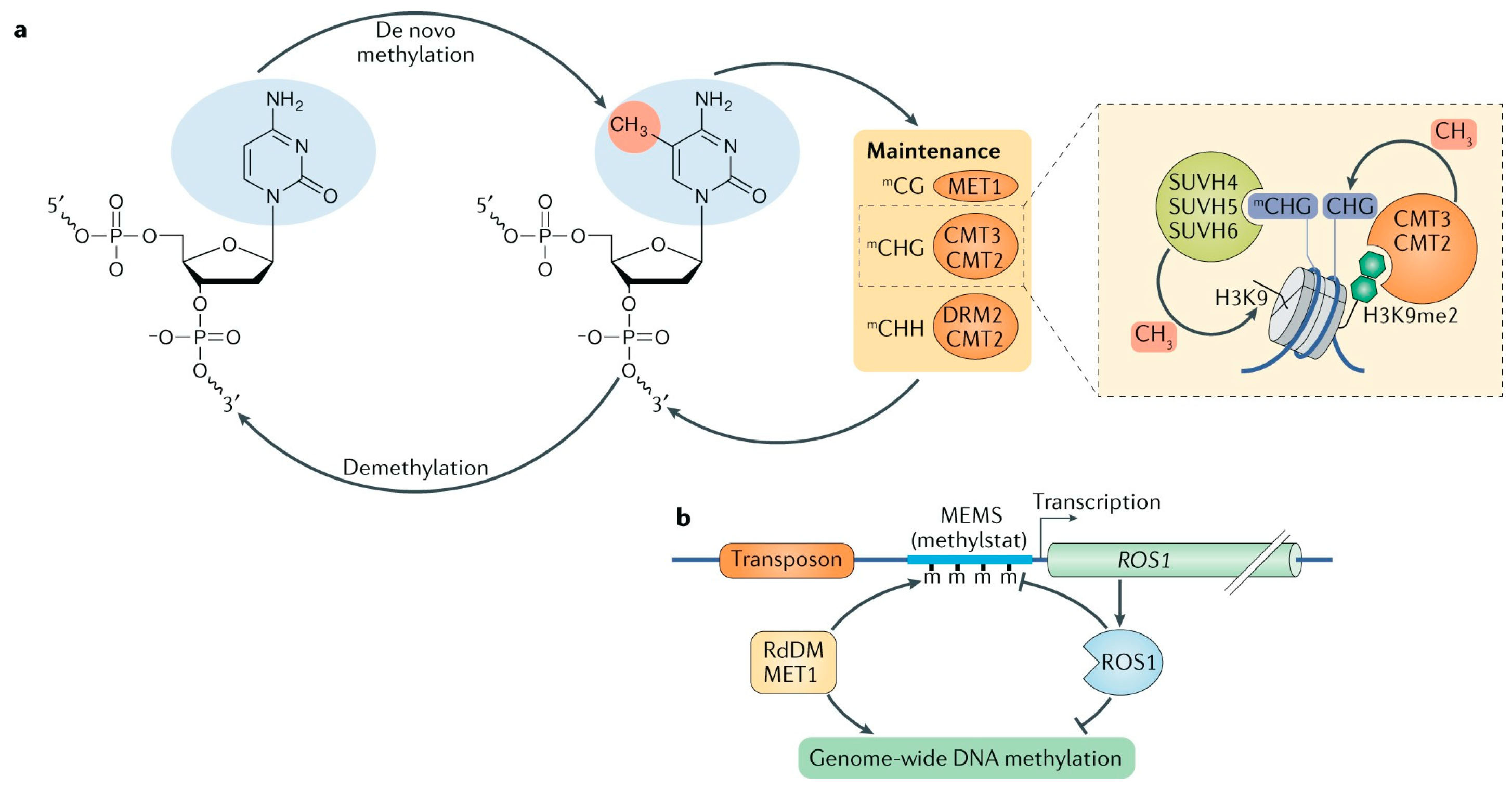
2.1. Establishment of DNA Methylation
2.2. Maintenance of DNA Methylation
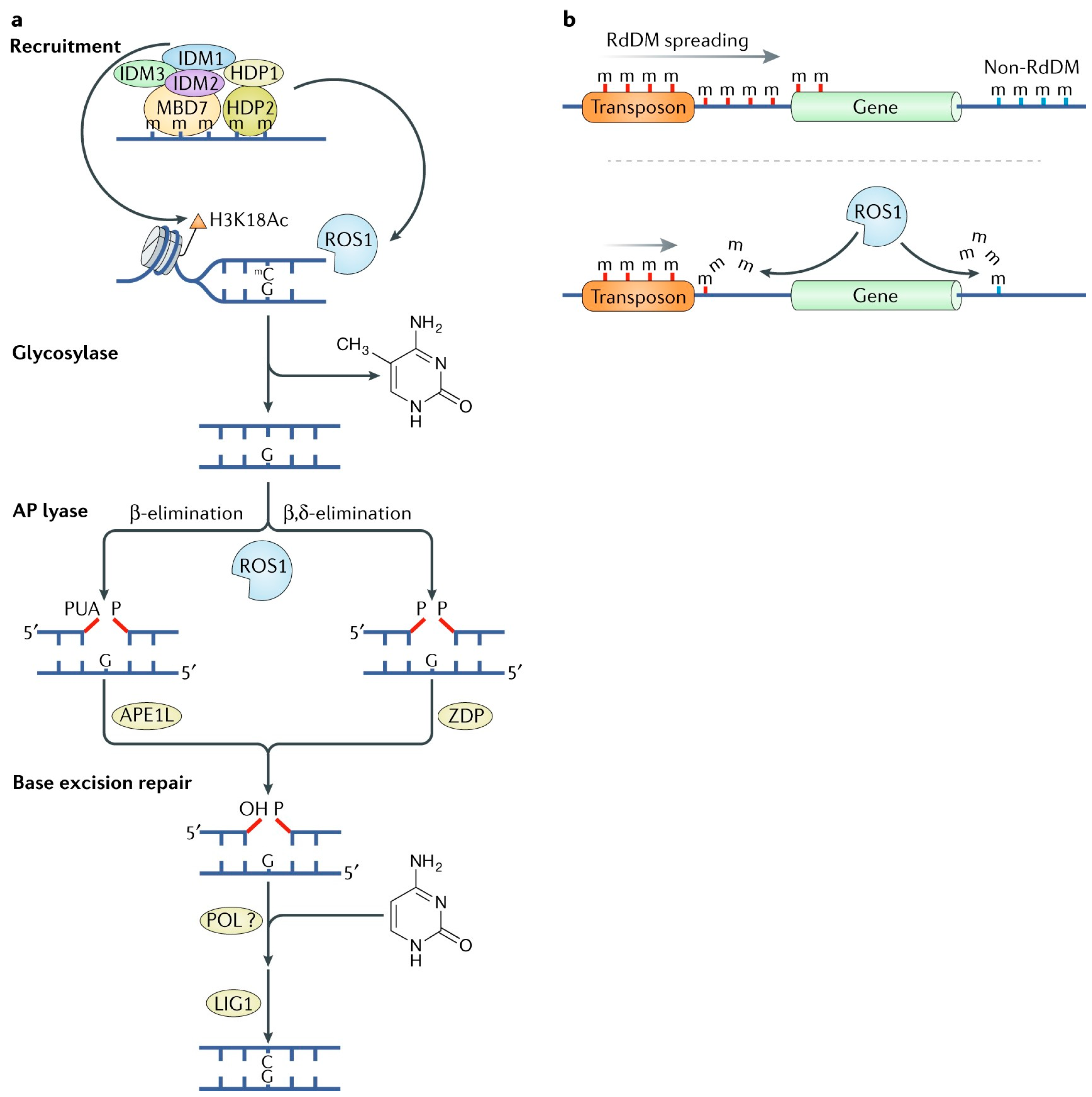
2.3. Demethylation of Active DNA
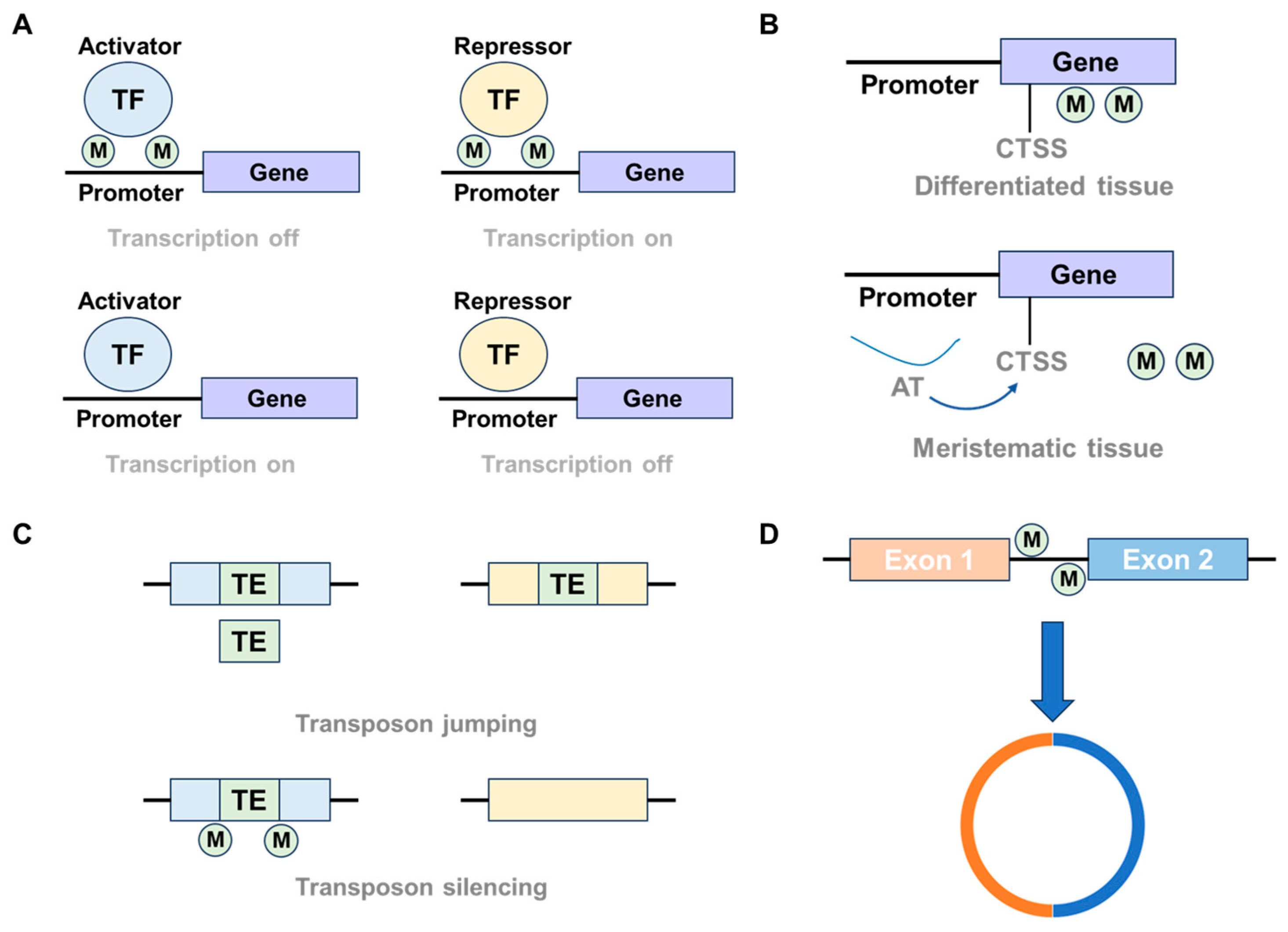
3. Molecular Functions of Plant DNA Methylation
3.1. Regulation of Gene Expression
3.2. TEs Silence
3.3. Chromosome Interaction and Biogenesis of circRNAs
4. The Role of DNA Methylation in Plant Development
4.1. Genomic Imprinting and Seed Development
4.2. Role of DNA Methylation in Plant Meristem and Leaf Epidermal Development
4.3. Role of DNA Methylation in Flower Development
4.4. Role of DNA Methylation in Fruit Ripening
5. The Role of Plant DNA Methylation in Abiotic Stress
5.1. High Temperature Stress
5.2. Drought Stress
5.3. Salt Stress
6. The Role of Plant DNA Methylation in Biological Stress
6.1. Dynamic Changes of DNA Methylation and Response to Biotic Stress
6.2. DNA Methylation Regulation of Disease Resistance Genes
6.3. Synergistic Effect of DNA Methylation with Plant Hormone Signaling
7. Prospects of DNA Methylation in Crop Breeding
8. Conclusions and Prospect
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Berger, S.L.; Kouzarides, T.; Shiekhattar, R.; Shilatifard, A. An operational definition of epigenetics. Genes Dev. 2009, 23, 781–783. [Google Scholar] [CrossRef] [PubMed]
- Henderson, I.R.; Jacobsen, S.E. Epigenetic inheritance in plants. Nature 2007, 447, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Maeji, H.; Nishimura, T. Epigenetic Mechanisms in Plants. Adv. Bot. Res. 2018, 88, 21–47. [Google Scholar]
- Hari Sundar, G.V.; Swetha, C.; Basu, D.; Pachamuthu, K.; Raju, S.; Chakraborty, T.; Mosher, R.A.; Shivaprasad, P.V. Plant polymerase IV sensitizes chromatin through histone modifications to preclude spread of silencing into protein-coding domains. Genome Res. 2023, 33, 715–728. [Google Scholar] [CrossRef]
- Fan, X.; Peng, L.; Zhang, Y. Plant DNA Methylation Responds to Nutrient Stress. Genes 2022, 13, 992. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Zhou, K.; Wu, T.; Zhao, B.S.; Sun, M.; Chen, Z.; Deng, X.; Xiao, G.; Auer, F.; et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 2019, 567, 414–419. [Google Scholar] [CrossRef]
- Lee, C.H.; Carroll, B.J. Evolution and diversification of small RNA pathways in flowering plants. Plant Cell Physiol. 2018, 59, 2169–2187. [Google Scholar] [CrossRef]
- Andrews, S.; Krueger, C.; Mellado-Lopez, M.; Hemberger, M.; Dean, W.; Perez-Garcia, V.; Hanna, C.W. Mechanisms and function of de novo DNA methylation in placental development reveals an essential role for DNMT3B. Nat. Commun. 2023, 14, 371. [Google Scholar] [CrossRef]
- Yaari, R.; Katz, A.; Domb, K.; Harris, K.D.; Zemach, A.; Ohad, N. Publisher Correction: RdDM-independent de novo and heterochromatin DNA methylation by plant CMT and DNMT3 orthologs. Nat. Commun. 2019, 6, 2552. [Google Scholar] [CrossRef]
- Qi, Y.; Ding, L.; Zhang, S.; Yao, S.; Ong, J.; Li, Y.; Wu, H.; Du, P. A plant immune protein enables broad antitumor response by rescuing microRNA deficiency. Cell 2022, 185, 1888–1904.e24. [Google Scholar] [CrossRef]
- Liu, Y.; Siejka-Zielińska, P.; Velikova, G.; Bi, Y.; Yuan, F.; Tomkova, M.; Bai, C.; Chen, L.; Schuster-Böckler, B.; Song, C.X. Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat. Biotechnol. 2019, 37, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Vidalis, A.; Živković, D.; Wardenaar, R.; Roquis, D.; Tellier, A.; Johannes, F. Methylome evolution in plants. Genome Biol. 2016, 17, 264. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Arai, Y.; Umehara, H.; Masuhara, M.; Kimura, T.; Taniguchi, H.; Sekimoto, T.; Ikawa, M.; Yoneda, Y.; Okabe, M.; et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 2007, 9, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wu, W.; Callen, E.; Pavani, R.; Zolnerowich, N.; Kodali, S.; Zong, D.; Wong, N.; Noriega, S.; Nathan, W.J.; et al. Active DNA demethylation promotes cell fate specification and the DNA damage response. Science 2022, 378, 983–989. [Google Scholar] [CrossRef]
- Lucibelli, F.; Valoroso, M.C.; Aceto, S. Plant DNA Methylation: An Epigenetic Mark in Development, Environmental Interactions, and Evolution. Int. J. Mol. Sci. 2022, 23, 8299. [Google Scholar] [CrossRef]
- He, X.J.; Chen, T.; Zhu, J.K. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011, 21, 442–465. [Google Scholar] [CrossRef]
- Huang, S.C.; Ecker, J.R. Piecing together cis-regulatory networks: Insights from epigenomics studies in plants. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018, 10, e1411. [Google Scholar] [CrossRef]
- Stroud, H.; Greenberg, M.V.; Feng, S.; Bernatavichute, Y.V.; Jacobsen, S.E. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 2013, 152, 352–364. [Google Scholar] [CrossRef]
- Cao, Q.; Wang, S.; Ma, H.; Luo, S.; He, H.; Zhang, Y. Hyper non-CG methylation of expanded plant disease resistance NLR genes. Plant Cell Rep. 2023, 42, 1251–1254. [Google Scholar] [CrossRef]
- Arora, H.; Singh, R.K.; Sharma, S.; Sharma, N.; Panchal, A.; Das, T.; Prasad, A.; Prasad, M. DNA methylation dynamics in response to abiotic and pathogen stress in plants. Plant Cell Rep. 2022, 41, 1931–1944. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.K. RNA-directed DNA methylation. Curr. Opin. Plant Biol. 2011, 14, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Pikaard, C.S.; Haag, J.R.; Pontes, O.M.; Blevins, T.; Cocklin, R. A transcription fork model for Pol IV and Pol V-dependent RNA-directed DNA methylation. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Hoareau, M.; Rincheval-Arnold, A.; Gaumer, S.; Guénal, I. DREAM a little dREAM of DRM: Model organisms and conservation of DREAM-like complexes: Model organisms uncover the mechanisms of DREAM-mediated transcription regulation. BioEssays 2024, 46, e2300125. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Liu, H.-L.; Daxinger, L.; Pontes, O.; He, X.; Qian, W.; Lin, H.; Xie, M.; Lorkovic, Z.J.; Zhang, S.; et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 2010, 465, 106–109. [Google Scholar] [CrossRef]
- Vaucheret, H.; Voinnet, O. The plant siRNA landscape. Plant Cell 2023, 36, 246–275. [Google Scholar] [CrossRef]
- Creasey, K.M.; Zhai, J.; Borges, F.; Van Ex, F.; Regulski, M.; Meyers, B.C.; Martienssen, R.A. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 2014, 508, 411–415. [Google Scholar] [CrossRef]
- Cuerda-Gil, D.; Slotkin, R.K. Non-canonical RNA-directed DNA methylation. Nat. Plants 2016, 2, 16163. [Google Scholar] [CrossRef]
- Dunker, F.; Lederer, B.; Weiberg, A. Plant ARGONAUTE Protein Immunopurification for Pathogen Cross Kingdom Small RNA Analysis. Bio-Protoc. 2021, 11, e3911. [Google Scholar]
- Bies-Etheve, N.; Pontier, D.; Lahmy, S.; Picart, C.; Vega, D.; Cooke, R.; Lagrange, T. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009, 10, 649–654. [Google Scholar] [CrossRef]
- Zhang, H.; Tang, K.; Qian, W.; Duan, C.G.; Wang, B.; Zhang, H.; Wang, P.; Zhu, X.; Lang, Z.; Yang, Y.; et al. An Rrp6-like protein positively regulates noncoding RNA levels and DNA methylation in Arabidopsis. Mol. Cell 2014, 54, 418–430. [Google Scholar] [CrossRef]
- Ausin, I.; Mockler, T.C.; Chory, J.; Jacobsen, S.E. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat. Struct. Mol. Biol. 2009, 16, 1325–1327. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, R.M.; Picard, C.L. RNA-directed DNA Methylation. PLoS Genet. 2020, 16, e1009034. [Google Scholar] [CrossRef] [PubMed]
- Ausin, I.; Greenberg, M.V.C.; Simanshu, D.K.; Hale, C.J.; Vashisht, A.A.; Simon, S.A.; Lee, T.-F.; Feng, S.; Española, S.D.; Meyers, B.C.; et al. INVOLVED IN DE NOVO 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 8374–8381. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ou, X.; Han, D.; He, Z.; Liu, S.; Mao, N.; Zhang, Z.; Peng, C.L.; Lai, J.; Yang, C. A diRNA-protein scaffold module mediates SMC5/6 recruitment in plant DNA repair. Plant Cell 2022, 34, 3899–3914. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, Z.-Y.; Zeng, L.; Tanaka, K.; Zhang, C.-J.; Ma, J.; Bai, G.; Wang, P.; Zhang, S.-W.; Liu, Z.-W.; et al. DTF1 is a core component of RNA-directed DNA methylation may assist in the recruitment of Pol IV. Proc. Natl. Acad. Sci. USA 2013, 110, 8290–8295. [Google Scholar] [CrossRef]
- Fang, C.; Huang, K.; Wu, X.; Zhang, H.; Gu, Z.; Wang, J.; Zhang, Y. Transcription elongation of the plant RNA polymerase IV is prone to backtracking. Sci Adv. 2024, 10, eadq3087. [Google Scholar] [CrossRef]
- Wang, Y.; Le, B.H.; Wang, J.; You, C.; Zhao, Y.; Galli, M.; Xu, Y.; Gallavotti, A.; Eulgem, T.; Mo, B.; et al. ZMP recruits and excludes Pol IV-mediated DNA methylation in a site-specific manner. Sci. Adv. 2022, 8, eadc9454. [Google Scholar] [CrossRef]
- Zhou, M.; Coruh, C.; Xu, G.; Martins, L.M.; Bourbousse, C.; Lambolez, A.; Law, J.A. The CLASSY family controls tissue-specific DNA methylation patterns in Arabidopsis. Nat. Commun. 2022, 13, 244. [Google Scholar] [CrossRef]
- Zhou, M.; Palanca, A.M.S.; Law, J.A. Locus-specific control of the de novo DNA methylation pathway in Arabidopsis by the CLASSY family. Nat. Genet. 2018, 50, 865–873. [Google Scholar] [CrossRef]
- Kanno, T.; Mette, M.F.; Kreil, D.P.; Aufsatz, W.; Matzke, M.; Matzke, A.J. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 2004, 14, 801–805. [Google Scholar] [CrossRef]
- Kanno, T.; Bucher, E.; Daxinger, L.; Huettel, B.; Böhmdorfer, G.; Gregor, W.; Kreil, D.P.; Matzke, M.; Matzke, A.J.M. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat. Genet. 2008, 40, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Du, X.; Hu, H.; Li, S.; Cao, X.; Jacobsen, S.E.; Du, J. Structure and mechanism of the plant RNA polymerase V. Science 2023, 379, 1209–1213. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Hale, C.J.; Law, J.A.; Johnson, L.M.; Feng, S.; Tu, A.; Jacobsen, S.E. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat. Struct. Mol. Biol. 2012, 19, 870–875. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.W.; Shao, C.R.; Zhang, C.J.; Zhou, J.X.; Zhang, S.W.; Li, L.; Chen, S.; Huang, H.-W.; Cai, T.; He, X.-J.; et al. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 2014, 10, e1003948. [Google Scholar] [CrossRef]
- Johnson, L.M.; Du, J.; Hale, C.J.; Bischof, S.; Feng, S.; Chodavarapu, R.K.; Zhong, X.; Marson, G.; Pellegrini, M.; Segal, D.J.; et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 2014, 507, 124–128. [Google Scholar] [CrossRef]
- Wierzbicki, A.T.; Haag, J.R.; Pikaard, C.S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 2008, 135, 635–648. [Google Scholar] [CrossRef]
- Xie, G.; Du, X.; Hu, H.; Du, J. Molecular mechanisms of the RNA polymerases in plant RNA-directed DNA methylation. Trends Biochem. Sci. 2024, 49, 247–256. [Google Scholar] [CrossRef]
- Li, S.; Vandivier, L.E.; Tu, B.; Gao, L.; Won, S.Y.; Li, S.; Zheng, B.; Gregory, B.D.; Chen, X. Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 2015, 25, 235–245. [Google Scholar] [CrossRef]
- Zhai, J.; Bischof, S.; Wang, H.; Feng, S.; Lee, T.-F.; Teng, C.; Chen, X.; Park, S.Y.; Liu, L.; Gallego-Bartolome, J.; et al. A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 2015, 163, 445–455. [Google Scholar] [CrossRef]
- Yang, D.L.; Zhang, G.; Tang, K.; Li, J.; Yang, L.; Huang, H.; Zhang, H.; Zhu, J.K. Dicer-independent RNA-directed DNA methylation in Arabidopsis. Cell Res. 2016, 26, 1264. [Google Scholar] [CrossRef]
- Ye, R.; Chen, Z.; Lian, B.; Rowley, M.J.; Xia, N.; Chai, J.; Li, Y.; He, X.-J.; Wierzbicki, A.T.; Qi, Y. A Dicer-independent route for biogenesis of siRNAs that direct DNA methylation in Arabidopsis. Mol. Cell 2016, 61, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Wang, Z.; Li, S.; Yu, B.; Liu, J.Y.; Chen, X. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009, 23, 2850–2860. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Shoji, K.; Naganuma, M.; Tomari, Y.; Iwakawa, H.O. The mechanisms of siRNA selection by plant Argonaute proteins triggering DNA methylation. Nucleic Acids Res. 2022, 50, 12997–13010. [Google Scholar] [CrossRef] [PubMed]
- McCue, A.D.; Panda, K.; Nuthikattu, S.; Choudury, S.G.; Thomas, E.N.; Slotkin, R.K. ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 2015, 34, 20–35. [Google Scholar] [CrossRef]
- Marí-Ordóñez, A.; Marchais, A.; Etcheverry, M.; Martin, A.; Colot, V.; Voinnet, O. Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 2013, 45, 1029–1039. [Google Scholar] [CrossRef]
- Huang, C.F.; Miki, D.; Tang, K.; Zhou, H.R.; Zheng, Z.; Chen, W.; Ma, Z.-Y.; Yang, L.; Zhang, H.; Liu, R.; et al. A pre-mRNA-splicing factor is required for RNA-directed DNA methylation in Arabidopsis. PLoS Genet. 2013, 9, e1003779. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Du, J.; Johnson, L.M.; Jacobsen, S.E.; Patel, D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015, 16, 519–532. [Google Scholar] [CrossRef]
- Riggs, A.D. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet. 1975, 14, 9–25. [Google Scholar] [CrossRef]
- Holliday, R.; Pugh, J.E. DNA modification mechanisms and gene activity during development. Science 1975, 187, 226–232. [Google Scholar] [CrossRef]
- Bashtrykov, P.; Jankevicius, G.; Smarandache, A.; Jurkowska, R.Z.; Ragozin, S.; Jeltsch, A. Specificity of Dnmt1 for methylation of hemimethylated CpG sites resides in its catalytic domain. Chem. Biol. 2012, 19, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Teplova, M.; Ishibe-Murakami, S.; Patel, D.J. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science 2012, 335, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Adam, S.; Klingel, V.; Radde, N.E.; Bashtrykov, P.; Jeltsch, A. On the accuracy of the epigenetic copy machine: Comprehensive specificity analysis of the DNMT1 DNA methyltransferase. Nucleic Acids Res. 2023, 51, 6622–6633. [Google Scholar] [CrossRef] [PubMed]
- Bostick, M.; Kim, J.K.; Estève, P.O.; Clark, A.; Pradhan, S.; Jacobsen, S.E. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 2007, 317, 1760–1764. [Google Scholar] [CrossRef]
- Sharif, J.; Muto, M.; Takebayashi, S.-I.; Suetake, I.; Iwamatsu, A.; Endo, T.A.; Shinga, J.; Mizutani-Koseki, Y.; Toyoda, T.; Okamura, K.; et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007, 450, 908–912. [Google Scholar] [CrossRef]
- Woo, H.R.; Dittmer, T.A.; Richards, E.J. Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet. 2008, 4, e1000156. [Google Scholar] [CrossRef]
- Petryk, N.; Bultmann, S.; Bartke, T.; Defossez, P.A. Staying true to yourself: Mechanisms of DNA methylation maintenance in mammals. Nucleic Acids Res. 2021, 49, 3020–3032. [Google Scholar] [CrossRef]
- Rocha, P.S.; Sheikh, M.; Melchiorre, R.; Fagard, M.; Boutet, S.; Loach, R.; Moffatt, B.; Wagner, C.; Vaucheret, H.; Furner, I. The Arabidopsis HOMOLOGY-DEPENDENT GENE SILENCING1 gene codes for an S-adenosyl-L-homocysteine hydrolase required for DNA methylation-dependent gene silencing. Plant Cell 2005, 17, 404–417. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, X.; Miki, D.; Cutler, S.; La, H.; Hou, Y.-J.; Oh, J.; Zhu, J.-K. Sulfamethazine suppresses epigenetic silencing in Arabidopsis by impairing folate synthesis. Plant Cell 2012, 24, 1230–1241. [Google Scholar] [CrossRef]
- Zhou, H.R.; Zhang, F.F.; Ma, Z.Y.; Huang, H.W.; Jiang, L.; Cai, T.; Zhu, J.K.; Zhang, C.; He, X.J. Folate polyglutamylation is involved in chromatin silencing by maintaining global DNA methylation and histone H3K9 dimethylation in Arabidopsis. Plant Cell 2013, 25, 2545–2559. [Google Scholar] [CrossRef]
- Gong, Z.; Morales-Ruiz, T.; Ariza, R.R.; Roldán-Arjona, T.; David, L.; Zhu, J.K. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 2002, 111, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Gehring, M.; Huh, J.H.; Hsieh, T.F.; Penterman, J.; Choi, Y.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 2006, 124, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Agius, F.; Kapoor, A.; Zhu, J.K. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl. Acad. Sci. USA 2006, 103, 11796–11801. [Google Scholar] [CrossRef]
- Morales-Ruiz, T.; Ortega-Galisteo, A.P.; Ponferrada-Marín, M.I.; Martínez-Macías, M.I.; Ariza, R.R.; Roldán-Arjona, T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl. Acad. Sci. USA 2006, 103, 6853–6858. [Google Scholar] [CrossRef]
- Penterman, J.; Zilberman, D.; Huh, J.H.; Ballinger, T.; Henikoff, S.; Fischer, R.L. DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. USA 2007, 104, 6752–6757. [Google Scholar] [CrossRef]
- Huh, J.H.; Bauer, M.J.; Hsieh, T.F.; Fischer, R.L. Cellular programming of plant gene imprinting. Cell 2008, 132, 735–744. [Google Scholar] [CrossRef]
- Martínez-Macías, M.I.; Qian, W.; Miki, D.; Pontes, O.; Liu, Y.; Tang, K.; Liu, R.; Morales-Ruiz, T.; Ariza, R.R.; Roldán-Arjona, T.; et al. A DNA 3’ phosphatase functions in active DNA demethylation in Arabidopsis. Mol. Cell 2012, 45, 357–370. [Google Scholar] [CrossRef]
- Lee, J.; Jang, H.; Shin, H.; Choi, W.L.; Mok, Y.G.; Huh, J.H. AP endonucleases process 5-methylcytosine excision intermediates during active DNA demethylation in Arabidopsis. Nucleic Acids Res. 2014, 42, 11408–11418. [Google Scholar] [CrossRef]
- Raja, P.; Sanville, B.C.; Buchmann, R.C.; Bisaro, D.M. Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 2008, 82, 8997–9007. [Google Scholar] [CrossRef]
- Zhang, X.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.W.; Chen, H.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E.; et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 2006, 126, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Miki, D.; Zhang, H.; Liu, Y.; Zhang, X.; Tang, K.; Kan, Y.; La, H.; Li, X.; Li, S.; et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 2012, 336, 1445–1448. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.P.; Pignatta, D.; Henikoff, S.; Gehring, M. Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLOS Genet. 2015, 11, e1005142. [Google Scholar] [CrossRef] [PubMed]
- Domcke, S.; Bardet, A.F.; Adrian Ginno, P.; Hartl, D.; Burger, L.; Schübeler, D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 2015, 528, 575–579. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, G.H.; Qian, J. Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet. 2016, 17, 551–565. [Google Scholar] [CrossRef]
- Tang, K.; Lang, Z.; Zhang, H.; Zhu, J.K. The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nat. Plants 2016, 2, 16169. [Google Scholar] [CrossRef]
- Lei, M.; Zhang, H.; Julian, R.; Tang, K.; Xie, S.; Zhu, J.K. Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 3553–3557. [Google Scholar] [CrossRef]
- Feng, Z.; Zhan, X.; Pang, J.; Liu, X.; Zhang, H.; Lang, Z.; Zhu, J.K. Genetic analysis implicates a molecular chaperone complex in regulating epigenetic silencing of methylated genomic regions. J. Integr. Plant Biol. 2021, 63, 1451–1461. [Google Scholar] [CrossRef]
- Qian, W.; Miki, D.; Lei, M.; Zhu, X.; Zhang, H.; Liu, Y.; Li, Y.; Lang, Z.; Wang, J.; Tang, K.; et al. Regulation of active DNA demethylation by an alpha-crystallin domain protein in Arabidopsis. Mol. Cell 2014, 55, 361–371. [Google Scholar] [CrossRef]
- Boone, B.A.; Ichino, L.; Wang, S.; Gardiner, J.; Yun, J.; Jami-Alahmadi, Y.; Sha, J.; Mendoza, C.P.; Steelman, B.J.; van Aardenne, A.; et al. ACD15, ACD21, and SLN regulate the accumulation and mobility of MBD6 to silence genes and transposable elements. Sci. Adv. 2023, 9, eadi9036. [Google Scholar] [CrossRef]
- Duan, C.G.; Wang, X.; Xie, S.; Pan, L.; Miki, D.; Tang, K.; Hsu, C.C.; Lei, M.; Zhong, Y.; Hou, Y.J.; et al. A pair of transposon-derived proteins function in a histone acetyltransferase complex for active DNA demethylation. Cell Res. 2017, 27, 226–240. [Google Scholar] [CrossRef]
- Haslbeck, M.; Weinkauf, S.; Buchner, J. Small heat shock proteins: Simplicity meets complexity. J. Biol. Chem. 2019, 294, 2121–2132. [Google Scholar] [CrossRef] [PubMed]
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhong, X.; Bernatavichute, Y.V.; Stroud, H.; Feng, S.; Caro, E.; Vashisht, A.A.; Terragni, J.; Chin, H.G.; Tu, A.; et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 2012, 151, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Gent, J.I.; Zynda, G.; Song, J.; Makarevitch, I.; Hirsch, C.D.; Hirsch, C.N.; Dawe, R.K.; Madzima, T.F.; McGinnis, K.M.; et al. RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc. Natl. Acad. Sci. USA 2015, 112, 14728–14733. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewski, F.; Schmidt, M.; Van Lijsebettens, M.; Schmidt, T. DNA methylation of retrotransposons, DNA transposons and genes in sugar beet (Beta vulgaris L.). Plant J. 2017, 90, 1156–1175. [Google Scholar] [CrossRef] [PubMed]
- Gouil, Q.; Baulcombe, D.C. DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet. 2016, 12, e1006526. [Google Scholar] [CrossRef]
- Grob, S.; Schmid, M.W.; Grossniklaus, U. Hi-C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco Locus of Drosophila. Mol. Cell 2014, 55, 678–693. [Google Scholar] [CrossRef]
- Feng, S.; Cokus, S.J.; Schubert, V.; Zhai, J.; Pellegrini, M.; Jacobsen, S.E. Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol. Cell 2014, 55, 694–707. [Google Scholar] [CrossRef]
- Rowley, M.J.; Rothi, M.H.; Bohmdorfer, G.; Kucinski, J.; Wierzbicki, A.T. Long-range control of gene expression via RNA-directed DNA methylation. PLoS Genet. 2017, 13, e1006749. [Google Scholar] [CrossRef]
- Gehring, M.; Bubb, K.L.; Henikoff, S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 2009, 324, 1447–1451. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.F.; Ibarra, C.A.; Silva, P.; Zemach, A.; Eshed-Williams, L.; Fischer, R.L.; Zilberman, D. Genome-wide demethylation of Arabidopsis endosperm. Science 2009, 324, 1451–1454. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, C.A.; Feng, X.; Schoft, V.K.; Hsieh, T.-F.; Uzawa, R.; Rodrigues, J.A.; Zemach, A.; Chumak, N.; Machlicova, A.; Nishimura, T.; et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 2012, 337, 1360–1364. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Kim, M.Y.; Vickers, M.; Park, J.-S.; Hyun, Y.; Okamoto, T.; Zilberman, D.; Fischer, R.L.; Feng, X.; Choi, Y.; et al. DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc. Natl. Acad. Sci. USA 2016, 113, 15138–15143. [Google Scholar] [CrossRef]
- Mierziak, J.; Wojtasik, W.; Kulma, A.; Dziadas, M.; Kostyn, K.; Dymińska, L.; Hanuza, J.; Żuk, M.; Szopa, J. 3-Hydroxybutyrate Is Active Compound in Flax That Upregulates Genes Involved in DNA Methylation. Int. J. Mol. Sci. 2020, 21, 2887. [Google Scholar] [CrossRef]
- Martínez, G.; Panda, K.; Köhler, C.; Slotkin, R.K. Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat. Plants. 2016, 2, 16030. [Google Scholar] [CrossRef]
- Ingouff, M. Live-cell analysis of DNA methylation during sexual reproduction in Arabidopsis reveals context and sex-specific dynamics controlled by noncanonical RdDM. Genes Dev. 2017, 31, 72–83. [Google Scholar] [CrossRef]
- Walker, J. Sexual-lineage-specific DNA methylation regulates meiosis in Arabidopsis. Nat. Genet. 2018, 50, 130–137. [Google Scholar] [CrossRef]
- Zhang, M.; Xie, S.; Dong, X.; Zhao, X.; Zeng, B.; Chen, J.; Li, H.; Yang, W.; Zhao, H.; Wang, G.; et al. Genome-wide high resolution parental-specific DNA and histone methylation maps uncover patterns of imprinting regulation in maize. Genome Res. 2014, 24, 167–176. [Google Scholar] [CrossRef]
- Klosinska, M.; Picard, C.L.; Gehring, M. Conserved imprinting associated with unique epigenetic signatures in the Arabidopsis genus. Nat. Plants 2016, 2, 16145. [Google Scholar] [CrossRef]
- Rodrigues, J.A.; Ruan, R.; Nishimura, T.; Sharma, M.K.; Sharma, R.; Ronald, P.C.; Fischer, R.L.; Zilberman, D. Imprinted expression of genes and small RNA is associated with localized hypomethylation of the maternal genome in rice endosperm. Proc. Natl. Acad. Sci. USA 2013, 110, 7934–7939. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, Y.; Saze, H.; Kinoshita, T.; Miura, A.; Soppe, W.J.; Koornneef, M.; Kakutani, T. Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J. 2007, 49, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.-F.; Shin, J.; Uzawa, R.; Silva, P.; Cohen, S.; Bauer, M.J.; Hashimoto, M.; Kirkbride, R.C.; Harada, J.J.; Zilberman, D.; et al. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. USA 2011, 108, 1755–1762. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Miura, A.; Choi, Y.; Kinoshita, Y.; Cao, X.; Jacobsen, S.E.; Fischer, R.L.; Kakutani, T. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 2004, 303, 521–523. [Google Scholar] [CrossRef]
- Bratzel, F.; Yang, C.; Angelova, A.; López-Torrejón, G.; Koch, M.; del Pozo, J.C.; Calonje, M. Regulation of the new Arabidopsis imprinted gene AtBMI1C requires the interplay of different epigenetic mechanisms. Mol. Plant 2012, 5, 260–269. [Google Scholar] [CrossRef]
- Moreno-Romero, J.; Jiang, H.; Santos-Gonzalez, J.; Kohler, C. Parental epigenetic asymmetry of PRC2-mediated histone modifications in the Arabidopsis endosperm. EMBO J. 2016, 35, 1298–1311. [Google Scholar] [CrossRef]
- Dong, X.; Zhang, M.; Chen, J.; Peng, L.; Zhang, N.; Wang, X.; Lai, J. Dynamic and antagonistic allele-Specific epigenetic modifications controlling the expression of imprinted genes in maize endosperm. Mol. Plant 2017, 10, 442–455. [Google Scholar] [CrossRef]
- Baubec, T.; Finke, A.; Scheid, O.M.; Pecinka, A. Meristem-specific expression of epigenetic regulators safeguards transposon silencing in Arabidopsis. EMBO Rep. 2014, 15, 446–452. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Stuart, T.; Valdes, M.; Breakfield, N.; Schmitz, R.J.; Nery, J.R.; Urich, M.A.; Han, X.; Lister, R.; Benfey, P.N.; et al. Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat. Plants 2016, 2, 16058. [Google Scholar] [CrossRef]
- Zhou, S.; Liu, X.; Zhou, C.; Zhou, Q.; Zhao, Y.; Li, G.; Zhou, D.X. Cooperation between the H3K27me3 chromatin mark and non-CG methylation in epigenetic regulation. Plant Physiol. 2016, 172, 1131–1141. [Google Scholar]
- Candaele, J.; Demuynck, K.; Mosoti, D.; Beemster, G.T.; Inzé, D; Nelissen, H. Differential methylation during maize leaf growth targets developmentally regulated genes. Plant Physiol. 2014, 164, 1350–1364. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, C.; Miki, D.; Zheng, Z.; Ma, J.; Wang, J.; Yang, Z.; Dong, J.; Zhu, J.-K. Overproduction of stomatal lineage cells in Arabidopsis mutants defective in active DNA demethylation. Nat. Commun. 2014, 5, 4062. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Xue, X.Y.; Zhu, J.K.; Dong, J. Demethylation of ERECTA receptor genes by IBM1 histone demethylase affects stomatal development. Development 2016, 143, 4452–4461. [Google Scholar] [CrossRef] [PubMed]
- Hieber, A.D.; Mudalige-Jayawickrama, R.G.; Kuehnle, A.R. Color genes in the orchid Oncidium Gower Ramsey: Identification, expression, and potential genetic instability in an interspecific cross. Planta 2006, 223, 521–531. [Google Scholar] [CrossRef]
- Chiou, C.Y.; Pan, H.A.; Chuang, Y.N.; Yeh, K.W. Differential expression of carotenoid-related genes determines diversified carotenoid coloration in floral tissues of Oncidium cultivars. Planta 2010, 232, 937–948. [Google Scholar] [CrossRef]
- Liu, X.J.; Chuang, Y.N.; Chiou, C.Y.; Chin, D.C.; Shen, F.Q.; Yeh, K.W. Methylation effect on chalcone synthase gene expression determines anthocyanin pigmentation in floral tissues of two Oncidium orchid cultivars. Planta 2012, 236, 401–409. [Google Scholar] [CrossRef]
- Han, M.L.; Yin, J.; Zhao, Y.H.; Sun, X.W.; Meng, J.X.; Zhou, J.; Shen, T.; Li, H.H.; Zhang, F. How the Color Fades from Malus halliana Flowers: Transcriptome Sequencing and DNA Methylation Analysis. Front. Plant Sci. 2020, 11, 576054. [Google Scholar] [CrossRef]
- Yuan, X.; Ma, K.; Zhang, M.; Wang, J.; Zhang, Q. Integration of Transcriptome and Methylome Analyses Provides Insight Into the Pathway of Floral Scent Biosynthesis in Prunus mume. Front. Genet. 2021, 12, 779557. [Google Scholar] [CrossRef]
- Zhong, S.; Fei, Z.; Chen, Y.R.; Zheng, Y.; Huang, M.; Vrebalov, J.; McQuinn, R.; Gapper, N.; Liu, B.; Xiang, J.; et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013, 31, 154–159. [Google Scholar] [CrossRef]
- Liu, R.; How-Kit, A.; Stammitti, L.; Teyssier, E.; Rolin, D.; Mortain-Bertrand, A.; Liu, M.; Kong, J.; Wu, C.; Degraeve-Guibaul, C.; et al. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl. Acad. Sci. USA 2015, 112, 10804–10809. [Google Scholar] [CrossRef]
- Lang, Z.; Wang, Y.; Tang, K.; Tang, D.; Datsenka, T.; Cheng, J.; Zhang, Y.; Handa, A.K.; Zhu, J.K. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. USA 2017, 114, E4511–E4519. [Google Scholar] [CrossRef] [PubMed]
- Telias, A.; Lin-Wang, K.; Stevenson, D.E.; Cooney, J.M.; Hellens, R.P.; Allan, A.C.; Hoover, E.E.; Bradeen, J.M. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol. 2011, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- El-Sharkawy, I.; Liang, D.; Xu, K.N. Transcriptome analysis of an apple (Malus x domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. J. Exp. Bot. 2015, 66, 7359–7376. [Google Scholar] [CrossRef] [PubMed]
- Daccord, N.; Celton, J.M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef]
- Tollefson, J. How hot will earth get by 2100? Nature 2020, 580, 443–445. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Mittler, R.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plant 2018, 162, 2–12. [Google Scholar] [CrossRef]
- Gao, G.; Li, J.; Li, H.; Li, F.; Xu, K.; Yan, G.; Chen, B.; Qiao, J.; Wu, X. Comparison of the heat stress induced variations in DNA methylation between heat-tolerant and heat-sensitive rapeseed seedlings. Breed Sci. 2014, 64, 125–133. [Google Scholar] [CrossRef]
- Singh, R.K.; Jaishankar, J.; Muthamilarasan, M.; Shweta, S.; Dangi, A.; Prasad, M. Genome-wide analysis of heat shock proteins in C4 model, foxtail millet identifies potential candidates for crop improvement under abiotic stress. Sci. Rep. 2016, 6, 32641. [Google Scholar] [CrossRef]
- Qian, Y.; Hu, W.; Liao, J.; Zhang, J.; Ren, Q. The dynamics of DNA methylation in the maize (Zea mays L.) inbred line B73 response to heat stress at the seedling stage. Biochem. Biophys. Res. Commun. 2019, 512, 742–749. [Google Scholar] [CrossRef]
- Lamelas, L.; Valledor, L.; Escandón, M.; Pinto, G.; Cañal, M.J.; Meijón, M. Integrative analysis of the nuclear proteome in Pinus radiata reveals thermopriming coupled to epigenetic regulation. J. Exp. Bot. 2020, 71, 2040–2057. [Google Scholar] [CrossRef]
- Liu, H.; Able, A.J.; Able, J.A. Priming crops for the future: Rewiring stress memory. Trends Plant Sci. 2021, 11, 1360–1385. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, S.; Wang, X.; Zhu, J.; Wei, Y.; Wang, Y.; Wen, Y.; Wang, L.; Huang, Y.; Zhang, B.; et al. DNA methylation dynamics: Identification and functional annotation. Brief Funct. Genom. 2016, 15, 470–484. [Google Scholar] [CrossRef] [PubMed]
- Sallam, N.; Moussa, M. DNA methylation changes stimulated by drought stress in ABA-deficient maize mutant vp10. Plant Physiol. Biochem. 2021, 160, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Fan, M.; He, Y. DNA methylation analysis of the Citrullus lanatus response to cucumber green mottle mosaic virus infection by whole-genome bisulfite sequencing. Genes 2019, 10, 344. [Google Scholar] [CrossRef]
- Herman, J.J.; Sultan, S.E. DNA methylation mediates stress-induced transgenerational plasticity in plants. Proc. Natl. Acad. Sci. USA 2016, 113, 8804–8809. [Google Scholar]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, S.D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef]
- Godwin, J.; Farrona, S. Plant Epigenetic Stress Memory Induced by Drought: A Physiological and Molecular Perspective. Methods Mol. Biol. 2020, 2093, 243–259. [Google Scholar]
- Wang, X.; Wang, H.; Liu, S.; Ferjani, A.; Li, J.; Yan, J.; Yang, X.; Qin, F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 2016, 48, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, D.; Zhang, Y.; Li, G.; Sun, D.; Zhou, B.; Li, J. Insights into the Epigenetic Basis of Plant Salt Tolerance. Int. J. Mol. Sci. 2024, 25, 11698. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef]
- Sahu, P.P.; Pandey, G.; Sharma, N.; Puranik, S.; Muthamilarasan, M.; Prasad, M. Epigenetic mechanisms of plant stress responses and adaptation. Plant Cell Rep. 2013, 32, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- Dowen, R.H.; Pelizzola, M.; Schmitz, R.J.; Lister, R.; Dowen, J.M.; Nery, J.R.; Dixon, J.E.; Ecker, J.R. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 2012, 109, E2183–E2191. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.; Lepère, G.; Jay, F.; Wang, J.; Bapaume, L.; Wang, Y.; Abraham, A.-L.; Penterman, J.; Fischer, R.L.; Voinnet, O.; et al. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. USA 2013, 110, 2389–2394. [Google Scholar] [CrossRef] [PubMed]
- López, A.; Ramírez, V.; García-Andrade, J.; Flors, V.; Vera, P. The RNA silencing enzyme RNA polymerase V is required for plant immunity. PLoS Genet. 2011, 7, e1002434. [Google Scholar] [CrossRef]
- Wang, W.; Pan, Y.; Zhao, X.; Zhu, L.; Fu, B.; Li, Z. DNA methylation is critical for defense against the fungal pathogen Verticillium dahliae in Arabidopsis. Plant Cell 2016, 28, 1237–1251. [Google Scholar]
- Gohlke, J.; Scholz, C.J.; Kneitz, S.; Weber, D.; Fuchs, J.; Hedrich, R.; Deeken, R. DNA methylation mediated control of gene expression is critical for development of crown gall tumors. PLoS Genet. 2013, 9, e1003267. [Google Scholar] [CrossRef]
- Singh, N.; Titov, V.; Ayemere, I.; Byeon, B.; Kovalchuk, I. Multigenerational exposure to heat stress induces phenotypic resilience, and genetic and epigenetic variations in Arabidopsis thaliana offspring. Front. Plant Sci. 2020, 13, 728167. [Google Scholar]
- Cubas, P.; Vincent, C.; Coen, E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 1999, 401, 157–161. [Google Scholar] [CrossRef]
- Gallego-Bartolome, J. DNA methylation in plants: Mechanisms and tools for targeted manipulation. New Phytol. 2020, 227, 38–44. [Google Scholar] [CrossRef]
- Le, T.N.; Schumann, U.; Smith, N.A.; Tiwari, S.; Au, P.C.; Zhu, Q.H.; Taylor, J.M.; Kazan, K.; Llewellyn, D.J.; Zhang, R.; et al. DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 2014, 15, 458. [Google Scholar] [CrossRef]
- Manning, K.; Tör, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Zhang, T.; Stelly, D.M.; Chen, Z.J. Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons. Genome Biol. 2017, 18, 99. [Google Scholar] [CrossRef] [PubMed]
- Pilu, R.; Panzeri, D.; Cassani, E.; Badone, F.C.; Landoni, M.; Nielsen, E. A paramutation phenomenon is involved in the genetics of maize low phytic acid1-241 (lpa1-241) trait. Heredity 2009, 102, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Kawanabe, T.; Ishikura, S.; Miyaji, N.; Sasaki, T.; Wu, L.M.; Itabashi, E.; Takada, S.; Shimizu, M.; Takasaki-Yasuda, T.; Osabe, K.; et al. Role of DNA methylation in hybrid vigor in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2016, 113, E6704–E6711. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Y.; Xu, T.; Srivastava, A.K.; Wang, D.; Zeng, L.; Yang, L.; He, L.; Zhang, H.; Zheng, Z. The chromatin remodeler DDM1 promotes hybrid vigor by regulating salicylic acid metabolism. Cell Discov. 2016, 2, 16027. [Google Scholar] [CrossRef]
- Brachi, B.; Morris, G.P.; Borevitz, J.O. Genome-wide association studies in plants: The missing heritability is in the field. Genome Biol. 2011, 12, 232. [Google Scholar] [CrossRef]
- Dubin, M.J.; Zhang, P.; Meng, D.; Remigereau, M.S.; Osborne, E.J.; Paolo Casale, F.; Drewe, P.; Kahles, A.; Jean, G.; Vilhjalmsson, B.; et al. DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. Elife 2015, 4, e05255. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Huang SS, C.; Jupe, F.; Sasaki, E.; Schmitz, R.J.; Urich, M.A.; Schork, N.J. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 2016, 166, 492–505. [Google Scholar]
- Eichten, S.R.; Briskine, R.; Song, J.; Li, Q.; Swanson-Wagner, R.; Hermanson, P.J.; Waters, A.J.; Starr, E.; West, P.T.; Tiffin, P.; et al. Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 2013, 25, 2783–2797. [Google Scholar] [CrossRef]
- Xu, G.; Lyu, J.; Li, Q.; Liu, H.; Wang, D.; Zhang, M.; Springer, N.M.; Ross-Ibarra, J.; Yang, J. Evolutionary and functional genomics of DNA methylation in maize domestication and improvement. Nat. Commun. 2020, 11, 5539. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, J.; Liu, Y.; Liu, S.; Liu, Z.; Duan, Z.; Wang, Z.; Zhu, B.; Guo, Y.-L.; Tian, Z. DNA methylation footprints during soybean domestication and improvement. Genome Biol. 2018, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Xie, L.; Zhang, Q.; Ouyang, W.; Deng, L.; Guan, P.; Ma, M.; Li, Y.; Zhang, Y.; Xiao, Q.; et al. Integrative analysis of reference epigenomes in 20 rice varieties. Nat. Commun. 2020, 11, 2658. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, L.-J.; Joynson, R.; Omony, J.; Rusholme-Pilcher, R.; Olohan, L.; Lang, D.; Bai, C.; Hawkesford, M.; Salt, D.; Spannagl, M.; et al. Hidden variation in polyploid wheat drives local adaptation. Genome Res. 2018, 28, 1319–1332. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chen, G.; Hermanson, P.J.; Xu, Q.; Sun, C.; Chen, W.; Kan, Q.; Li, M.; Crisp, P.A.; Yan, J.; et al. Population-level analysis reveals the widespread occurrence and phenotypic consequence of DNA methylation variation not tagged by genetic variation in maize. Genome Biol. 2019, 20, 243. [Google Scholar] [CrossRef]
- Schmitz, R.J.; Schultz, M.D.; Urich, M.A.; Nery, J.R.; Pelizzola, M.; Libiger, O.; Alix, A.; McCosh, R.B.; Chen, H.; Schork, N.J.; et al. Patterns of population epigenomic diversity. Nature 2013, 495, 193–198. [Google Scholar] [CrossRef]
- Kou, H.P.; Li, Y.; Song, X.X.; Ou, X.F.; Xing, S.C.; Ma, J.; Von Wettstein, D.; Liu, B. Heritable alteration in DNA methylation induced by nitrogen-deficiency stress accompanies enhanced tolerance in rice (Oryza sativa L.). J. Plant Pyhsiol. 2011, 65, 655–664. [Google Scholar] [CrossRef]
- Gouil, Q.; Baulcombe, D.C. Paramutation-like features of multiple natural epialleles in tomato. BMC Genom. 2018, 19, 203. [Google Scholar] [CrossRef]
- Cao, S.; Wang, L.; Han, T.; Ye, W.; Liu, Y.; Sun, Y.; Moose, S.P.; Song, Q.; Chen, Z.J. Small RNAs mediate transgenerational inheritance of genome-wide trans-acting epialleles in maize. Genome Biol. 2022, 23, 53. [Google Scholar] [CrossRef]
- Cortijo, S.; Wardenaar, R.; Colomé-Tatché, M.; Gilly, A.; Etcheverry, M.; Labadie, K.; Caillieux, E.; Hospital, F.; Aury, J.-M.; Wincker, P.; et al. Mapping the epigenetic basis of complex traits. Science 2014, 343, 1145–1148. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Latzel, V.; Fischer, M.; Bossdorf, O. Understanding the evolutionary potential of epigenetic variation: A comparison of heritable phenotypic variation in epiRILs, RILs, and natural ecotypes of Arabidopsis thaliana. Heredity 2018, 121, 257–265. [Google Scholar] [CrossRef]
- Schmidt, M.; Byzova, M.; Martens, C.; Peeters, M.; Raj, Y.; Shukla, S.; Verwulgen, T.; De Block, M.; Van Lijsebettens, M. Methylome and epialleles in rice epilines selected for energy use efficiency. Agronomy 2018, 8, 163. [Google Scholar] [CrossRef]
- Hauben, M.; Haesendonckx, B.; Standaert, E.; Van Der Kelen, K.; Azmi, A.; Akpo, H.; Van Breusegem, F.; Guisez, Y.; Bots, M.; Lambert, B.; et al. Energy use efficiency is characterized by an epigenetic component that can be directed through artificial selection to increase yield. Proc. Natl. Acad. Sci. USA 2009, 106, 20109–20114. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Li, G.; Malzahn, A.A.; Cheng, Y.; Leyson, B.; Sretenovic, S.; Gurel, F.; Coleman, G.D.; Qi, Y. Boosting plant genome editing with a versatile CRISPR-Combo system. Nat. Plants 2022, 8, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Bartolome, J.; Gardiner, J.; Liu, W.; Papikian, A.; Ghoshal, B.; Kuo, H.Y.; Zhao, J.M.-C.; Segal, D.J.; Jacobsen, S.E. Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc. Natl. Acad. Sci. USA 2018, 115, E2125–E2134. [Google Scholar] [CrossRef]
- Ji, L.; Jordan, W.T.; Shi, X.; Hu, L.; He, C.; Schmitz, R.J. TET-mediated epimutagenesis of the Arabidopsis thaliana methylome. Nat. Commun. 2018, 9, 895. [Google Scholar] [CrossRef]
- Papikian, A.; Liu, W.; Gallego-Bartolome, J.; Jacobsen, S.E. Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat. Commun. 2019, 10, 729. [Google Scholar] [CrossRef]
- Ghoshal, B.; Picard, C.L.; Vong, B.; Feng, S.; Jacobsen, S.E. CRISPR-based targeting of DNA methylation in Arabidopsis thaliana by a bacterial CG-specific DNA methyltransferase. Proc. Natl. Acad. Sci. USA 2021, 118, e2125016118. [Google Scholar] [CrossRef]
- Galonska, C.; Charlton, J.; Mattei, A.L.; Donaghey, J.; Clement, K.; Gu, H.; Mohammad, A.W.; Stamenova, E.K.; Cacchiarelli, D.; Klages, S.; et al. Genome-wide tracking of dCas9-methyltransferase footprints. Nat. Commun. 2018, 9, 597. [Google Scholar] [CrossRef]
- Pflueger, C.; Tan, D.; Swain, T.; Nguyen, T.; Pflueger, J.; Nefzger, C.; Polo, J.M.; Ford, E.; Lister, R. A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs. Genome Res. 2018, 28, 1193–1206. [Google Scholar] [CrossRef]
- Chan, W.F.; Coughlan, H.D.; Chen, Y.; Keenan, C.R.; Smyth, G.K.; Perkins, A.C.; Johanson, T.M.; Allan, R.S. Activation of stably silenced genes by recruitment of a synthetic de-methylating module. Nat. Commun. 2022, 13, 5582. [Google Scholar] [CrossRef]
- Swain, T.; Pflueger, C.; Freytag, S.; Poppe, D.; Pflueger, J.; Nguyen, T.V.; Li, J.K.; Lister, R. A modular dCas9-based recruitment platform for combinatorial epigenome editing. Nucleic Acids Res. 2024, 52, 474–491. [Google Scholar] [CrossRef]
| Arabidopsis Mutants | Pathogen | Phenotype | Defense Response | References |
|---|---|---|---|---|
| DNA Hypomethylation | ||||
| drd1 | Pseudomonas syringae pv. tomato DC3000 (Pst) | Resistant | Enhancement of SA-dependent defense | Dowen et al. [152] |
| Plectosphaerella cucumerina | Susceptible | Suppression of JA-dependent defense | López et al. [154] | |
| ago4 | Botrytis cinerea | Susceptible | Suppression of JA-dependent defense | López et al. [154] |
| rdr2 | Plectosphaerella cucumerina | Susceptible | Suppression of JA-dependent defense | López et al. [154] |
| Pst | Resistant | Enhancement of SA-dependent defense | Dowen et al. [152] | |
| rdr6 | Botrytis cinerea | Susceptible | Loss of transfer siRNAs that target pathogen genes | Cai et al. [146] |
| Pst | Resistant | − | Dowen et al. [152] | |
| nrpd1 | Pst | Resistant | Enhancement of SA-dependent defense | Dowen et al. [152] |
| nrpe1 | Plectosphaerella cucumerina | Susceptible | Suppression of JA-dependent defense | López et al. [154] |
| Botrytis cinerea | Susceptible | Suppression of JA-dependent defense | López et al. [154] | |
| Pst | Resistant | Enhancement of SA-dependent defense | López et al. [154] | |
| nrpd2 | Plectosphaerella cucumerina | Susceptible | Suppression of JA-dependent defense | López et al. [154] |
| Botrytis cinerea | Susceptible | Suppression of JA-dependent defense | López et al. [154] | |
| Pst | Resistant | Enhancement of SA-dependent defense | López et al. [154] | |
| nrpd1/nrpe1 | Plectosphaerella cucumerina | Susceptible | Suppression of JA-dependent defense | López et al. [154] |
| Pst | Resistant | Enhancement of SA-dependent defense | López et al. [154] | |
| drm1/drm2 | Plectosphaerella cucumerina | Susceptible | Suppression of JA-dependent defense | López et al. [154] |
| Pst | Resistant | Enhancement of SA-dependent defensePrimed state of defense response | Yu et al. [142] | |
| Cabbage leaf curl virus | Susceptible | − | Raja et al. [80] | |
| Beet curly top virus | Susceptible | − | Raja et al. [80] | |
| drm1/drm2/cmt3 (ddc) | Agrobacterium tumefaciens | Susceptible | Enhancement of ABA-dependent response | Gohlke et al. [156] |
| Pst | Resistant | Enhancement of SA-dependent defense | Dowen et al. [152] | |
| dcl2/3/4 | Botrytis cinerea | Susceptible | Loss of siRNAs that move into fungal cells and suppress virulence genes | Cai et al. [146] |
| Pst | Resistant | Enhancement of SA-dependent defense | Dowen et al. [152] | |
| Cabbage leaf curl virus | Susceptible | − | Raja et al. [80] | |
| Beet curly top virus | Susceptible | − | Raja et al. [80] | |
| DNA Hypermethylation | ||||
| ros1 | Pst | Susceptible | Methylation at the promoter of RMG1 and RLP43 | Yu et al. [142] |
| ros1/dml2/dml3 (rdd) | Fusarium oxysporum | Susceptible | Suppression of defense-related genes | Le et al. [160] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, S.; Song, W.; Hu, W.; Wang, F.; Liao, A.; Tan, W.; Yang, S. The Role of Plant DNA Methylation in Development, Stress Response, and Crop Breeding. Agronomy 2025, 15, 94. https://doi.org/10.3390/agronomy15010094
Qiao S, Song W, Hu W, Wang F, Liao A, Tan W, Yang S. The Role of Plant DNA Methylation in Development, Stress Response, and Crop Breeding. Agronomy. 2025; 15(1):94. https://doi.org/10.3390/agronomy15010094
Chicago/Turabian StyleQiao, Shuai, Wei Song, Wentao Hu, Fang Wang, Anzhong Liao, Wenfang Tan, and Songtao Yang. 2025. "The Role of Plant DNA Methylation in Development, Stress Response, and Crop Breeding" Agronomy 15, no. 1: 94. https://doi.org/10.3390/agronomy15010094
APA StyleQiao, S., Song, W., Hu, W., Wang, F., Liao, A., Tan, W., & Yang, S. (2025). The Role of Plant DNA Methylation in Development, Stress Response, and Crop Breeding. Agronomy, 15(1), 94. https://doi.org/10.3390/agronomy15010094





