Abstract
This study aimed to develop an aromatic thermosensitive genic male sterile (TGMS) line in indica rice using CRISPR/Cas9 technology. The TMS5 and FGR in the high-quality conventional rice variety Huahang 48 were targeted for editing using CRISPR/Cas9 technology. CRISPR/Cas9 vectors designed for TMS5 and FGR were constructed and introduced into rice calli through Agrobacterium-mediated transformation. Transgenic seedlings were subsequently regenerated, and the target sites of the edited plants were analyzed via sequencing. A total of fifteen T0 double mutants were successfully obtained. Three mutants without T-DNA insertion were screened in the T1 generation by the PCR detection of hygromycin gene fragments, and homozygous mutants without T-DNA insertion were screened in the T2 generation by the sequencing analysis of the mutation sites, named Huahang 48s. Huahang 48s exhibited complete sterility at 24 °C and pollen transfer at 23 °C. The 2-acetyl-1-pyrroline (2-AP) content was detected in the young panicles, leaves, and stems of Huahang 48s. The leaves of Huahang 48s had the highest 2-AP content, contrasting with the absence of 2-AP in HuaHang 48. F1 hybrids that crossed Huahang 48s with two high-quality restorer lines were superior to the two parents in terms of yield per plant and 1000-grain weight. Huahang 48s has a certain combining ability and application potential in two-line cross breeding. The successful application of CRISPR/Cas9 technology in Huahang 48 established a foundation for developing aromatic TGMS lines, providing both theoretical insights and practical materials for breeding efforts.
1. Introduction
Rice is a critical global food crop facing increasing challenges as the world population grows and arable land decreases due to environmental degradation. Addressing food production issues remains a key priority [1]. Hybrid rice represents a significant advancement in agricultural science, offering a 10–20% yield increase over conventional rice. In China, approximately 60% of the rice cultivated is hybrid, underscoring its importance for food security [2]. Hybrid rice breeding methods are divided into three-line hybrids and two-line hybrids. The two-line hybrid lines include sterile and restorative lines. According to the sensitivity of sterile lines to light and temperature, they can be divided into photosensitive and thermosensitive genic male sterile (TGMS) lines [3]. Photosensitive genic male sterile lines exhibit controlled fertility based on the duration of light exposure. Photosensitive genic male sterile lines display male sterility under prolonged daylight and high temperatures, which are vital in producing hybrid rice seeds through crosses with restorer lines. Photosensitive genic male sterile lines revert to fertility under short-day and low-temperature conditions for self-breeding [4]. TGMS lines are controlled by temperature and remain unaffected by photoperiod conditions, exhibiting sterility under high temperatures and fertility under low temperatures [5]. Two-line hybrid rice overcomes the constraints of restoration relationships, leading to greater genetic diversity between the parental lines. On average, two-line hybrid rice shows a noticeable increase in yield compared to three-line hybrid rice [6].
The reported photosensitive and TGMS genes mainly include tms1-tms10, pms1-pms3, and ptgms2-1. Notably, the key genes that have been precisely mapped to photosensitive and TGMS lines are pms3, tms9-1, tms2, ptgms2-1, and tms5 [7,8]. In China, a range of thermosensitive genic sterile line materials have been developed through the functional deletion of the tms5. Using Annong S-1 and Zhu 1S materials, tms5 was localized and its molecular mechanism was revealed for the first time [9,10]. Among the 25 PTGMS lines, 24 varieties carried the same tms5 mutation type.
The loss-of-function mutant of tms5 has significantly contributed to selecting and breeding two-line hybrid rice, with approximately 71% of two-line sterile varieties harboring the tms5 recessive gene [9]. Consumer expectations for rice quality, particularly its fragrance, are rising as economies develop. Fragrance, a critical quality attribute of cooked rice, primarily originates from the volatile compound 2-acetyl-1-pyrroline (2-AP). The metabolic pathway that synthesizes 2-AP involves betaine aldehyde dehydrogenase. The loss of function of fgr disrupts the metabolic pathway of the substrate of beet aldehyde dehydrogenase, thus increasing the content of 2-acetylpyrroline in rice to produce fragrance [9,11,12,13].
The quality of hybrid rice is influenced by the endosperm produced in the F1 generation, resulting from fertilization involving two polar nuclei from a sterile line and one sperm nucleus from a restorer line [14]. Consequently, the selection of high-quality sterile lines is crucial for developing superior hybrid rice. Establishing two-line sterile lines with fragrance traits is particularly valuable in production. Traditional cross-breeding methods, typically involving cross-transformation, are time consuming and labor intensive, often failing to preserve the desirable traits of the parent lines completely during the selection process [4]. High-quality two-line sterile lines with fragrance are currently scarce, making their development critically important for rice breeding. Gene editing is a technique for precise modifications at specific genomic DNA loci, offering a streamlined approach to genome alteration. This technology enables the rapid and efficient production of high-quality two-line sterile lines due to its short development times, high efficiency, and simplicity of operation [15,16]. The CRISPR/Cas9 system, a third-generation gene editing tool, has been extensively used in various crops, including Arabidopsis, wheat, maize, and rice [17,18,19]. In rice, the CRISPR/Cas9 system has been widely used to enhance the breeding efficiency and conduct functional gene studies [20,21].
This study used the high-quality indica rice cultivar Huahang 48 as our primary material. Employing the CRISPR/Cas9 system, we specifically targeted the editing of the TMS5 and FGR. Subsequently, we identified the fragrant indica rice line Huahang 48s, which exhibited thermosensitive genic male sterility, from the edited offspring. Our investigation then focused on determining the sterility-inducing temperature of Huahang 48s, quantifying the content of the fragrance substance 2-AP in various tissues, and identifying its application potential and value, which provided research ideas and laid the groundwork for the development of a novel TGMS line germplasm with fragrance.
2. Materials and Methods
2.1. Experimental Materials
The indica rice variety Huahang 48 was used as the experimental material in this study. Huahang 48 is a high-quality conventional rice variety that successfully underwent validation in Guangdong Province in 2016. Since its validation, it has been extensively utilized in production and has become the principal cultivar in Guangdong Province.
2.2. Construction of Cas9 Expression Vector
In this study, the Cas9 expression vector system of pRGEB32 was used for gene editing [22]. The corresponding fragments were amplified from the pGTR vector using the designed target primers Cas9-tms5-F/R and Cas9-fgr-F/R (Table 1). The primers contained the homologous sequences recomb-F and recomb-R, respectively. Subsequently, the PTG fragment (sgRNA containing the target gene) was created through the enzymatic ligation of the target fragment using Golden Gate assembly (GG assembly). The Cas9 expression vector pRGEB32 was enzymatically cleaved with BsaI endonuclease, and the PTG fragment was recombinantly cloned into the enzymatically cleaved Cas9 expression vector using homologous recombination ligase. Following this, E. coli DH5α was transformed, and a single colony was selected and agitated to extract the plasmid. The accurate recombinant vector was identified through Sanger sequencing. The bacterial broth and plasmid were preserved for subsequent genetic transformation experiments.

Table 1.
Primers for vector construction and transgenic detection.
2.3. Agrobacterium-Mediated Genetic Transformation
The plasmids were introduced into the Agrobacterium tumefaciens EH105 strain via transformation. The transformed Agrobacterium tumefaciens strain EH105, carrying the desired genetic construct, was utilized to infect the callus tissue of the rice variety Huahang 48. The infection process involved co-cultivating the callus with the Agrobacterium suspension for a specified period under controlled conditions to facilitate the transfer of the T-DNA region into the plant cells. After the co-cultivation phase, the callus was thoroughly washed to remove any residual Agrobacterium and then transferred to a selection medium supplemented with hygromycin, an antibiotic used to select for successfully transformed cells. The hygromycin-resistant callus tissues were carefully monitored and periodically subcultured onto fresh selection medium to ensure the continued growth of transformed cells. The calli were transferred to a regeneration medium to induce the formation of shoots and roots, ultimately leading to the development of the T0 generation plants [23]. DNA was extracted from the leaves of the T0 generation plants by the cetyltrimethylammonium bromide (CTAB) method [24] and tested with hygromycin-specific primers hyg-F/R (Table 1). The hygromycin-positive transgenic plants were retained for subsequent research.
2.4. Mutation Detection at Target Loci of Transgenic Plants
To identify mutations in the TMS5 and FGR in transgenic plants, gene-specific primers (tms5-F/tms5-R and fgr-F/fgr-R) were designed using Primer 5.0 software (Table 1), optimizing parameters such as length (18–22 bp), GC content (40–60%), and Tm (55–65 °C). PCR amplification was performed in a 50 µL reaction containing 50–100 ng template DNA, 0.2 µM primers, 200 µM dNTPs, 1 × PCR buffer, 1.5 mM MgCl2, and 1 U Taq DNA polymerase. The cycling conditions included initial denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 57 °C for 30 s, and 72 °C for 1 min, with a final extension at 72 °C for 10 min. Amplified products were verified on a 1.5% agarose gel and purified. The purified PCR products were then subjected to Sanger sequencing. The mutation genotypes of the transgenic plants were analyzed using Degenerate Sequence Decode (DSDecode, http://skl.scau.edu.cn/dsdecode/, accessed on 25 September 2020) [25].
2.5. Off-Target Site Analysis
Potential off-target sites were predicted using the CRISPR-GE online tool (http://skl.scau.edu.cn/, accessed on 20 January 2020) [26]. Genomic DNA from T0 generation plants was extracted via the CTAB method, and predicted off-target regions were amplified by PCR and sequenced using Sanger sequencing. Off-target mutations were distinguished from natural polymorphisms by comparison with wild-type controls.
2.6. Acquisition of T-DNA Element-Free Mutants
Seeds from the self-crosses of T0 generation plants were grown into seedlings. DNA was extracted from the young leaves of each plant using the CTAB method [24]. PCR was conducted using the hygromycin-specific primer Hyg-F/R and the Cas9 gene-specific primer Cas9-F/R (Table 1) to screen for T1 generation mutants without T-DNA elements. These selected lines were further propagated through self-crossing into the T2 generation. All rice populations were planted in the experimental field of South China Agricultural University (Guangzhou, N 23.16°, E 113.36°) with a planting density of 20 cm × 20 cm. Water and fertilizer were managed regularly.
2.7. Analysis of Temperature-Sensitive Breeding Conversion of Pure Mutants Without T-DNA Elements
Huahang 48s and Huahang 48 were planted in an artificial climate box in the early season of 2019. The light conditions were set at 12 h light and 12 h dark at 30 °C. As the young spikelets of the main stem of Huahang 48s reached the fifth to sixth developmental stage, the temperature was adjusted to 24 °C, 23 °C, and 22 °C, respectively. After 14 days of treatment at each temperature, the temperature was adjusted to 30 °C. The pollen fertility of the spikelet was observed by microscopy after staining with 1% I2-KI solution [27].
2.8. Detection of Fragrance Content of Pure Mutants Without T-DNA Elements
The young spikelets (50 g), stems (100 g), and leaves (100 g) of Huahang 48s and Huahang 48 were pulverized using liquid nitrogen. The 2-AP content was extracted using simultaneous distillation extraction (SDE) and subsequently quantified by GCMS-QP 2010 Plus [28].
2.9. Acquisition and Planting of Huahang 48s Hybrid
TGMS line Huahang 48s was crossed with two indica restorer lines, YXZ and SBSM. To prevent unintended pollination, the panicles were carefully covered with paper bags. After 25 days of grain maturation, the F1 seeds were harvested. The resulting F1 hybrids were subsequently cultivated in field plots under controlled conditions, with each plot consisting of six rows, each containing six plants, and with a planting density of 20 cm × 20 cm.
3. Results
3.1. Identification of T0 Generation Plants
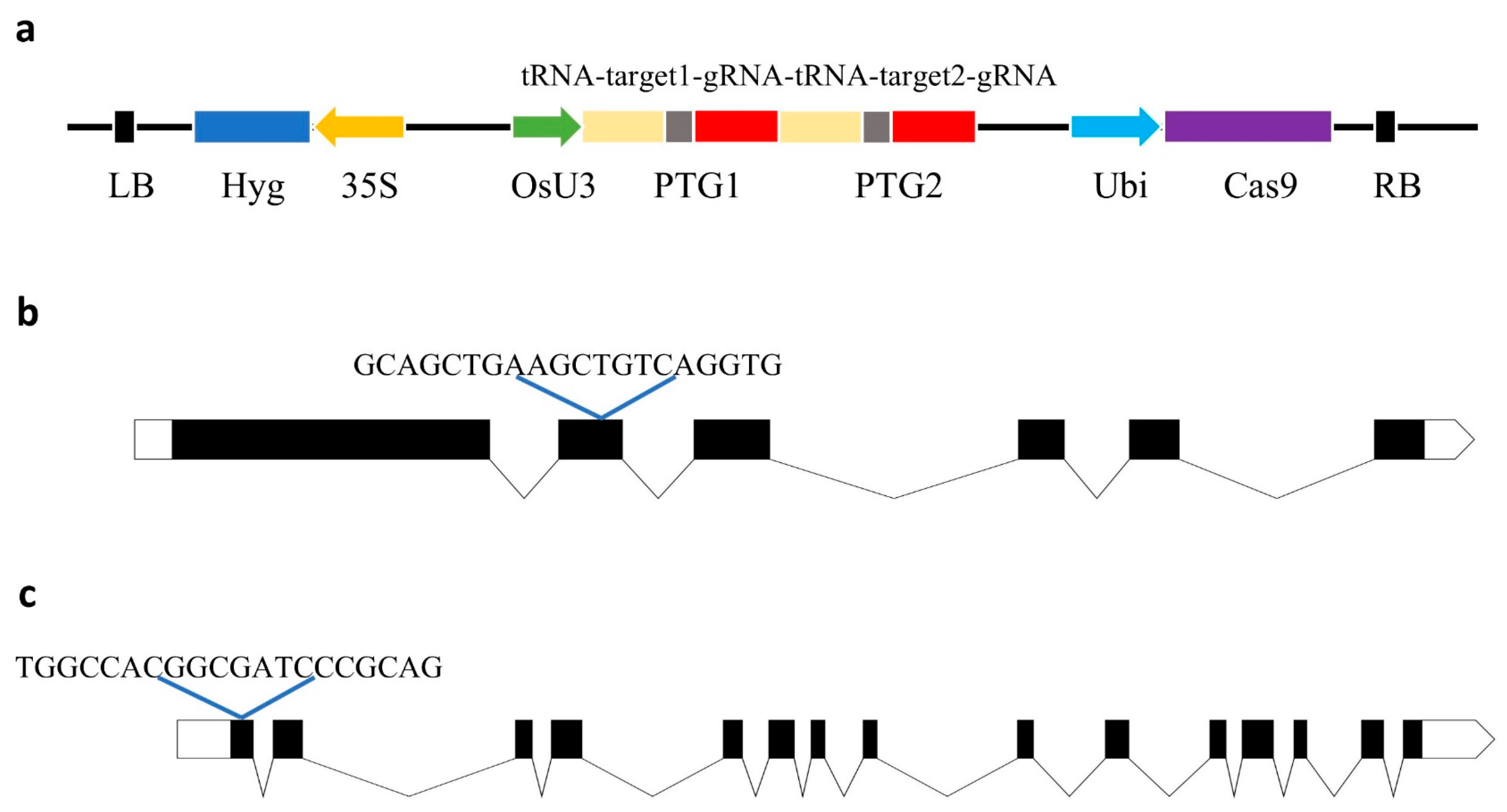
The functions of TMS5 and FGR have been extensively studied and well characterized, and their transgenic materials exhibit stable phenotypic traits [10,29]. To develop a two-line sterile line of Huahang 48s with fragrance, sgRNAs were designed using the CRISPR-GE online tool, with one sgRNA targeting the second exon of TMS5 and another sgRNA targeting the first exon of FGR (Figure 1b,c). A CRISPR/Cas9 vector incorporating these two target-specific sgRNAs was subsequently constructed (Figure 1a). Through Agrobacterium-mediated genetic transformation, we successfully obtained twenty-one T0 generation plants, as confirmed by hygromycin-specific primer (Hyg-F/R) assays. Finally, sixteen plants were identified as positive transgenic lines.
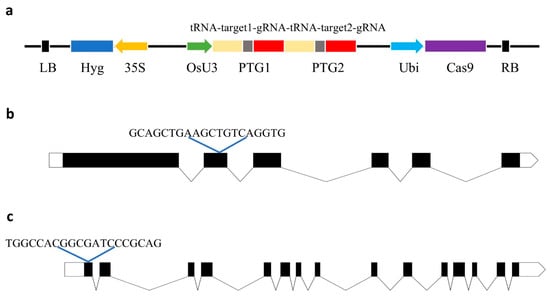
Figure 1.
Schematic diagram of the CRISPR/Cas9-TMS5-FGR vector construction. (a) Schematic diagram of the pRGEB32-Cas9-TMS5-FGR construct; (b) schematic diagram of the location of targeted sites in TMS5; and (c) schematic diagram of the location of targeted sites in FGR.
3.2. Analysis of Target Site Sequencing Results of T0 Generation Plants
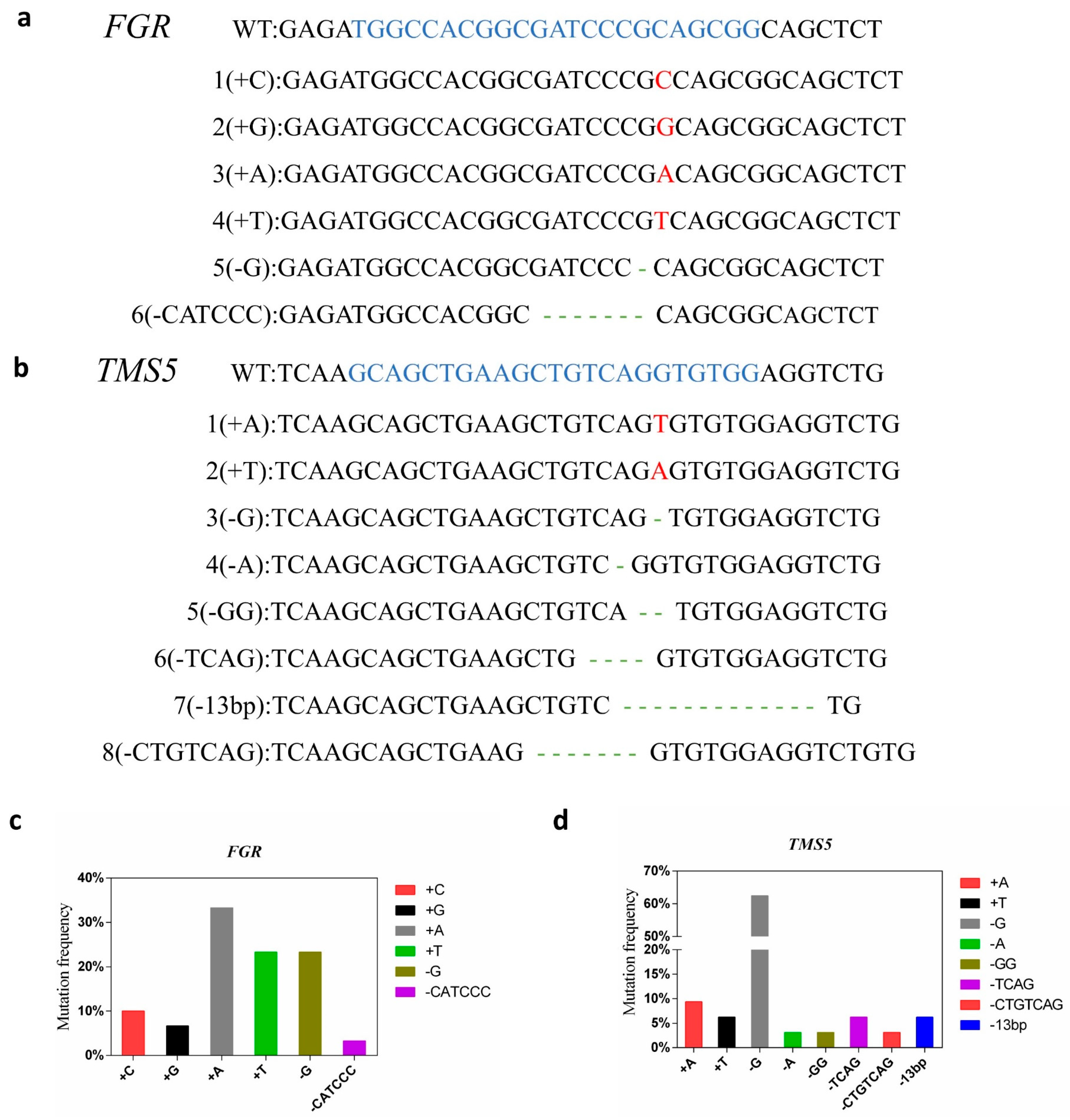
The positive plants were subjected to PCR amplification and sequencing using the TMS5 and FGR target site detection primers tms5-F/R and fgr-F/R, respectively. Analysis via the DSDecode revealed that fifteen positive plants had mutations in fgr, containing eight deletion mutations (73.33%) and twenty-two insertion mutations (26.67%). All sixteen positive plants had mutations in tms5, including twenty-eight deletion mutations (87.50%) and four insertion mutations (12.50%). A total of fifteen double knockout plants and one single knockout plant were identified (Table 2). Among these, fifteen fgr mutants exhibited double allelic mutations, and ten tms5 mutants also displayed double allelic mutations, while the remaining six mutants showed pure mutations. The mutations induced by the Cas9 technology predominantly resulted in double allelic mutations, with six different genotypes identified in fgr and eight in tms5 (Figure 2a,b). Mutation analysis revealed that A-base insertions and G-base deletions were the most common mutation types in fgr and tms5 mutants, respectively (Figure 2c,d). These results underscore the high efficiency of the CRISPR/Cas9 system in editing multiple genes simultaneously, primarily resulting in single-base mutations.

Table 2.
Mutation detection of T0 plants.
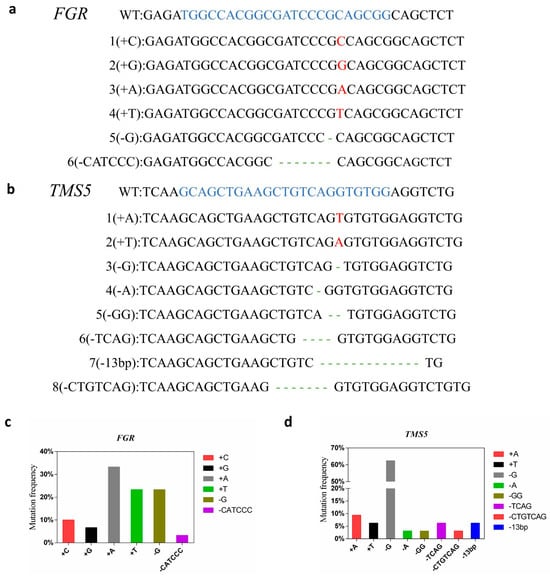
Figure 2.
Analysis of TMS5 and FGR mutation types. (a) The mutant genotypes of FGR; (b) the mutant genotypes of TMS5. The blue letters are the target sequences (including the PAM site), the red letters are insert bases, and the dashes show deletions. (c) Frequency of the mutation types of FGR; (d) frequency of the mutation types of TMS5.
3.3. Detection and Analysis of Off-Target Sites
Off-target effects may arise during gene editing procedures utilizing Cas9 technology [30]. Therefore, CRISPR-GE [26] was used to predict the potential off-target sites of the targets. Sites with higher scores were selected as potential off-target locations (Table 3). Specific primers were then designed for PCR amplification and sequencing to analyze the off-target effects in the T0 generation plants. Notably, no off-target effects were detected in the T0 generation plants.

Table 3.
Mutation detections in the putative off-target site.
3.4. Identification of T-DNA-Free T1 Generation Plants
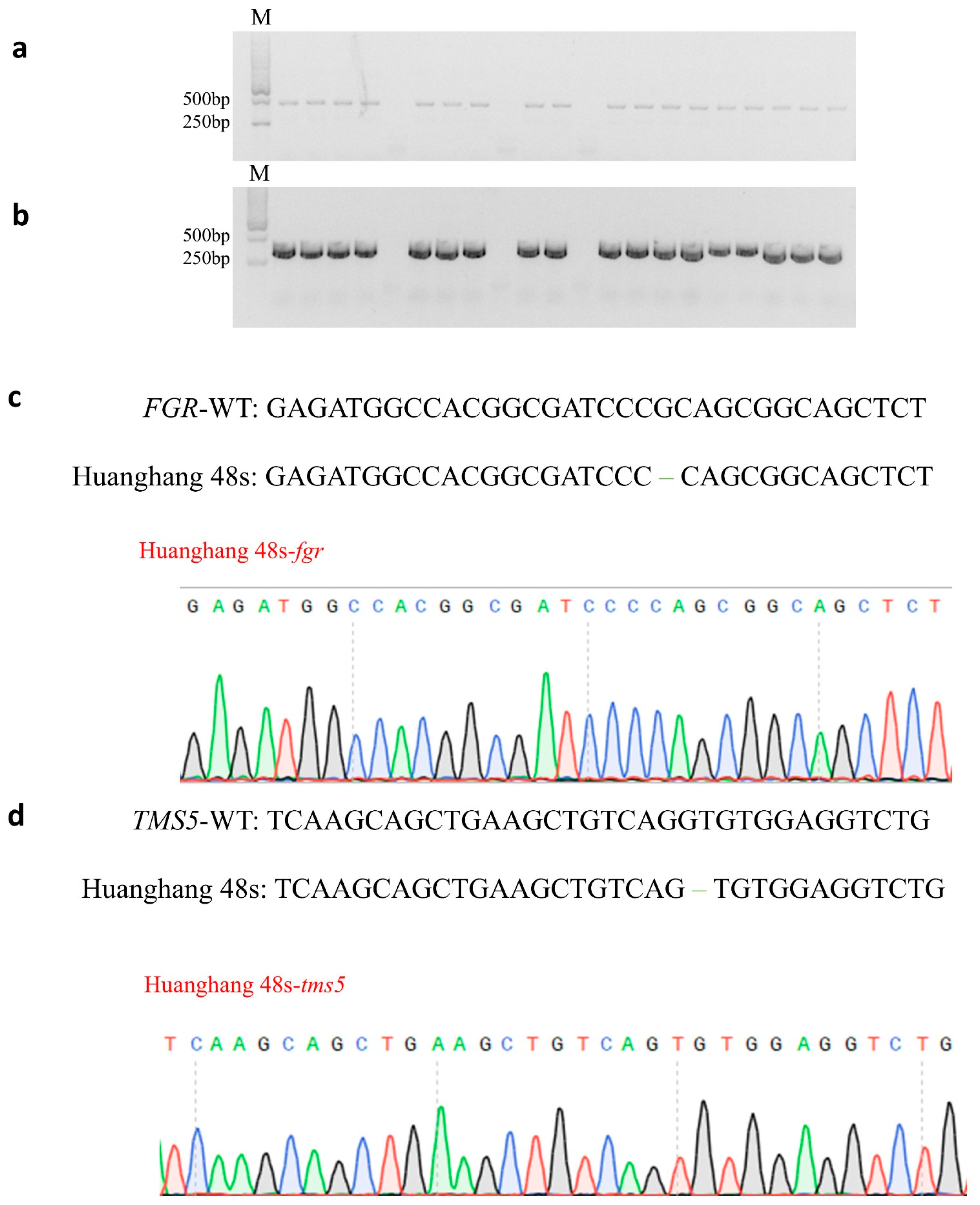
The seeds obtained from T0 generation double knockout plants were cultivated to produce T1 generation plants. In order to further obtain transgenic plants without T-DNA elements, twenty-one T1 generation plants were detected using the primers Hyg-F/R for hygromycin-specific detection and Cas9-F/R for Cas9 protein-specific detection. Most of the detected T1 generation plants carried hygromycin and Cas9 protein elements. Finally, three double mutant plants without T-DNA elements were obtained (Figure 3a,b). Three T-DNA-Free T1 generation plants were subjected to low temperature treatment at the booting stage to obtain seeds.
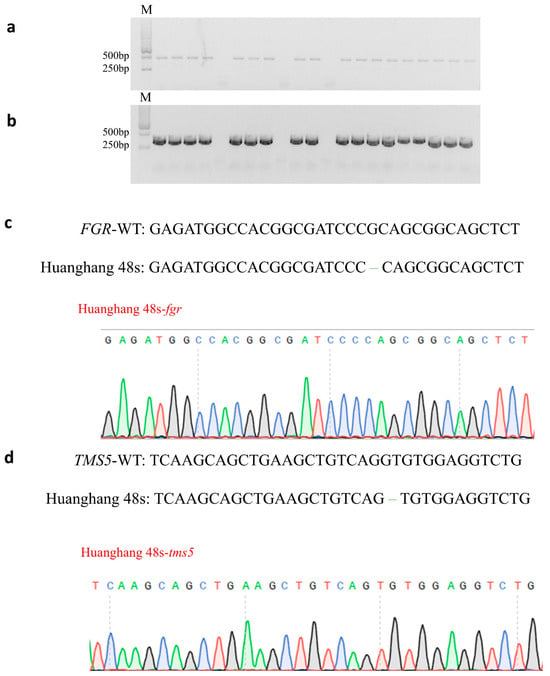
Figure 3.
Genotyping of homozygous mutants without T-DNA elements. (a) PCR detection of Cas9 in T1 generation mutants; (b) PCR detection of hygromycin in T1 generation mutants with M. DL2000 DNA markers; (c) mutation types of FGR in Huahang 48s; and (d) mutation types of TMS5 in Huahang 48s.
3.5. Fertility Identification and Fragrance Substance Detection of Pure Mutant Plants of the T2 Generation
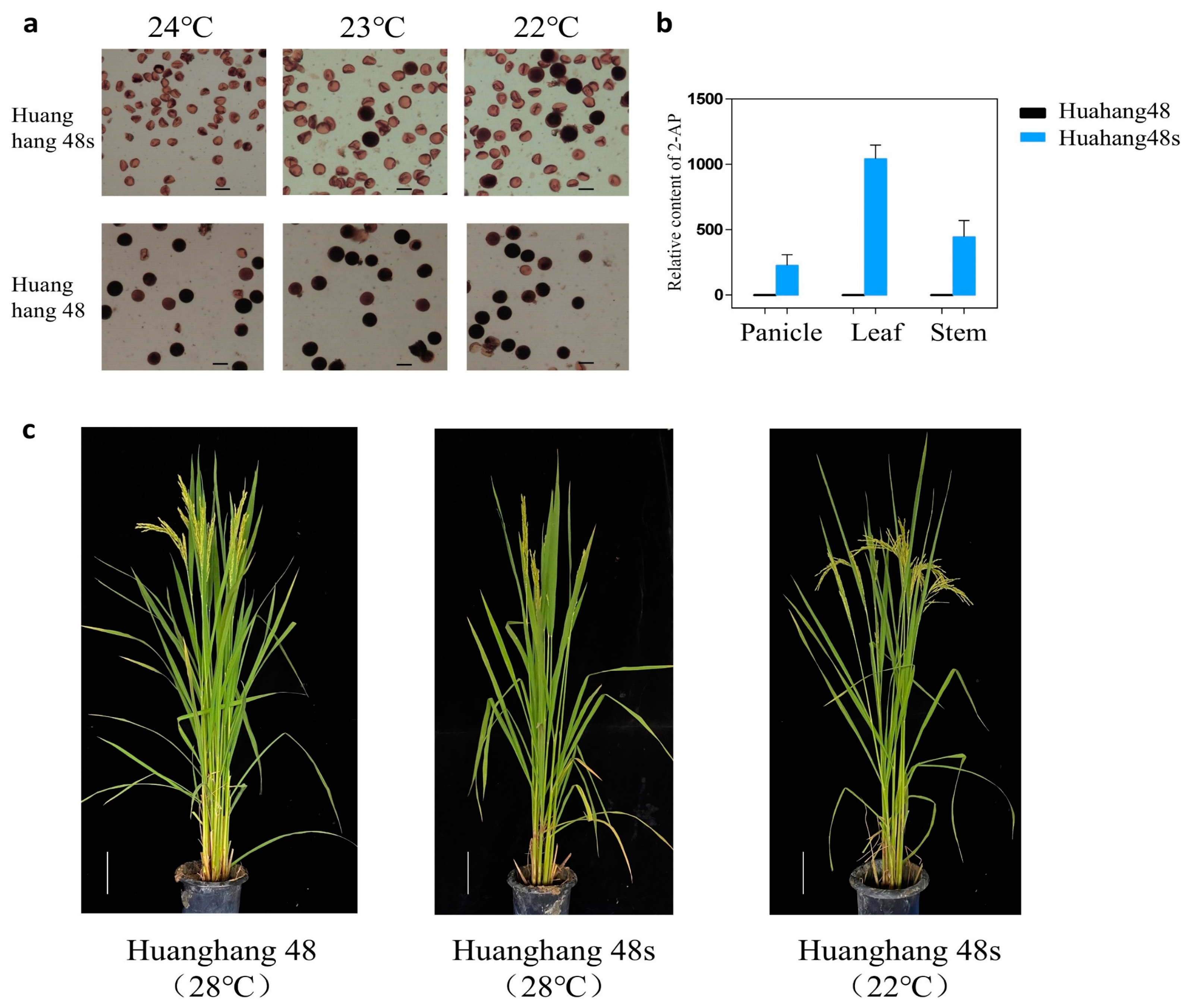
The T2 generation double gene mutant plants without T-DNA elements were detected using the TMS5 target detection primer tms5-F/R and the FGR target detection primer fgr-F/R. The sanger sequencing results were compared with the wild-type Huahang 48. Ultimately, five homozygous double gene mutant plants were identified without T-DNA elements (Figure 3c,d), and the genotypic strain was designated as Huahang 48s. The pollen of Huahang 48s exhibited complete sterility at 24 °C, partial fertility at 23 °C, and further partial fertility at 22 °C (Figure 4a,c). Due to the low fruiting rate of the sterile lines and the limited number of pure mutant plants, the young spikelets, leaves, and stems of the T2 generation pure mutant plants were selected for fragrance substance analysis, with the wild-type Huahang 48 serving as the control. The results demonstrated the absence of 2-AP content in the young spikelets, leaves, and stems of the wild-type plants, whereas 2-AP was detected in the corresponding tissues of the Huahang 48s, with concentrations of 327.27, 1143.05, and 545.37 ng–g−1, respectively, with the highest level present in the leaves (Figure 4b).
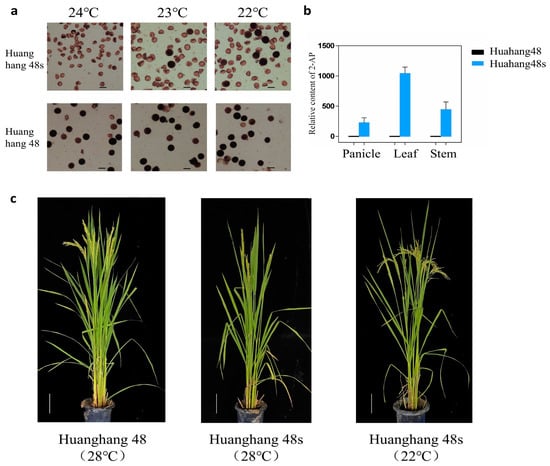
Figure 4.
Identification of pollen fertility and detection of 2-AP content. (a) Pollen fertility of T2 generation mutants under different temperatures; (b) relative content of 2-AP in wild-type and mutants; and (c) photograph of Huahang 48 and Huahang 48s.
3.6. Application of Huahang 48s in Hybrid Rice Breeding
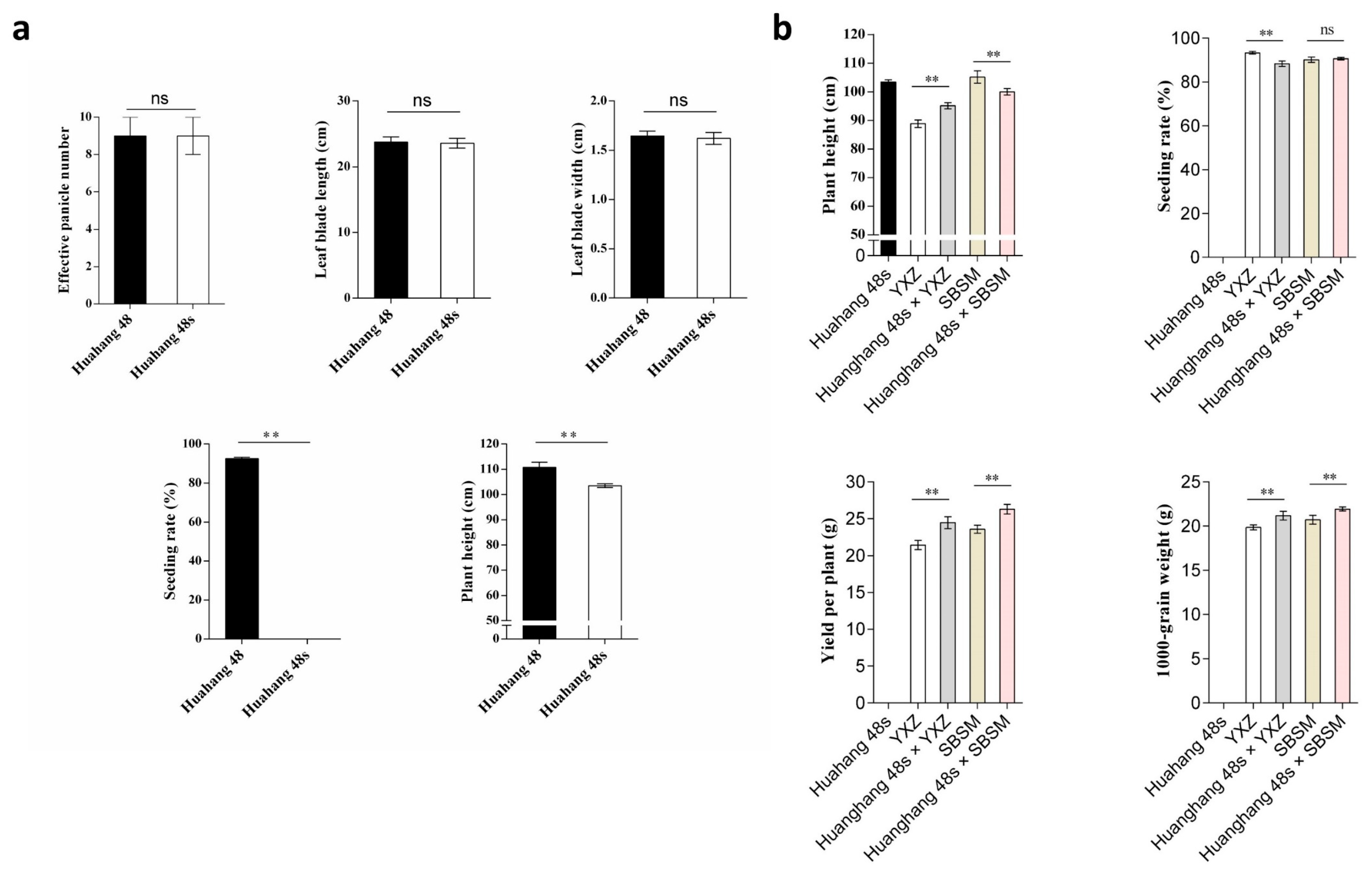
The main agronomic traits of Huahang 48s and Huahang 48 were determined. The results showed that there was no significant difference in the effective panicle number, leaf blade length, and leaf blade width, and the plant height of Huahang 48s decreased significantly (Figure 5a). To test the combinatorial capacity of Huahang 48s, we crossed it with YZX and SBSM. The hybrids of Huahang 48s/YXZ and Huahang 48s/SBSM were significantly higher than the parents in terms of yield per plant and 1000-grain weight (Figure 5b), and had excellent heterosis effects, indicating that Huahang 48s was a TGMS line with application value.
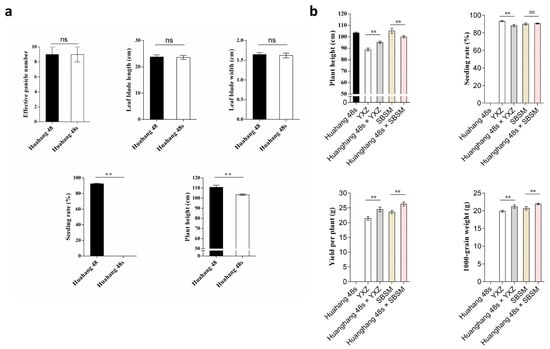
Figure 5.
Agronomic traits of Huahang 48, Huahang 48s, YXZ, SBSM, and hybrids. (a) Agronomic trait of Huahang 48 and Huahang 48s; and (b) agronomic traits of hybrids and their parents. ** p < 0.01; ns, no significance (Student’s t-test).
4. Discussion
CRISPR/Cas9 technology has emerged as the predominant method for gene editing due to its straightforward operation, high specificity, and low off-target rate [31]. The insertion of T-DNA may be lost through self-cross separation. Consequently, pure mutants lacking the T-DNA insertion fragment can be obtained in selfing progeny. This capability significantly reduces the breeding cycle duration, making CRISPR/Cas9 technology a popular choice in enhancing breeding programs for a wide range of crops [16]. CRISPR/Cas9 technology has been successfully applied to the improvement of various crop traits. Compared with traditional breeding, its efficient and simple characteristics have great application potential and value [32].
The TMS5 encodes an RNA enzyme, and its loss of function results in thermosensitive male sterility [10]. The FGR encodes betaine aldehyde dehydrogenase, and its loss of function leads to the accumulation of 2-AP, the key compound responsible for the characteristic fragrance in rice [33]. The targeted editing of the TMS5 gene successfully induced thermosensitive male sterility, while the editing of the FGR gene effectively introduced desirable aroma traits, thereby enhancing both the quality and market value of the rice. This study utilized CRISPR/Cas9 technology to edit the TMS5 and FGR in the high-quality indica rice variety, Huahang 48. The results showed that 37.5% of pure mutations occurred at the tms5 target site in the T0 generation plants (Table 2). Both target sites of the fgr exhibited double allelic mutations (Table 2), suggesting that the selection of the target site influenced the mutation type. The majority of mutations for both targets were situated three to four bases before the PAM site (Figure 2), primarily involving single-base insertions or deletions. The researchers used the CRISPR/Cas9 gene editing technology to knock out the TMS5 of multiple rice varieties [34]. These sterile lines showed different sterility-inducing temperatures due to differences in the genetic background. Among them, the sterility-inducing temperature of most japonica rice materials is higher, at about 26 °C, while the starting temperature of sterility of indica rice materials is about 24 °C. Indica rice exhibits a lower sterility-inducing temperature than japonica rice in TGMS lines, with the transfer temperature occurring at or below 24 °C [10,34]. This study investigated sterility-inducing temperature in the TGMS line Huahang 48s of fragrant rice, revealing a fertility transition at 23 °C and sterility at 24 °C (Figure 4a,c). The sterility-inducing temperature of Huahang 48s was identified as 23 °C. The sterility-inducing temperature of the TGMS line is an important factor to determine the safety of two-line hybrid rice seed production. Breeding the TGMS line with a sterility-inducing temperature of sterility can significantly improve the purity of two-line hybrid rice seed production.
Furthermore, the deletion of a single base in the FGR of Huahang 48s resulted in a shift code mutation, leading to increased 2-AP content in the young spikelets, leaves, and stems of Huahang 48s for the fragrance substance 2-AP (Figure 4b). Notably, the highest 2-AP content was detected in leaves, possibly due to the stronger expression of genes related to 2-AP synthesis in leaves.
With the change of the global climate, the safety of grain production has gained more and more attention [35]. Rice is the most important food crop. More than half of the world’s people live on rice, especially in Asian countries. Improving the yield of rice is significant to ensuring food security. Hybrid rice, yielding approximately 20% more than conventional rice varieties, significantly addresses global food shortages and ensures food security [36]. The quality of sterile lines, serving as parents in hybrid rice production, is crucial for the overall quality of the hybrid rice [14]. Furthermore, as people’s living standards continue to improve, fragrant rice is more popular in the market. Developing high-quality sterile lines with fragrance is pivotal for enhancing the desirability and market value of hybrid rice [29,33]. Using Huahang 48s crossed with YXZ and SBSM, the hybrid showed that the yield per plant and 1000-grain weight were better than those of the parents, indicating that Huahang 48s had a certain combining ability in heterosis utilization (Figure 5b).
5. Conclusions
This study utilized CRISPR/Cas9 technology to enhance the high-quality conventional rice variety Huahang 48, transforming it into the high-quality, fragrant, TGMS line Huahang 48s. Huahang 48 shows good compatibility in the application of two-line hybrid rice. Our findings demonstrate the feasibility and efficiency of utilizing CRISPR/Cas9 technology to simultaneously edit TMS5 and FGR loci for converting conventional rice varieties into superior male sterile lines. This genome editing-based approach significantly shortens the breeding cycle by several years compared to conventional breeding methods.
Author Contributions
Conceptualization, W.X., Z.C., X.H. and H.W.; Methodology, W.X., Z.C. and H.W.; Software, T.C. and Z.L.; Formal analysis, T.C., N.P., M.N., H.X., Z.Z. and J.H.; Investigation, H.X., Z.Z. and J.H.; Data curation, N.P., M.N., H.X., Z.Z. and J.H.; Writing—original draft, T.C. and N.P.; Writing—review & editing, T.C., X.H. and H.W.; Visualization, M.N.; Supervision, Z.L., W.X., Z.C., X.H. and H.W.; Funding acquisition, T.C., Z.L. and X.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Science and Technology Talent Introduction Project of Guangdong Academy of Agricultural Sciences (R2024YJ-YB3001), the Guangdong Academy of Agricultural Sciences Young and Middle-aged Discipline Leader (Jinying Star) Training Program (R2023PY-JX003), and the Key Laboratory of Rice Science and Technology of Guangdong Province (2023B1212060042).
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| TGMS | Thermosensitive genic male sterile |
| GG assembly | Golden Gate assembly |
| SDE | simultaneous distillation extraction |
References
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef]
- Yuan, L.P. Development of hybrid rice to ensure food security. Rice Sci. 2014, 21, 1. [Google Scholar] [CrossRef]
- Zhou, Y.B.; Zhao, X.H.; Tang, X.D.; Zhou, Z.W.; Zhuang, C.X.; Yang, Y.Z. Acquisition of Mutants of the Reverse Photoperiod-sensitive Genic Male Sterility Gene csa in Rice Based on CRISPR/Cas9 Technology. Hybrid Rice 2018, 33, 64–70. [Google Scholar]
- Fang, Y.; Yang, J.; Guo, X.; Qin, Y.; Zhou, H.; Liao, S.; Liu, F.; Qin, B.; Zhuang, C.; Li, R. CRISPR/Cas9-Induced Mutagenesis of TMS5 Confers Thermosensitive Genic Male Sterility by Influencing Protein Expression in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2022, 23, 8354. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Cao, X.; Zhang, Q. Progress on photoperiod thermo-sensitive genic male sterile rice. Chin. Sci. Bull. 2016, 61, 3822–3832. [Google Scholar] [CrossRef]
- Yang, Q.; Liang, C.; Zhuang, W.; Li, J.; Deng, H.; Deng, Q.; Wang, B. Characterization and identification of the candidate gene of rice thermo-sensitive genic male sterile gene tms5 by mapping. Planta 2007, 225, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Wang, L.; Wang, J.; Zeng, Y.; Xu, Y.; Li, S. The transcription factor OsbHLH138 regulates thermosensitive genic male sterility in rice via activation of TMS5. Theor. Appl. Genet. 2019, 132, 1721–1732. [Google Scholar] [CrossRef]
- Wang, Y.G.; Xing, Q.H.; Deng, Q.Y.; Liang, F.S.; Yuan, L.P.; Weng, M.L.; Wang, B. Fine mapping of the rice thermo-sensitive genic male-sterile gene TMS5. Theor. Appl. Genet. 2003, 107, 917–921. [Google Scholar] [CrossRef]
- Jiang, D.; Lu, S.; Zhou, H.; Wu, X.; Zhuang, C.; Liu, Y.; Mei, M. Mapping of the rice (Oryza sativa L.) thermo-sensitive genic male sterile gene tms5 with EST and SSR markers. Chin. Sci. Bull. 2006, 51, 148–151. [Google Scholar] [CrossRef]
- Zhou, H.; Zhou, M.; Yang, Y.; Li, J.; Zhu, L.; Jiang, D.; Dong, J.; Liu, Q.; Gu, L.; Zhou, L. RNase ZS1 processes Ub L40 mRNAs and controls thermosensitive genic male sterility in rice. Nat. Commun. 2014, 5, 4884. [Google Scholar] [CrossRef]
- Sakthivel, K.; Sundaram, R.M.; Rani, N.S.; Balachandran, S.M.; Neeraja, C.N. Genetic and molecular basis of fragrance in rice. Biotechnol. Adv. 2009, 27, 468–473. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, L.M.T.; Fitzgerald, T.L.; Henry, R.J.; Jin, Q.; Waters, D.L.E. The gene for fragrance in rice. Plant Biotechnol. J. 2005, 3, 363–370. [Google Scholar] [CrossRef]
- Kovach, M.J.; Calingacion, M.N.; Fitzgerald, M.A.; McCouch, S.R. The origin and evolution of fragrance in rice (Oryza sativa L.). Proc. Natl. Acad. Sci. USA 2009, 106, 14444–14449. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wu, J.; Deng, X.W.; Tang, X. Establishment and Advances of Third-Generation Hybrid Rice Technology: A Review. Rice 2023, 16, 56. [Google Scholar] [CrossRef] [PubMed]
- Samanta, M.K.; Dey, A.; Gayen, S. CRISPR/Cas9: An advanced tool for editing plant genomes. Transgenic Res. 2016, 25, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Patron, N.J.; Nekrasov, V. Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 2015, 32, 76–84. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, R.; Huang, G.; Li, Y.; Melaku, G.; Zhang, S.; Chen, H.; Zhao, Y.; Zhang, J.; Zhang, Y. Developing superior alleles of yield genes in rice by artificial mutagenesis using the CRISPR/Cas9 system. Crop J. 2018, 6, 475–481. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genom. 2014, 41, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhang, B.; Ding, W.; Liu, X.; Yang, D.-L.; Wei, P.; Cao, F.; Zhu, S.; Zhang, F.; Mao, Y. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013, 23, 1229–1232. [Google Scholar] [CrossRef]
- Xu, R.F.; Li, H.; Qin, R.Y.; Li, J.; Qiu, C.H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 2015, 5, 11491. [Google Scholar] [CrossRef]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar] [CrossRef] [PubMed]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef]
- Stewart, C.N.J.; Via, L.E. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 1993, 14, 748–750. [Google Scholar]
- Liu, W.; Xie, X.; Ma, X.; Li, J.; Chen, J.; Liu, Y.G. DSDecode: A web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol. Plant 2015, 8, 1431–1433. [Google Scholar] [CrossRef]
- Xie, X.; Ma, X.; Zhu, Q.; Zeng, D.; Li, G.; Liu, Y.G. CRISPR-GE: A convenient software toolkit for CRISPR-based genome editing. Mol. Plant 2017, 10, 1246–1249. [Google Scholar] [CrossRef]
- Barman, H.N.; Sheng, Z.; Fiaz, S.; Zhong, M.; Wu, Y.; Cai, Y.; Wang, W.; Jiao, G.; Tang, S.; Wei, X.; et al. Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system. BMC Plant Biol. 2019, 19, 109. [Google Scholar] [CrossRef]
- Ashokkumar, S.; Jaganathan, D.; Ramanathan, V.; Rahman, H.; Palaniswamy, R.; Kambale, R.; Muthurajan, R. Creation of novel alleles of fragrance gene OsBADH2 in rice through CRISPR/Cas9 mediated gene editing. PLoS ONE 2020, 15, e0237018. [Google Scholar] [CrossRef]
- Hui, S.; Li, H.; Mawia, A.M.; Zhou, L.; Cai, J.; Ahmad, S.; Lai, C.; Wang, J.; Jiao, G.; Xie, L.; et al. Production of aromatic three-line hybrid rice using novel alleles of BADH2. Plant Biotechnol. J. 2022, 20, 59–74. [Google Scholar] [CrossRef]
- Lawrenson, T.; Shorinola, O.; Stacey, N.; Li, C.; Østergaard, L.; Patron, N.; Uauy, C.; Harwood, W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015, 16, 1–13. [Google Scholar] [CrossRef]
- Romero, F.M.; Gatica-Arias, A. CRISPR/Cas9: Development and Application in Rice Breeding. Rice Sci. 2019, 26, 265–281. [Google Scholar] [CrossRef]
- Fiaz, S.; Ahmad, S.; Noor, M.A.; Wang, X.; Younas, A.; Riaz, A.; Riaz, A.; Ali, F. Applications of the CRISPR/Cas9 System for Rice Grain Quality Improvement: Perspectives and Opportunities. Int. J. Mol. Sci. 2019, 20, 888. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Zhang, Y.; Chen, K.; Zhang, K.; Gao, C. Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol. J. 2015, 13, 791–800. [Google Scholar] [CrossRef]
- Zhou, H.; He, M.; Li, J.; Chen, L.; Huang, Z.; Zheng, S.; Zhu, L.; Ni, E.; Jiang, D.; Zhao, B. Development of commercial thermo-sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated TMS5 editing system. Sci. Rep. 2016, 6, 37395. [Google Scholar] [CrossRef]
- Khush, G.S. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 2005, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Gaballah, M.M.; Attia, K.A.; Ghoneim, A.M.; Khan, N.; EL-Ezz, A.F.; Yang, B.; Xiao, L.; Ibrahim, E.I.; Al-Doss, A.A. Assessment of Genetic Parameters and Gene Action Associated with Heterosis for Enhancing Yield Characters in Novel Hybrid Rice Parental Lines. Plants 2022, 11, 266. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).