MPG-YOLO: Enoki Mushroom Precision Grasping with Segmentation and Pulse Mapping
Abstract
1. Introduction
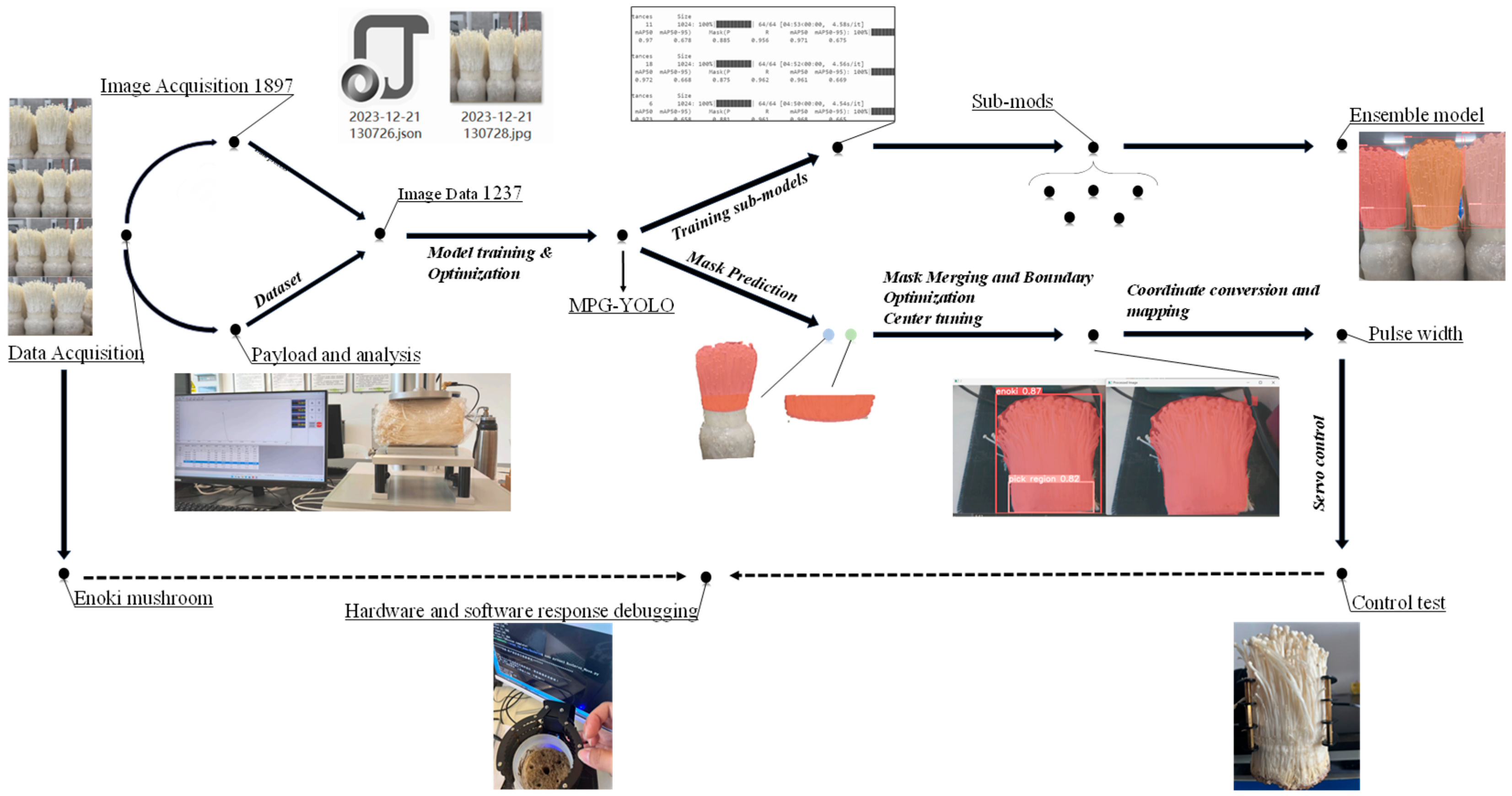
2. Materials and Methods
2.1. Plant Material
2.2. Image Data Acquisition
2.3. Production of Dataset
2.3.1. Analysis of the Optimal Stress Position of the Enoki Mushrooms
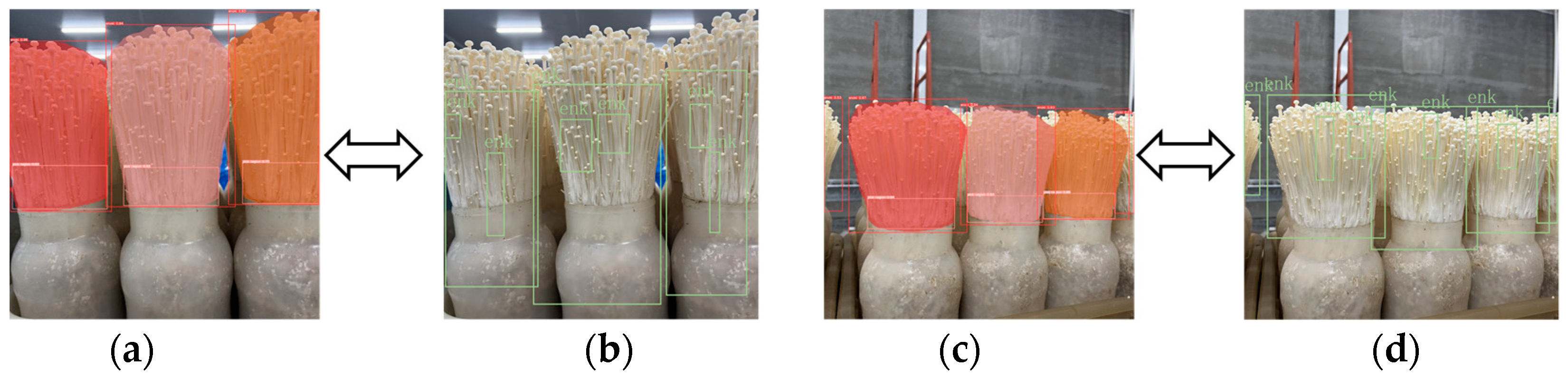
2.3.2. Labeled Segmentation and Analysis of Datasets
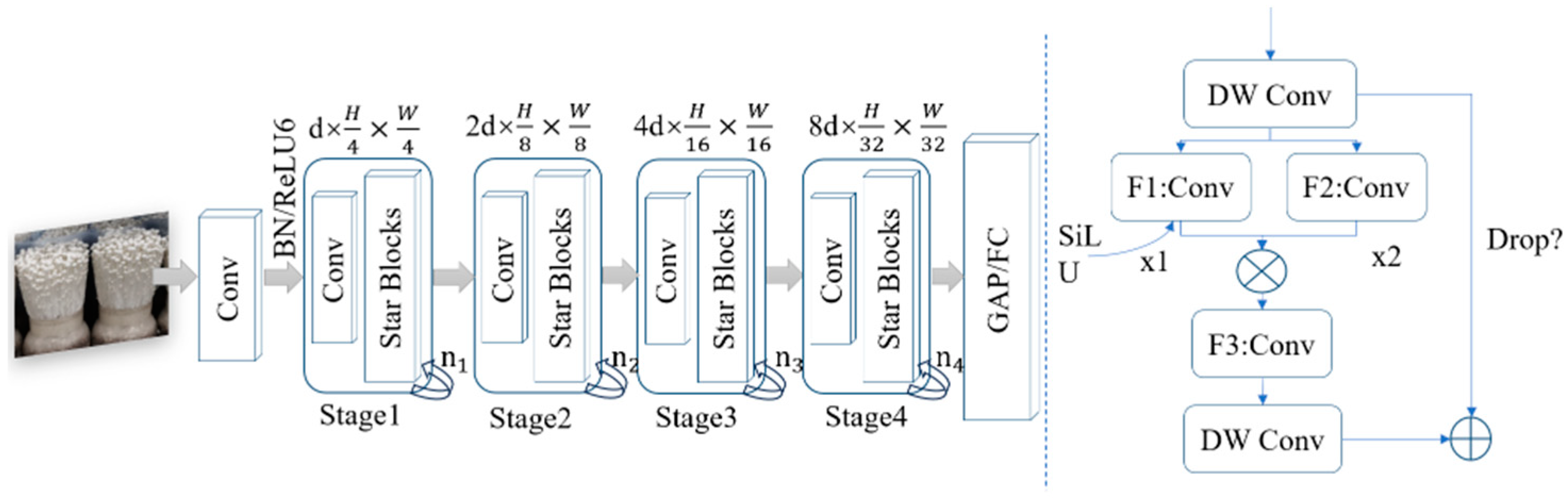
2.4. MPG-YOLOv8 Segmentation Model of Bottle-Planted Enoki Mushrooms
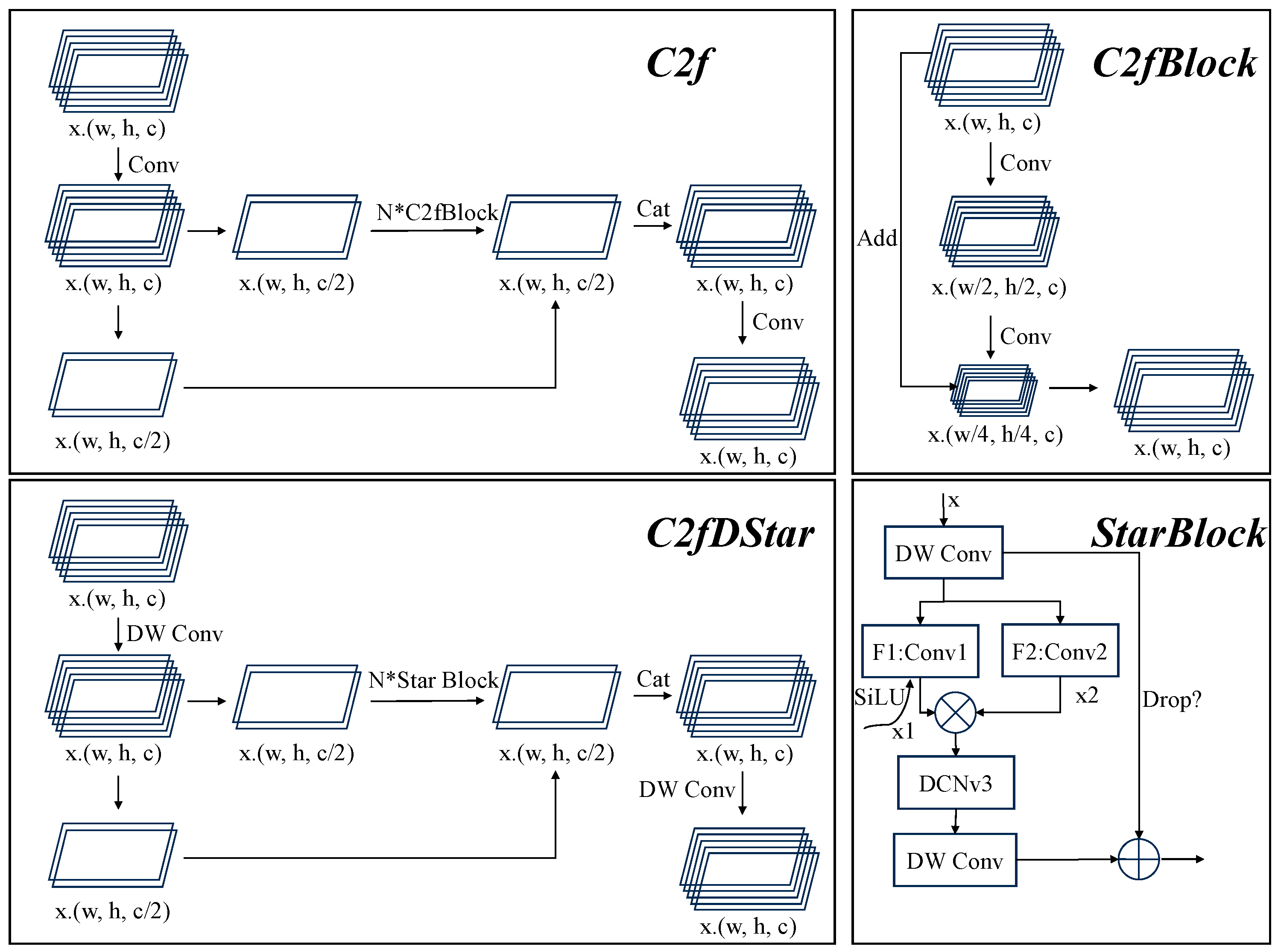
2.4.1. StarNet
2.4.2. SPPECAN
2.4.3. C2fDStar
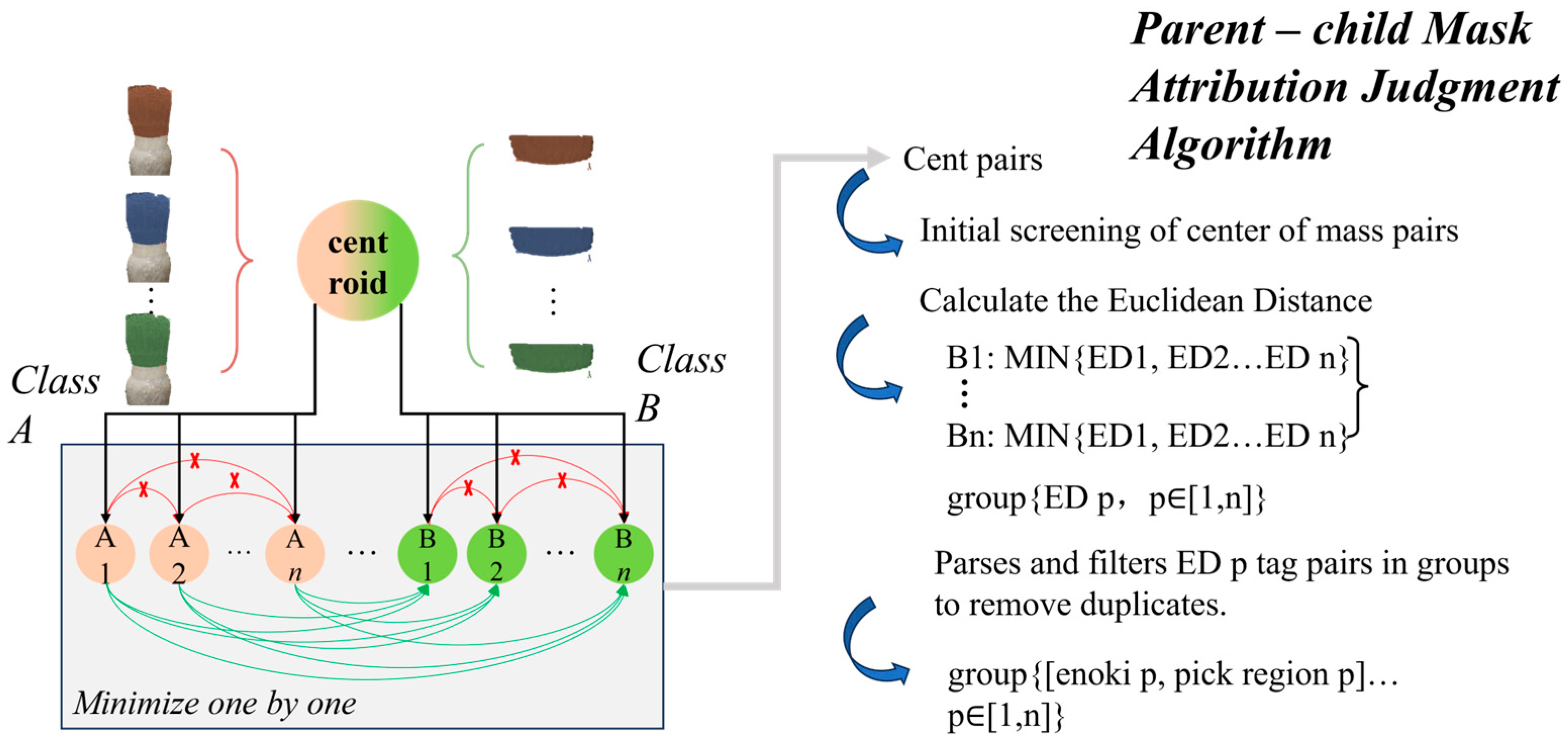
2.5. Mask Relation Determination and Merger Optimization Based on Euclidean Distance
2.5.1. Modeling Parent–Child Relationship
| Algorithm 1. Generate label pairs based on region distribution and center of mass distance |
| Input: enoki_tags: Set of “enoki” tags pick_region_tags: Set of “pick region” tags Output: group: Set of label pairs {enoki_tag, pick_region_tag} 1. Initialize group ← ∅ 2. For each enoki_tag ∈ enoki_tags do For each pick_region_tag ∈ pick_region_tags do // Step 2a: Read contour pixels corresponding to the labeled objects contour_pixels ← ReadContourPixels(enoki_tag, pick_region_tag) 3. Calculate center_of_mass(enoki_tag, pick_region_tag) for each contour 4. Filter pairs of centers of mass that meet region distribution conditions 5. For each pair of centers of mass (Class A, Class B) do // Step 5a: Compute Euclidean distance (ED) between centers of mass ED ← EuclideanDistance(center_of_mass_A, center_of_mass_B) // Step 5b: Track the minimum Euclidean distance for each pair min_ED ← min(min_ED, ED) 6. For each pair {enoki_tag, pick_region_tag} do // Step 6a: If minimum ED value is obtained, add label pair to group if ED = min_ED then group ← group ∪ {enoki_tag, pick_region_tag} 7. Remove duplicates from group Return group |
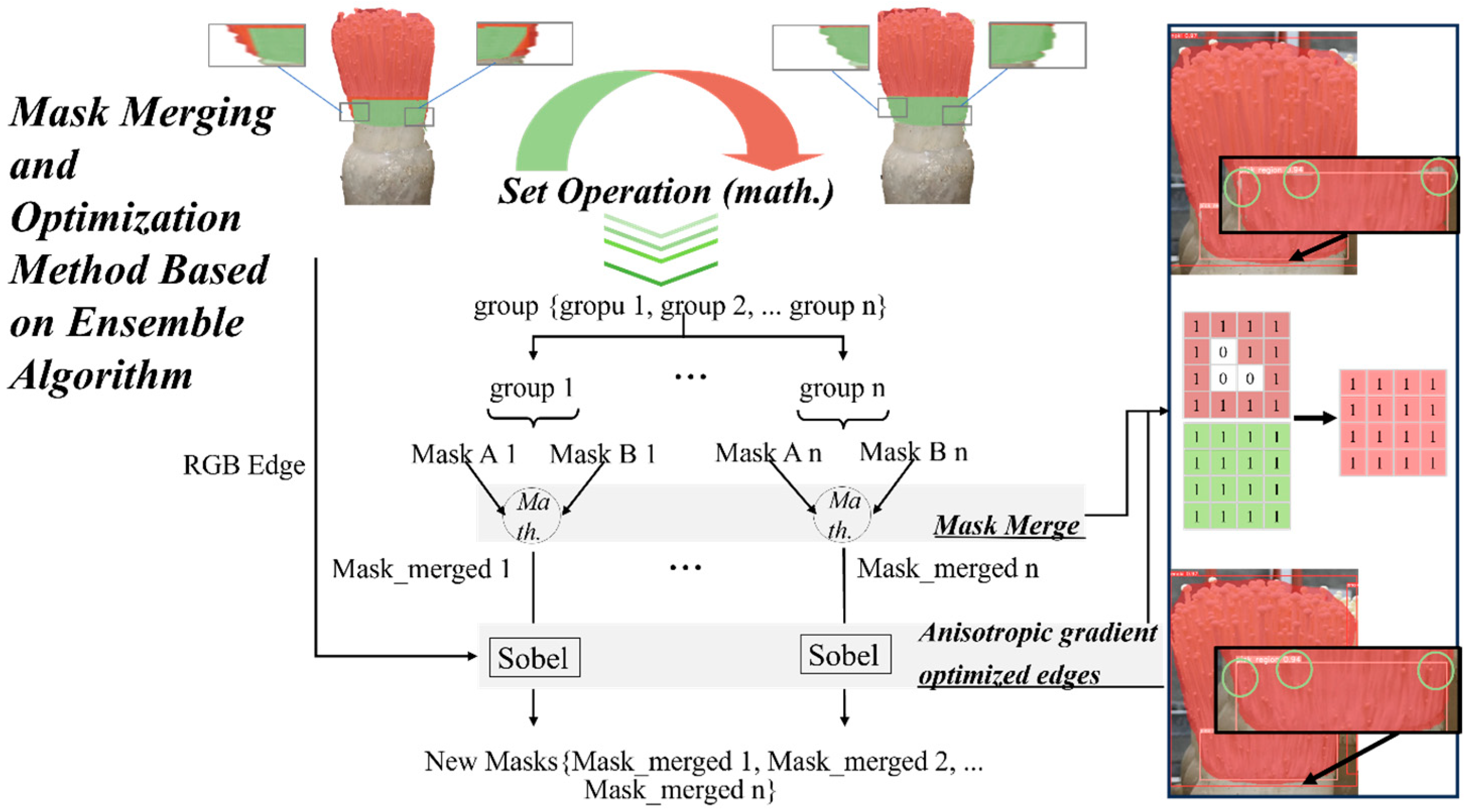
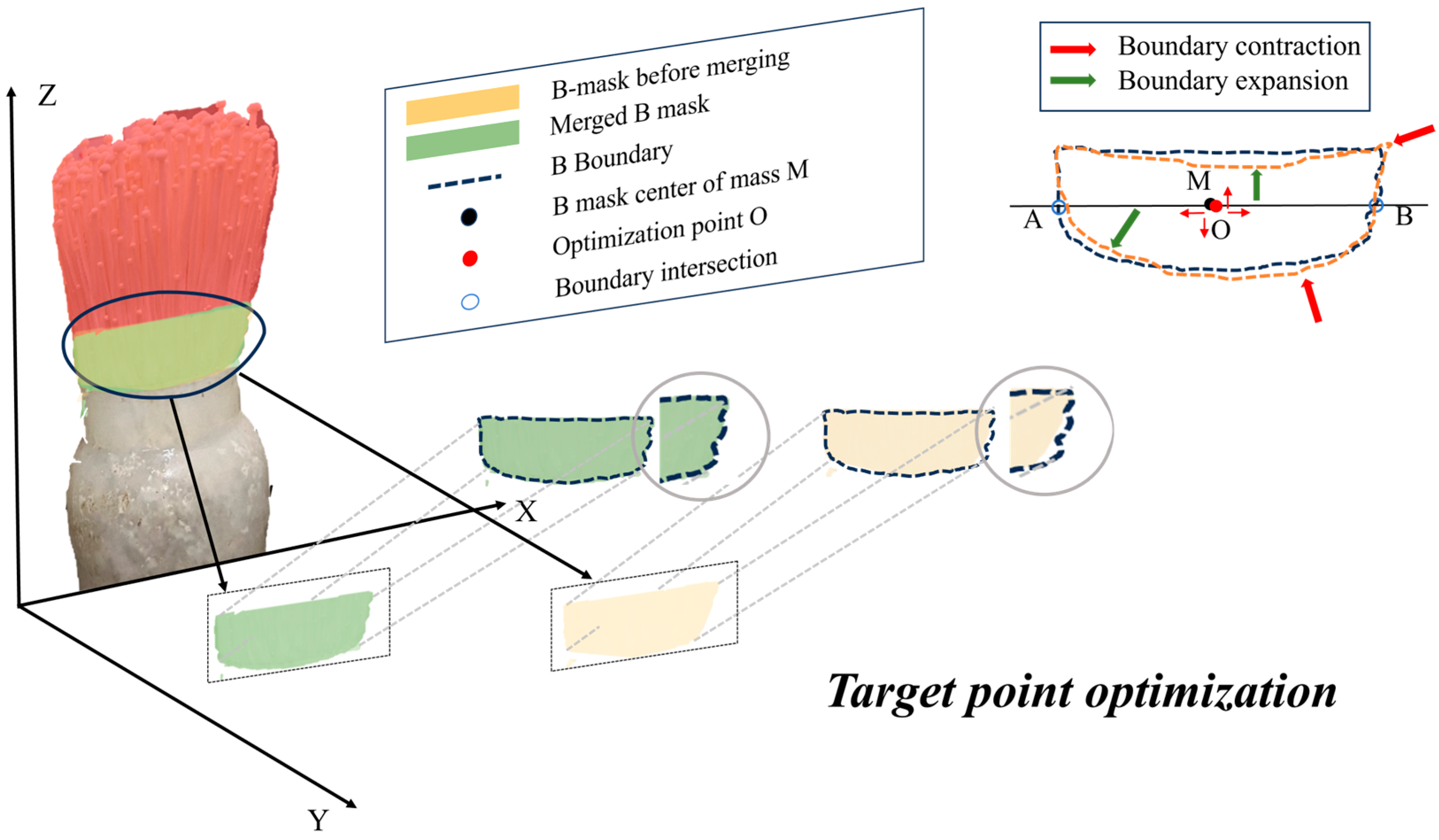
2.5.2. Mask Merging and Edge Optimization
2.5.3. Weighted Box Fusion (WBF)
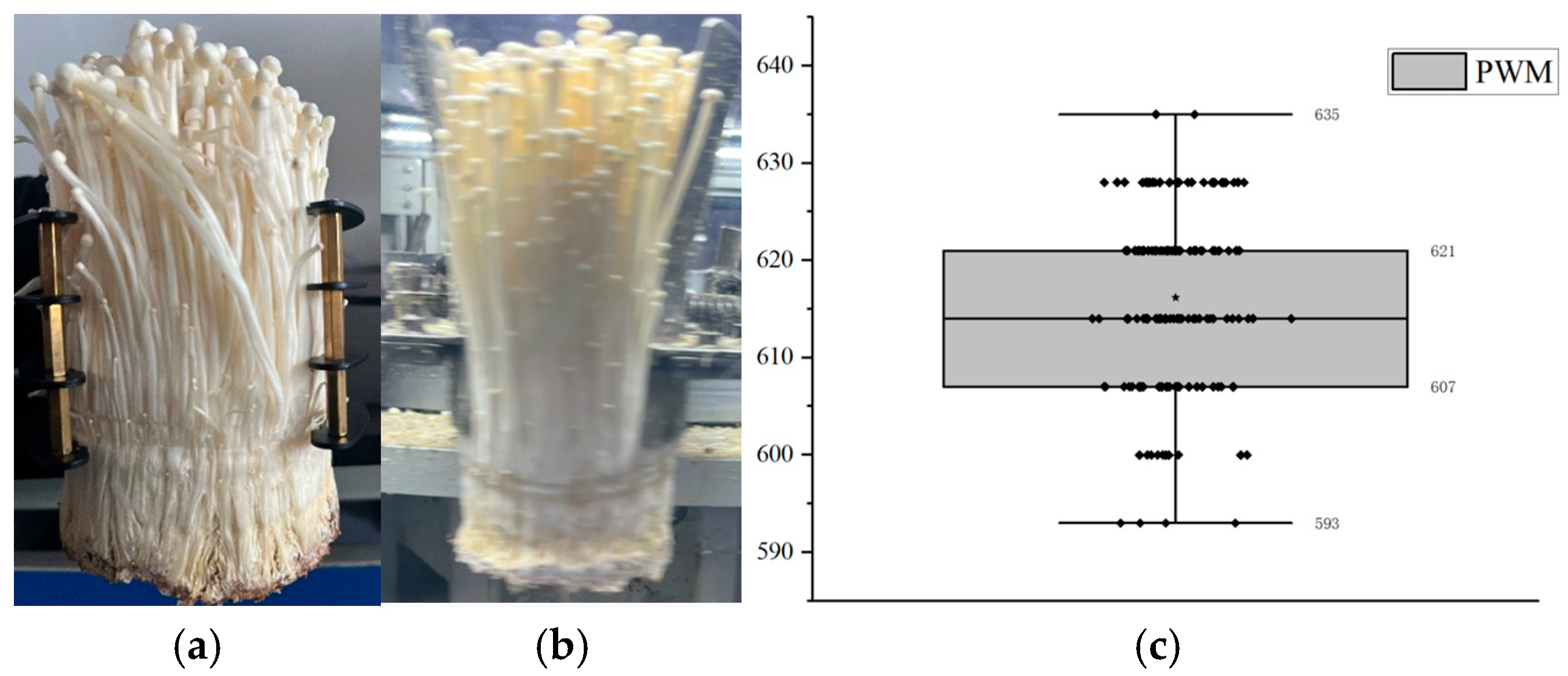
2.6. Optimization of Grabbing Parameters for Enoki Mushrooms
2.7. Communications and Control
2.8. Software and Hardware for Data Analysis
2.9. Model Evaluation Indicators
3. Results
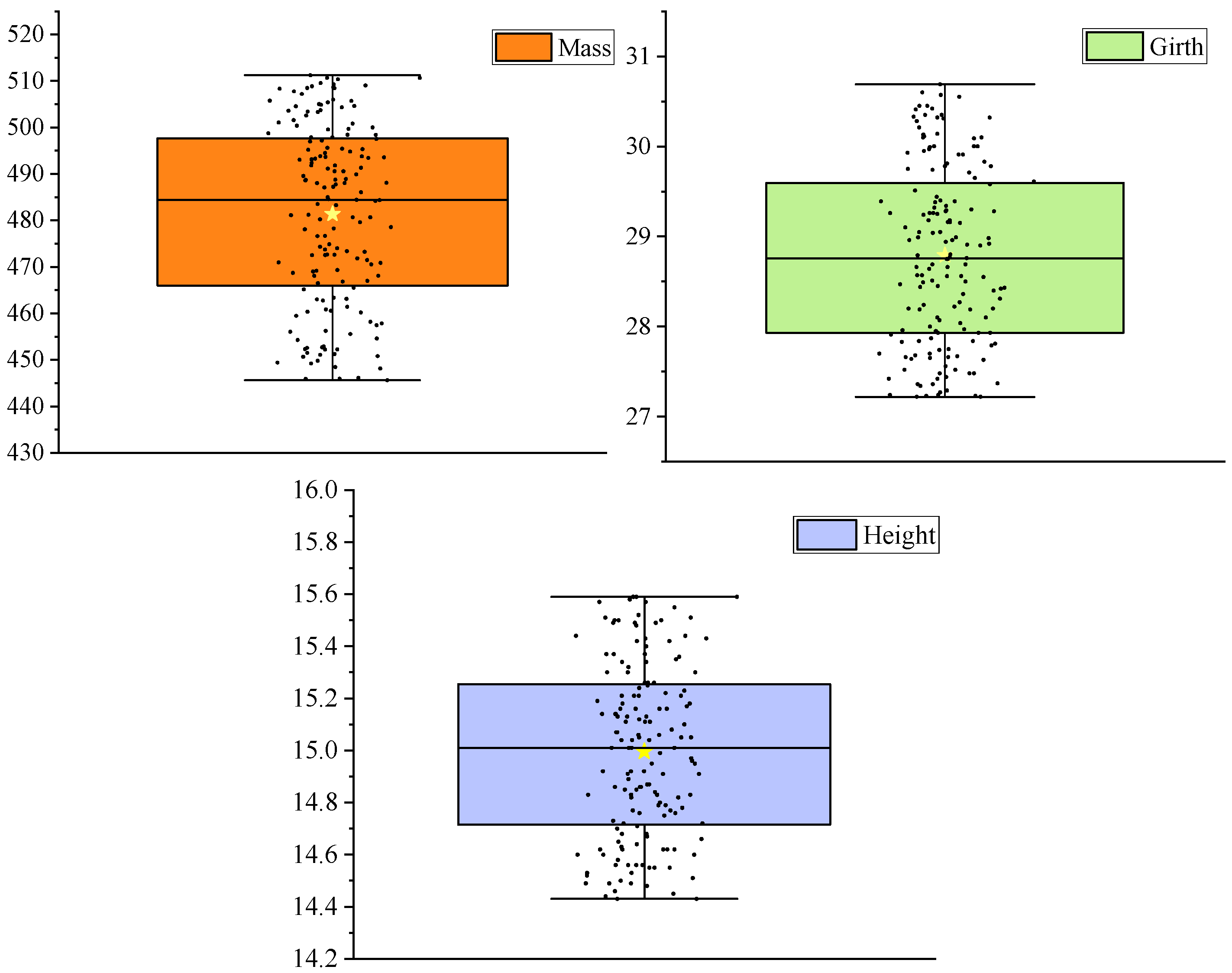
3.1. Results of Payload on Enoki Mushrooms
3.2. Model Training Details
3.3. Results of Ablation Analysis
- (1)
- StarNet: By comparing YOLOv8 and YOLOv8 + StarNet, it was found that the mAP50-95 of A and B masks were improved from 0.713 and 0.711 to 0.717 and 0.715, respectively, which improved the prediction ability of the model significantly. Regarding lightweighting, the model size was reduced from the original 7.6 to 6.1 M, the number of parameters was reduced from close to 3.2 to close to 2.8 M, and the number of network layers was also reduced to 173. Although the floating-point operation volume increased, the overall goal of lightweighting was achieved, confirming that StarNet has a lightweighting advantage over the original backbone network.
- (2)
- SPPECAN: When comparing YOLOv8 + StarNet + SPPECAN and YOLOv8 + StarNet, it was found that SPPECAN could effectively improve model performance compared with SPPELAN. After replacing SPPELAN with SPPECAN, the mAP50-95 for the A and B masks was improved from 0.717 and 0.715 to 0.734 and 0.724, respectively, and the model size was reduced by 0.5 M. The number of covariates was also reduced by approximately 0.2 M. When comparing YOLOv8 + StarNet + SPPELAN and YOLOv8 + StarNet + SPECAN, it was found that the mAP50-95 for the A and B masks was improved from 0.721 and 0.717 to 0.734 and 0.724, respectively, and the model size was reduced by 0.2 M. However, SPPECAN uses the two-branch feature idea; the left-branch feature was added to the right-branch before the convolution. Thus, the expected loss did not occur in the features. Similarly, an effective reduction in the number of parameters involved in the operation can be achieved by reducing the number of channels by half.
- (3)
- C2fDStar: By comparing YOLOv8 + StarNet + C2fDStar and YOLOv8 + StarNet, it was found that C2fDStar can be used to effectively reduce the model size and number of parameters without affecting the model performance. Similarly, a comparison of YOLOv8 + StarNet + C2fDStar + SPPECAN and YOLOv8 + StarNet + SPPECAN showed the advantage of C2fDStar in achieving model gain and lightweighting simultaneously.
- (4)
- Commonality: By adding three modules—StarNet, SPPECAN, and C2fDStar—to both YOLOv5 and YOLOv9, it was observed that the parameter count of YOLOv9 with StarNet, C2fDStar, and SPPECAN decreased significantly from 57,991,744 to 12,971,324. At the same time, the model size was reduced from 579 MB to 33.4 MB. Similarly, YOLOv5 with the same modules saw its parameter count drop from 7,621,277 to 1,741,257, shrinking the model size from 6.7 MB to 1.7 MB. These results demonstrate that the three proposed methods effectively reduce other models’ parameter count and model size while improving detection performance, thereby confirming the approach’s effectiveness.
3.4. Comparative Performance Analysis Against Alternative Models
3.5. Results of Multi-Scale Sub-Model Ensemble
3.6. Evaluations of Algorithm Effectiveness
3.6.1. Results Compared with Those Obtained Using the Traditional Method
3.6.2. Comparative Analysis Against Mask Quality
3.6.3. Mapping and Control Reliability Verification
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SPPELAN | Spatial pyramid pooling enhanced with efficient local aggregation network |
| ELAN | Efficient local aggregation network |
| R2 | Coefficient of determination |
| ECA | Efficient channel attention |
References
- Grocholl, J.; Ferguson, M.; Hughes, S.; Trujillo, S.; Burall, L.S. Listeria monocytogenes contamination leads to survival and growth during enoki mushroom cultivation. J. Food Prot. 2024, 87, 100290. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.Y.; Qi, J.; Wang, Z.R.; Zhou, H.Y.; Sun, B.; Zhao, H.C.; Wu, H.L.; Xu, Z.Q. Current situation and thinking of Flammulina filiformis in industrialized production in China. Edible Fungi China 2021, 40, 83–88+92. Available online: https://link.cnki.net/doi/10.13629/j.cnki.53-1054.2021.12.015 (accessed on 6 February 2025). (In Chinese).
- Li, C.; Xu, S. Edible mushroom industry in China: Current state and perspectives. Appl. Microbiol. Biotechnol. 2022, 106, 3949–3955. [Google Scholar] [CrossRef]
- Lu, Y.; Young, S. A survey of public datasets for computer vision tasks in precision agriculture. Comput. Electron. Agric. 2020, 178, 105760. [Google Scholar] [CrossRef]
- Yin, H.; Yi, W.; Hu, D. Computer vision and machine learning applied in the mushroom industry: A critical review. Comput. Electron. Agric. 2022, 198, 107015. [Google Scholar] [CrossRef]
- Genno, H.; Kobayashi, K. Apple growth evaluated automatically with high-definition field monitoring images. Comput. Electron. Agric. 2019, 164, 104895. [Google Scholar] [CrossRef]
- Li, Q.; Ma, W.; Li, H.; Zhang, X.; Zhang, R.; Zhou, W. Cotton-YOLO: Improved YOLOV7 for rapid detection of foreign fibers in seed cotton. Comput. Electron. Agric. 2024, 219, 108752. [Google Scholar] [CrossRef]
- Dang, F.; Chen, D.; Lu, Y.; Li, Z. YOLOWeeds: A novel benchmark of YOLO object detectors for multi-class weed detection in cotton production systems. Comput. Electron. Agric. 2023, 205, 107655. [Google Scholar] [CrossRef]
- Kim, W.-S.; Lee, D.-H.; Kim, Y.-J.; Kim, T.; Lee, W.-S.; Choi, C.-H. Stereo-vision-based crop height estimation for agricultural robots. Comput. Electron. Agric. 2021, 181, 105937. [Google Scholar] [CrossRef]
- Shen, Y.; Wang, L.; Jin, Y. AAFormer: A multi-modal transformer network for aerial agricultural images. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), New Orleans, LA, USA, 19–20 June 2022; pp. 1704–1710. [Google Scholar] [CrossRef]
- Kim, W.-S.; Lee, D.-H.; Kim, Y.-J. Machine vision-based automatic disease symptom detection of onion downy mildew. Comput. Electron. Agric. 2020, 168, 105099. [Google Scholar] [CrossRef]
- Thai, H.-T.; Le, K.-H.; Nguyen, N.L.-T. FormerLeaf: An efficient vision transformer for Cassava Leaf Disease detection. Comput. Electron. Agric. 2023, 204, 107518. [Google Scholar] [CrossRef]
- Tongcham, P.; Supa, P.; Pornwongthong, P.; Prasitmeeboon, P. Mushroom spawn quality classification with machine learning. Comput. Electron. Agric. 2020, 179, 105865. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Gao, Z.; Xie, Y.; Wang, H. Computer vision online measurement of shiitake mushroom (Lentinus edodes) surface wrinkling and shrinkage during hot air drying with humidity control. J. Food Eng. 2021, 292, 110253. [Google Scholar] [CrossRef]
- Wang, L.; Dong, P.; Wang, Q.; Jia, K.; Niu, Q. Dried shiitake mushroom grade recognition using D-VGG network and machine vision. Front. Nutr. 2023, 10, 1247075. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.P.; Liaw, J.J. A novel image measurement algorithm for common mushroom caps based on convolutional neural network. Comput. Electron. Agric. 2020, 171, 105336. [Google Scholar] [CrossRef]
- Sutayco, M.J.Y.; Caya, M.V.C. Identification of medicinal mushrooms using computer vision and convolutional neural network. In Proceedings of the 2022 6th International Conference on Electrical, Telecommunication and Computer Engineering (ELTICOM), Medan, Indonesia, 22–23 November 2022; pp. 167–171. [Google Scholar] [CrossRef]
- Tao, K.; Liu, J.; Wang, Z.; Yuan, J.; Liu, L.; Liu, X. ReYOLO-MSM: A novel evaluation method of mushroom stick for selective harvesting of shiitake mushroom sticks. Comput. Electron. Agric. 2024, 225, 109292. [Google Scholar] [CrossRef]
- Charisis, C.; Gyalai-Korpos, M.; Nagy, A.S.; Karantzalos, K.; Argyropoulos, D. Detecting and Locating Mushroom Clusters by a Mask R-CNN Model in Farm Environment; Brill|Wageningen Academic: Wageningen, The Netherlands, 2023; pp. 393–400. [Google Scholar] [CrossRef]
- Wang, X.; Yang, W.; Qi, W.; Wang, Y.; Ma, X.; Wang, W. STaRNet: A spatio-temporal and Riemannian network for high-performance motor imagery decoding. Neural Netw. 2024, 178, 106471. [Google Scholar] [CrossRef]
- Cao, M.-T. Drone-assisted segmentation of tile peeling on building façades using a deep learning model. J. Build. Eng. 2023, 80, 108063. [Google Scholar] [CrossRef]
- An, R.; Zhang, X.; Sun, M.; Wang, G. GC-YOLOv9: Innovative smart city traffic monitoring solution. Alex. Eng. J. 2024, 106, 277–287. [Google Scholar] [CrossRef]
- Duong, V.H.; Nguyen, D.Q.; Van Luong, T.; Vu, H.; Nguyen, T.C. Robust data augmentation and ensemble method for object detection in fisheye camera images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Seattle, WA, USA, 17–18 June 2024; pp. 7017–7026. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Yeh, I.-H.; Liao, H.-Y.M. YOLOv9: Learning What You Want to Learn Using Programmable Gradient Information. In Computer Vision—ECCV; Springer: Berlin/Heidelberg, Germany, 2024. [Google Scholar]
- Aboah, A.; Wang, B.; Bagci, U.; Adu-Gyamfi, Y. Real-time multi-class helmet violation detection using few-shot data sampling technique and YOLOv8. In Proceedings of the 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Vancouver, BC, Canada, 17–24 June 2023; pp. 5350–5358. [Google Scholar] [CrossRef]
- Li, H.; Wu, A.; Jiang, Z.; Liu, F.; Luo, M. Improving object detection in YOLOv8n with the C2f-f module and multi-scale fusion reconstruction. In Proceedings of the Electronic and Automation Control. Conference (IMCEC), Chongqing, China, 24–26 May 2024; pp. 374–379. [Google Scholar] [CrossRef]
- Chen, F.; Wu, F.; Xu, J.; Gao, G.; Ge, Q.; Jing, X.-Y. Adaptive deformable convolutional network. Neurocomputing 2021, 453, 853–864. [Google Scholar] [CrossRef]
- Sapkota, R.; Ahmed, D.; Karkee, M. Comparing YOLOv8 and Mask R-CNN for instance segmentation in complex orchard environments. Artif. Intell. Agric. 2024, 13, 84–99. [Google Scholar] [CrossRef]
- Liu, N.; Long, Y.; Zou, C.; Niu, Q.; Pan, L.; Wu, H. ADCrowdNet: An attention-injective deformable convolutional network for crowd understanding. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 3220–3229. [Google Scholar] [CrossRef]
- Wang, X.; Chan, K.C.K.; Yu, K.; Dong, C.; Loy, C.C. EDVR: Video restoration with enhanced deformable convolutional networks. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Long Beach, CA, USA, 16–17 June 2019; pp. 1954–1963. [Google Scholar] [CrossRef]
- Wang, W.; Dai, J.; Chen, Z.; Huang, Z.; Li, Z.; Zhu, X.; Hu, X.; Lu, T.; Lu, L.; Li, H.; et al. InternImage: Exploring large-scale vision foundation models with deformable convolutions. In Proceedings of the 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Vancouver, BC, Canada, 17–24 June 2023; pp. 14408–14419. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, W.; Li, X.; Rao, Q.; Jiang, T.; Han, M.; Fan, H.; Sun, J.; Liu, S. ADNet: Attention-guided deformable convolutional network for high dynamic range imaging. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Nashville, TN, USA, 19–25 June 2021; pp. 463–470. [Google Scholar] [CrossRef]
- Yin, H.; Zhao, J.; Lei, D. Detecting dried Oudemansiella raphanipies using RGB imaging: A case study. Dry. Technol. 2024, 42, 1466–1479. [Google Scholar] [CrossRef]
- Wang, J.; Lin, X.; Luo, L. Cognition of grape cluster picking point based on visual knowledge distillation in complex vineyard environment. Comput. Electron. Agric. 2024, 39, 166177. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, R.; Peng, J. YOLO-chili: An efficient lightweight network model for localization of pepper picking in complex environments. Appl. Sci. 2024, 14, 5524. [Google Scholar] [CrossRef]






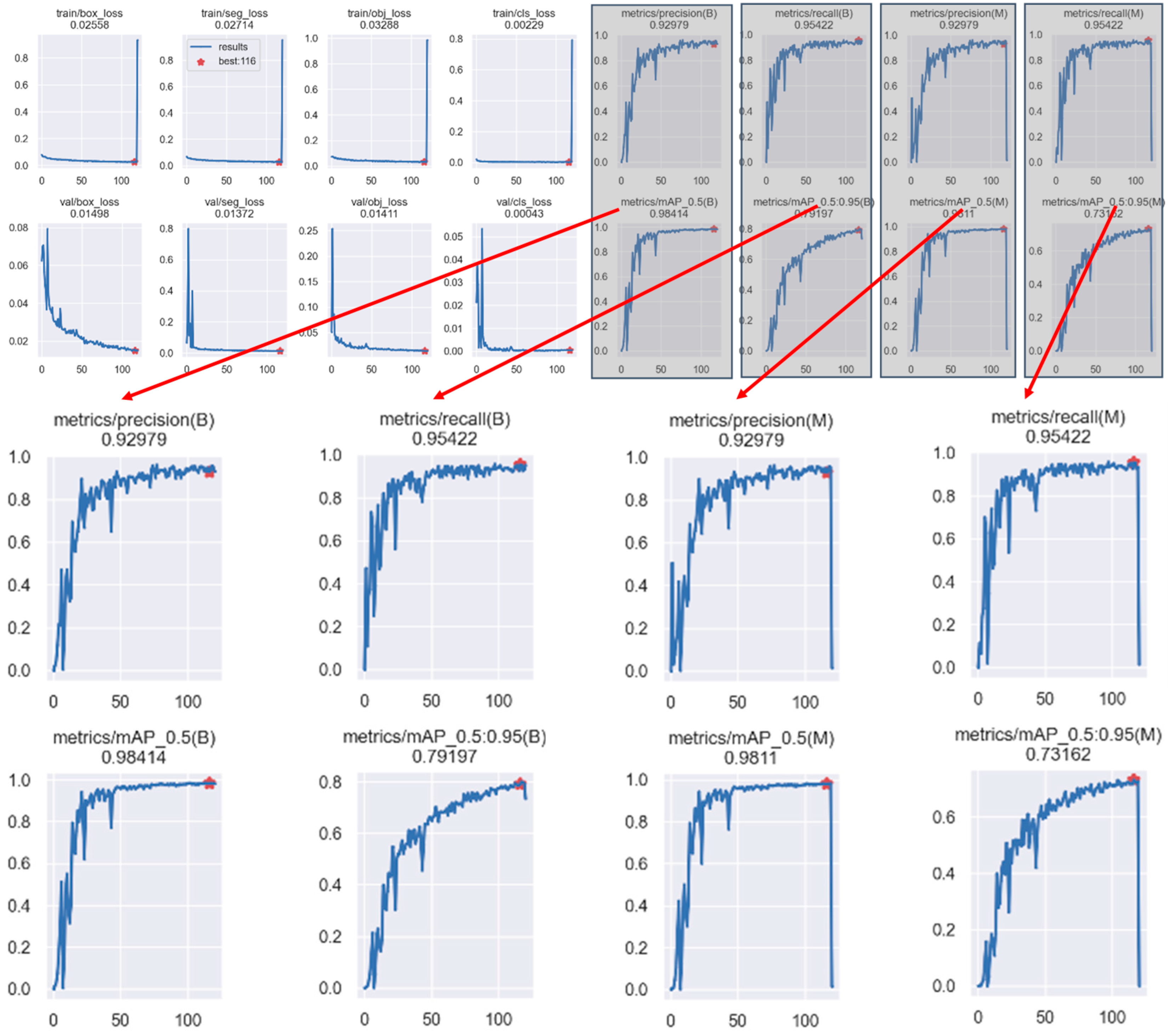




| Model | Category | P/% | R/% | mAP50:95/% | Model Size/M | GFLOPs | Parameters/K | Layers |
|---|---|---|---|---|---|---|---|---|
| YOLOv8 | A B | 0.927 0.923 | 0.944 0.938 | 0.713 0.711 | 7.6 | 8.9 | 3,157,200 | 225 |
| YOLOv8+ StarNet | A B | 0.921 0.912 | 0.937 0.943 | 0.717 0.715 | 6.1 | 14.8 | 2,836,671 | 173 |
| YOLOv8+ StarNet +SPPELAN | A B | 0.920 0.925 | 0.931 0.933 | 0.721 0.717 | 5.9 | 14.8 | 2,806,435 | 179 |
| YOLOv8+ StarNet +SPPECAN | A B | 0.919 0.928 | 0.954 0.950 | 0.734 0.724 | 5.6 | 14.6 | 2,696,625 | 177 |
| YOLOv8+ StarNet +C2fDStar | A B | 0.916 0.918 | 0.944 0.944 | 0.741 0.721 | 3.8 | 12.7 | 1,729,713 | 177 |
| YOLOv8+ StarNet+ C2fDStar+ SPPECAN | A B | 0.925 0.922 | 0.958 0.951 | 0.739 0.743 | 3.8 | 13.1 | 1,728,548 | 177 |
| YOLOv5 | A B | 0.921 0.927 | 0.934 0.934 | 0.708 0.710 | 6.7 | 26.6 | 7,621,277 | 225 |
| YOLOv5+ StaNet+ C2fDStar+ SPPECAN | A B | 0.929 0.933 | 0.940 0.937 | 0.727 0.725 | 1.7 | 12.8 | 1,741,257 | 195 |
| YOLOv9 | A B | 0.947 0.939 | 0.947 0.942 | 0.773 0.751 | 57.9 | 372.6 | 57,991,744 | 1204 |
| YOLOv9+ StaNet+ C2fDStar+ SPPECAN | A B | 0.947 0.933 | 0.944 0.948 | 0.775 0.751 | 33.4 | 103.4 | 12,971,324 | 844 |
| Model | Category | P/% | R/% | mAP50:95/% | Parameters/K | GFLOPs | Layers |
|---|---|---|---|---|---|---|---|
| YOLOv8 | A B | 0.927 0.923 | 0.944 0.938 | 0.713 0.711 | 3.2 | 8.9 | 225 |
| YOLOv7 | A B | 0.936 0.935 | 0.948 0.942 | 0.741 0.736 | 37.6 | 106.5 | 391 |
| YOLOv5 | A B | 0.920 0.922 | 0.944 0.937 | 0.707 0.706 | 2.6 | 7.8 | 262 |
| Swin Transformer | A B | 0.929 0.922 | 0.933 0.948 | 0.747 0.735 | 48.2 | 267.4 | \ |
| YOLACT++ | A B | 0.927 0.930 | 0.943 0.940 | 0.744 0.740 | 34.9 | 192.1 | \ |
| TPH-YOLOv5 | A B | 0.944 0.940 | 0.953 0.949 | 0.733 0.747 | 41.9 | 161.5 | 466 |
| MPG-YOLOv8 | A B | 0.925 0.922 | 0.958 0.951 | 0.739 0.743 | 1.7 | 13.1 | 177 |
| Model | Category | P/% | R/% | mAP50:95/% | Model Size/M | Training Time/h |
|---|---|---|---|---|---|---|
| A (576) | all | 0.967 | 0.914 | 0.718 | 3.8 | 5.27 |
| B (640) | all | 0.955 | 0.952 | 0.769 | 3.8 | 7.73 |
| C (768) | all | 0.934 | 0.944 | 0.724 | 3.9 | 5.87 |
| D (896) | all | 0.952 | 0.947 | 0.761 | 4.0 | 9.51 |
| E (1024) | all | 0.901 | 0.932 | 0.698 | 4.1 | 11.67 |
| Ensemble (by NMS) | all | 0.964 | 0.958 | 0.774 | \ | \ |
| Ensemble (by WBF) | all | 0.969 | 0.963 | 0.779 | \ | \ |
| Status | g1 | g2 | g3 | g4 | g5 | g6 | g7 |
|---|---|---|---|---|---|---|---|
| S | 10,200 | 11,019 | 11,143 | 10,568 | 11,250 | 11,024 | 10,563 |
| Sall merged | 47,586 | 44,778 | 46,823 | 47,694 | 44,857 | 45,869 | 46,230 |
| SM | 10,196 | 11,014 | 11,151 | 10,594 | 11,021 | 11,017 | 10,553 |
| IoU > 0.8? | √ | √ | √ | √ | √ | √ | √ |
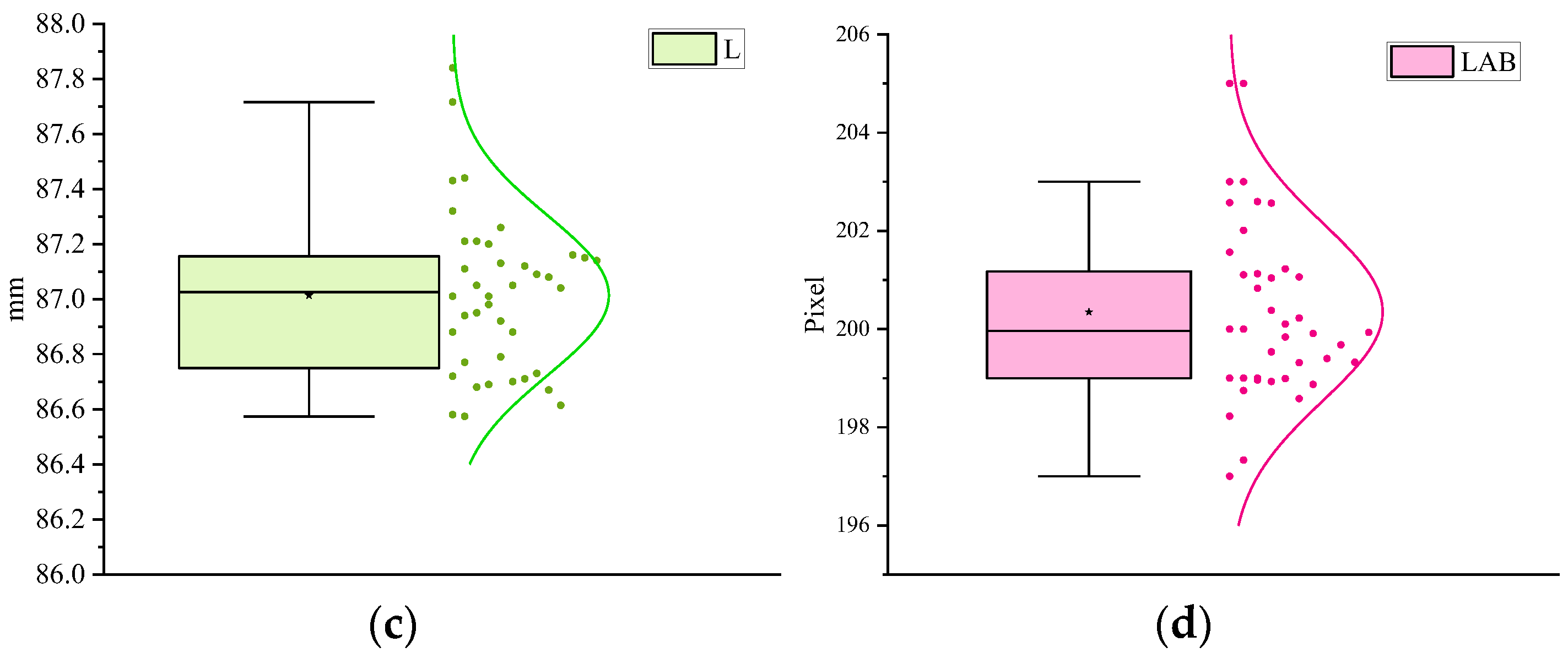
| Num | L/mm | LAB/Pixel | PWM | /mm | /% |
| 1 | 87.01 | 200 | 627 | 86.94 | 0.16 |
| 2 | 86.88 | 197 | 613 | 85.28 | 0.17 |
| 3 | 87.32 | 205 | 620 | 87.11 | 0.15 |
| 4 | 86.94 | 199 | 629 | 86.07 | 0.15 |
| 5 | 87.21 | 203 | 602 | 87.13 | 0.09 |
| 6 | 87.43 | 205 | 596 | 87.29 | 0.16 |
| 7 | 86.95 | 199 | 610 | 87.04 | 0.10 |
| 8 | 87.02 | 200 | 627 | 87.04 | 0.14 |
| 9 | 87.21 | 203 | 605 | 87.14 | 0.18 |
| 10 | 86.98 | 199 | 609 | 87.03 | 0.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, L.; Jing, J.; Wu, H.; Kang, Q.; Zhao, Y.; Ye, D. MPG-YOLO: Enoki Mushroom Precision Grasping with Segmentation and Pulse Mapping. Agronomy 2025, 15, 432. https://doi.org/10.3390/agronomy15020432
Xie L, Jing J, Wu H, Kang Q, Zhao Y, Ye D. MPG-YOLO: Enoki Mushroom Precision Grasping with Segmentation and Pulse Mapping. Agronomy. 2025; 15(2):432. https://doi.org/10.3390/agronomy15020432
Chicago/Turabian StyleXie, Limin, Jun Jing, Haoyu Wu, Qinguan Kang, Yiwei Zhao, and Dapeng Ye. 2025. "MPG-YOLO: Enoki Mushroom Precision Grasping with Segmentation and Pulse Mapping" Agronomy 15, no. 2: 432. https://doi.org/10.3390/agronomy15020432
APA StyleXie, L., Jing, J., Wu, H., Kang, Q., Zhao, Y., & Ye, D. (2025). MPG-YOLO: Enoki Mushroom Precision Grasping with Segmentation and Pulse Mapping. Agronomy, 15(2), 432. https://doi.org/10.3390/agronomy15020432





