Abstract
Through the identification and expression pattern analysis of potato Laccase (LAC) gene family members, the characteristics of the StLAC gene family were elucidated, and the biological function of potato StLACs was further analyzed. In this study, bioinformatics approaches were employed to identify the members of the potato LAC family at the whole-genome level. Subsequently, their physicochemical properties, chromosomal localizations, gene structures, gene duplication events, and expression patterns were thoroughly analyzed. By leveraging the RNA-seq data retrieved from the PGSC database, the expression patterns of StLACs in diploid monoploid (DM) potatoes under various tissue, stress, and hormone treatments were investigated. Moreover, real-time fluorescence quantitative polymerase chain reaction (qPCR) was utilized to analyze the relative expression levels of StLACs in the “Atlantic” potato cultivar under salt stress treatments at different time points (zero hours, one hour, three hours, twelve hours, and twenty-four hours), aiming to screen for the StLAC genes potentially involved in the potato’s response to salt stress. Forty-five members of the potato StLAC gene family were identified, unevenly distributed across 12 chromosomes. Through the analysis of their gene structures and phylogenetic characteristics, the 45 StLACs members were classified into five subgroups. Collinearity analysis indicated that segmental duplication and tandem repeats played major roles in the expansion of the StLAC genes. Using RNA-seq and qPCR analyses, two candidate StLAC genes (Soltu09G001990 and Soltu04G028320) involved in the potato’s response to salt stress were screened out. This study provides a theoretical basis for further understanding StLAC gene family characteristics and an in-depth analysis of StLAC gene function in potato.
1. Introduction
Laccase (LAC, EC1.10.3.2), belonging to the blue multicopper oxidase family, is capable of catalyzing the oxidation of polyphenolic substances. It was first discovered in the sap of the Japanese lacquer tree (Toxicodendron vernicifluum) [1,2]. In nature, laccase is widely distributed in plants, animals, and microorganisms. Fungal laccases are involved in processes such as lignin degradation, pigment synthesis, morphogenesis, infection, and pathogenesis [3]. However, plant laccases exhibit distinct functions compared to their fungal counterparts. They participate in the lignin synthesis process within plants. With their redox catalytic activity towards lignin, plant laccases play a central role in the biosynthesis and reinforcement mechanisms of the cell wall, which are crucial for maintaining the structural integrity and mechanical strength of plant cells as well as the plant’s ability to withstand stresses [4,5].
Wang et al. [6] found that when the GaLAC1 gene of cotton (Gossypium arboreum) was heterogeneously expressed in poplar (Populus deltoides), the transgenic plants showed significantly accelerated oxidation rate of phenolic substances and increased lignin content in stems. The mutation AtLAC15 in Arabidopsis thaliana showed that the seed coat was light brown or yellow, and the lignin content in the mutant seeds was reduced by nearly 30% compared with wild-type seeds, which was the first genetic evidence of laccase’s involvement in lignin synthesis in plants [7]. Cesarino et al. [8] discovered that SofLAC in sugarcane (Saccharum officinarum) is highly expressed in the young internodes of the stem, and its expression level decreases as the stem develops and matures. Furthermore, the heterologous overexpression of the SofLAC gene in the Arabidopsis lac17 mutant line allowed for the direct observation of the lignin content in the cells. In plants, lignin content is closely related to plant stress resistance. A study found that the heterologous expression of miR397-5p from Medicago sativa cv. ‘Pianguan’ in tobacco (Nicotiana tabacum K326) plants increased lignin content. Additionally, under drought stress conditions, miR397-5p from ‘Pianguan’ was able to down-regulate the expression of its target gene, LAC4 [9]. When subjected to drought and salt stress, rice (Oryza sativa) exhibits a high expression of the OsChI1 (Os01g61160) gene in its roots. When this gene was overexpressed in Arabidopsis, it was found that the root length of the transgenic plants significantly increased under NaCl and PEG stress, thereby enhancing their resistance [10]. The overexpression of GhLac1 in cotton leads to increased lignification in leaf parts, which improves cotton resistance to Verticillium dahliae, Helicoverpa armigera, and Aphis gospel. However, the suppression of GhLac1 expression leads to a redirection of metabolic flux in the phenylpropanoid pathway, thus promoting the biosynthesis of secondary metabolites and jasmonic acid [11].
Potato (Solanum tuberosum) belongs to the Solanaceae herb family. Because of its high and stable yield, rich nutritional value, and wide adaptability, it is widely cultivated throughout the world [12]. Abiotic stress has always been a major problem in potato industry development. It has seriously restricted the development of the potato industry [13]. Numerous studies have shown that laccase is involved in plant responses to bollworms, cotton aphids, salt, and drought [9,10,11]. In addition, numerous laccase gene families have been identified and analyzed in various plants to date, including 57 members in strawberry (Fragaria vesca) [14], 29 members in common bean (Phaseolus vulgaris) [15], 24 members in citrus (Camellia sinensis) [16], and 42 members in eggplant (Solanum melongena) [17]. However, the laccase gene family in potato has not been reported.
In this study, we identified the members of the LAC gene family in the potato genome and conducted a systematic analysis of their physicochemical properties, phylogeny, conserved domains, gene structures, and collinearity to elucidate the evolutionary history of the StLAC gene family. Meanwhile, we also investigated the expression patterns of potato LAC genes in different tissues and their responses to abiotic stresses. The findings of this study contribute to a deeper understanding of the relevant aspects and lay a solid foundation for the further exploration of the functions of StLAC genes in the growth and the development of potato.
2. Materials and Methods
2.1. Identification of StLAC Members in the Potato Genome
The potato genome, nucleotide sequence, and protein sequence can be downloaded from the online database PGSC (http://spuddb.uga.edu/) accessed on 21 November 2024. Two methods were used to identify potato LAC family members: (1) The three conserved domains of LACs (PF00394, PF07731, and PF07732) were downloaded from the Pfam database (http://pfam.xfam.org/, accessed on 26 November 2024) as template sequences, and HMM3.1 (http://hmmer.org/download.html, accessed on 22 November 2024) in the potato genome contained the conservative candidate member of the structure domain search. (2) The potato protein database was compared using the BLASTp method (threshold E ≤ 1 × 10−5) to obtain LAC candidate members. Combining the two methods of screening candidate members, the protein sequence was uploaded to the online website SMART (http://smart.emblheidelberg.de/, accessed on 29 November 2024) and NCBI (https://www.ncbi.nlm.nih.gov, accessed on 10 November 2024). The LACs gene family members of potato were selected by deleting the members with or without the LACs domain. The online website ExPasy (http://web.expasy.org/protparam/, accessed on 20 November 2024) [18] was utilized to analyze the number of amino acids, molecular weight (MW), and theoretical isoelectric point (pI) of the StLACs proteins. Additionally, CELLO (http://cello.life.nctu.edu.tw/, accessed on 16 November 2024) [19] was employed to predict the subcellular localization of LACs. The transcript ID was converted to the corresponding gene ID, and the “.DM.” in the ID was omitted for subsequent analysis.
2.2. Evolutionary Analysis of StLACs
MEGA7.0 software was applied to construct a phylogenetic tree of potato and Arabidopsis [20] to explore the evolutionary relationship of LACs protein in the two species by using the neighbor-joining method. Parameter settings were as follows: validation parameter bootstrap was repeated 1000 times in p-distance model mode. The gap was set as partial deletion [21]. Also, a phylogenetic tree of 45 StLAC members was built using MEGA7.0.
2.3. StLACs Sequence Analysis and Structural Characteristics
The MEME program (http://meme-suite.org/tools/meme, accessed on 10 November 2024) was used to analyze the conserved protein motif of StLACs. The parameters were set as follows: the maximum motif number was 20, the optimal motif width was 6–50 amino acid residues, and other parameters were kept as default values [22]. Using the Gene Structure Display Server (GSDS2.0, https://gsds.cbi.pku.edu.cn/, accessed on 10 November 2024) StLACs member exons and their contained substructure were drawn [23].
2.4. Duplication Events and Syntenic Patterns of LAC Genes
The MCScanX v1.5.1 [24] software was utilized to conduct duplication and collinearity analyses of the StLAC genes. Meanwhile, the Circus v0.69 [25] software was applied to visually represent the syntenic relationships among the LAC genes in different species, including Solanum tuberosum, Arabidopsis thaliana, Brassica oleracea, Oryza sativa, and Zea mays.
2.5. Functional Analysis of Genes in the Potato LAC Family
To elucidate the functional characteristics of LAC genes, we utilized the Metware Cloud (https://cloud.metware.cn, accessed on 11 November 2024), a freely accessible online data analysis platform for KEGG and GO enrichment analyses. We obtained potatoes’ Gene Ontology (GO) annotations by inputting the putative candidate protein or gene IDs. A p-value of less than 0.01 was adopted as the significance threshold for evaluating the enrichment level.
2.6. Expression Analysis of StLACs in Potato
Based on the RNA-seq data retrieved from PGSC, an analysis was carried out on the expression of StLACs genes in DM potato. This covered various scenarios: different tissue types such as leaves, roots, shoots, tubers, sepals, stamens, stolons, mature flowers, petioles, petals, carpels, callus, as well as mature and immature fruits; abiotic stress conditions, including salt treatment at 150 mmol NaCl for 24 h, mannitol treatment at 260 μmol for 24 h, and heat treatment at 35 °C for 24 h; simulated biological stress situations, like leaves infected with P. infestans for 24, 48, or 72 h, leaves treated with β-aminobutyric acid for the same time intervals, and leaves treated with Benzo-2,1,3-thiadiazole for 24, 48, or 72 h; and hormone-treated plants, specifically those treated with 50 μM of abscisic acid for 24 h, 10 μM of indole acetic acid for 24 h, 50 μM of gibberellin for 24 h, and 10 μM of 6-benzylaminopurine for 24 h [26]. Subsequently, the TBtools v2.030 software was used to draw the expression heat map [27].
2.7. Plant Materials and Treatments
The retained tetraploid “Atlantic” potato tissue culture seedlings served as experimental materials. These seedlings were placed into the Murashige and Skoog (MS) liquid medium for preliminary cultivation. In this medium, the sucrose mass–volume ratio was 3%, with the pH value between 6.0 ± 0.5. The cultivation environment was set at 23 ± 1 °C, under a 12 h light and 12 h dark cycle, lasting three consecutive weeks. During the salt stress treatment phase, the potato seedlings were exposed to an MS liquid medium supplemented with 150 mmol NaCl. Samples were collected at zero, one, three, twelve, and twenty-five hours, respectively. For each treatment, three biological replicates were prepared. Each replicate consisted of three conical flasks, each containing five potato seedlings. Once the whole plants were sampled, the samples were immediately put into liquid nitrogen and stored at −80 °C for subsequent qPCR analysis.
2.8. RNA Extraction and qPCR
Total RNA was extracted using an RNA extraction kit (Tiangen DP419), and the integrity and concentration of RNA were detected by agar–gel electrophoresis and Nanodrop ND-2000 (Nanodrop Technologies, Wilmington, DE, USA) spectrophotometer. The elimination of genomic DNA contamination and synthesis of first-strand cDNA were performed using a rapid RT kit with gDNase (Tiangen KR116). qPCR was performed on CFX96 (Bio-Rad, Hercules, CA, USA) using Tiangen’s Super RealPreMix Plus (SYBR Green FP217) kit with three biological replicates. qPCR conditions were as follows: 95 °C for 2 min, 95 °C for 5 s, 60 °C for 15 s, for 40 cycles. The melting point curve was detected at 65~95 °C. Using StEF-1α (AB061263) [28] as the internal reference gene, the relative expression of the gene was calculated using the 2−∆∆Ct method [29], and the mapping was carried out with Origin 2021. The primers are shown in Table S1.
3. Results
3.1. Identification of StLACs and Characteristic Analysis of Corresponding Proteins
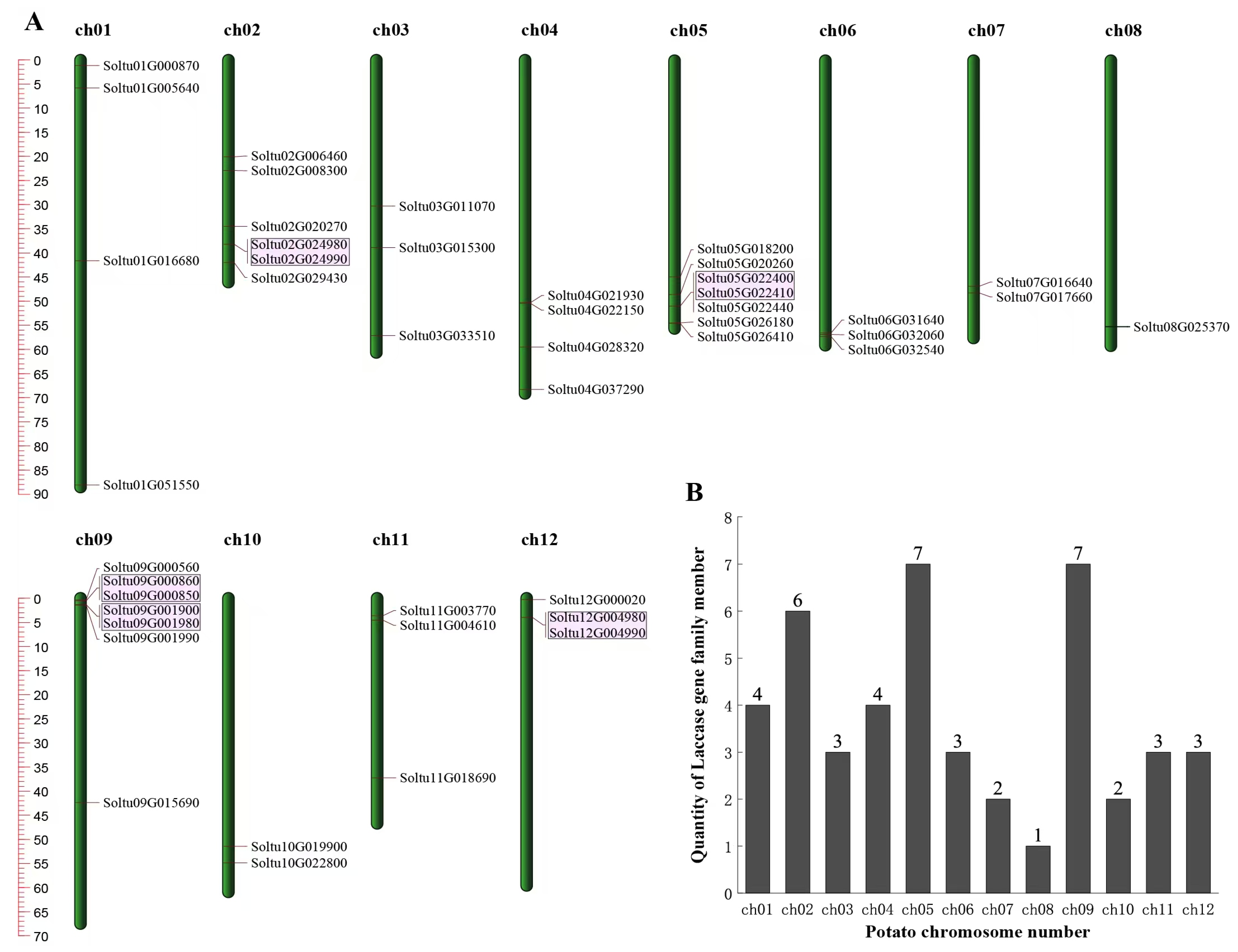
The candidate members of the StLACs gene family were initially screened using HMM3.1 and BLASTp, and the candidate members that did not contain the LACs domain were manually deleted by SMART and NCBI (CDD). Finally, 45 StLACs gene family members were obtained. They were unevenly distributed in 12 chromosomes. Chromosome 5 and 9 harbored the highest count of genes, encompassing seven StLAC genes (Figure 1). The physicochemical analysis showed that the length of StLACs protein was 533~1383 aa, the molecular weight was 59.43~152.75 kD, and the theoretical isoelectric point was 5.68~9.66. The subcellular localization of 45 members of the StLAC family was predicted using CELLO v.2.5, of which 24 are extracellular, 12 are lysosomes, and 9 are cell membranes (Table S2).
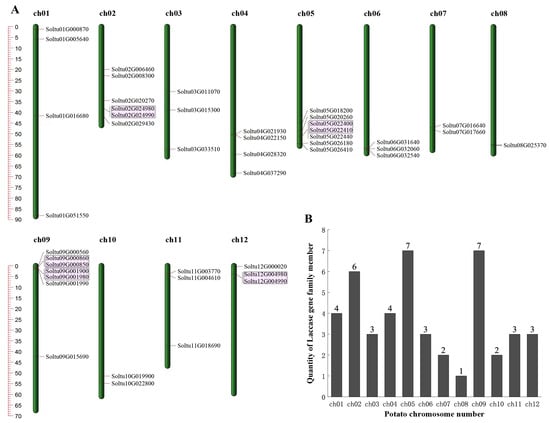
Figure 1.
Chromosome distribution and tandem replication analysis of StLACs. (A) Chromosome distribution. The pink area marked tandem duplication of the StLACs. (B) The chromosome number of potato.
3.2. Phylogenetic Analysis of StLACs
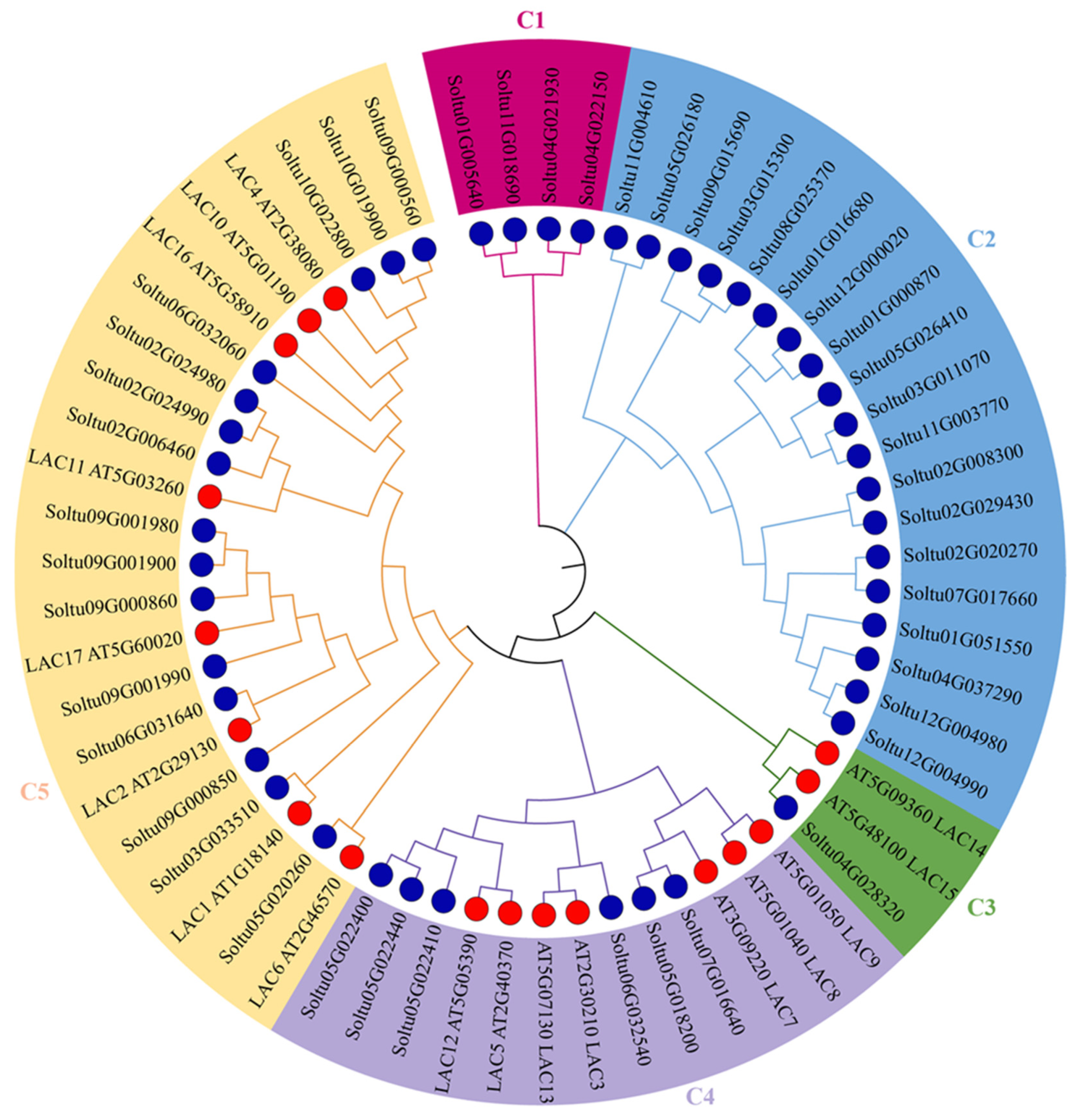
To further investigate the phylogenetic relationship between potato and Arabidopsis laccases, a phylogenetic tree was constructed using MEGA7.0. The 45 potato laccase members were divided into five groups. However, the distribution of potato laccase members within each group was uneven (Figure 2). Both the C1 and C2 groups consisted solely of potato laccase members, containing 4 and 19 StLACs, respectively. In contrast, the C3, C4, and C5 groups all included Arabidopsis LACs, with 1, 6, and 15 potato laccase members, respectively. This indicates that some potato laccases are closely related to Arabidopsis in terms of phylogenetic affinity.
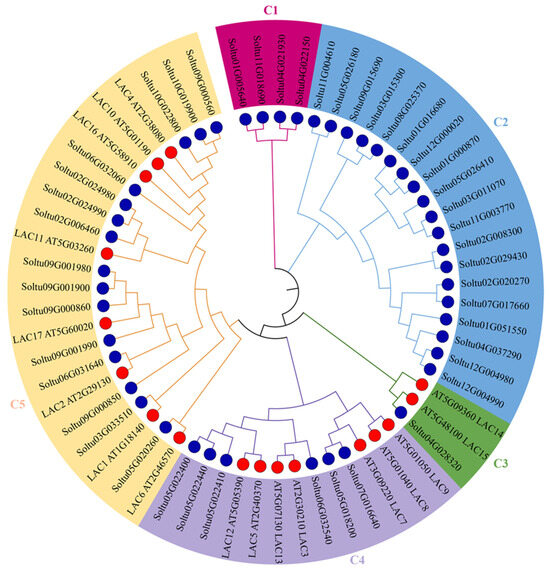
Figure 2.
Phylogenic tree of StLAC and AtLAC gene family. LAC genes of potato and Arabidopsis are represented by a blue circle and red circle, respectively. The blue and red circles represent StLACs and AtLACs, respectively.
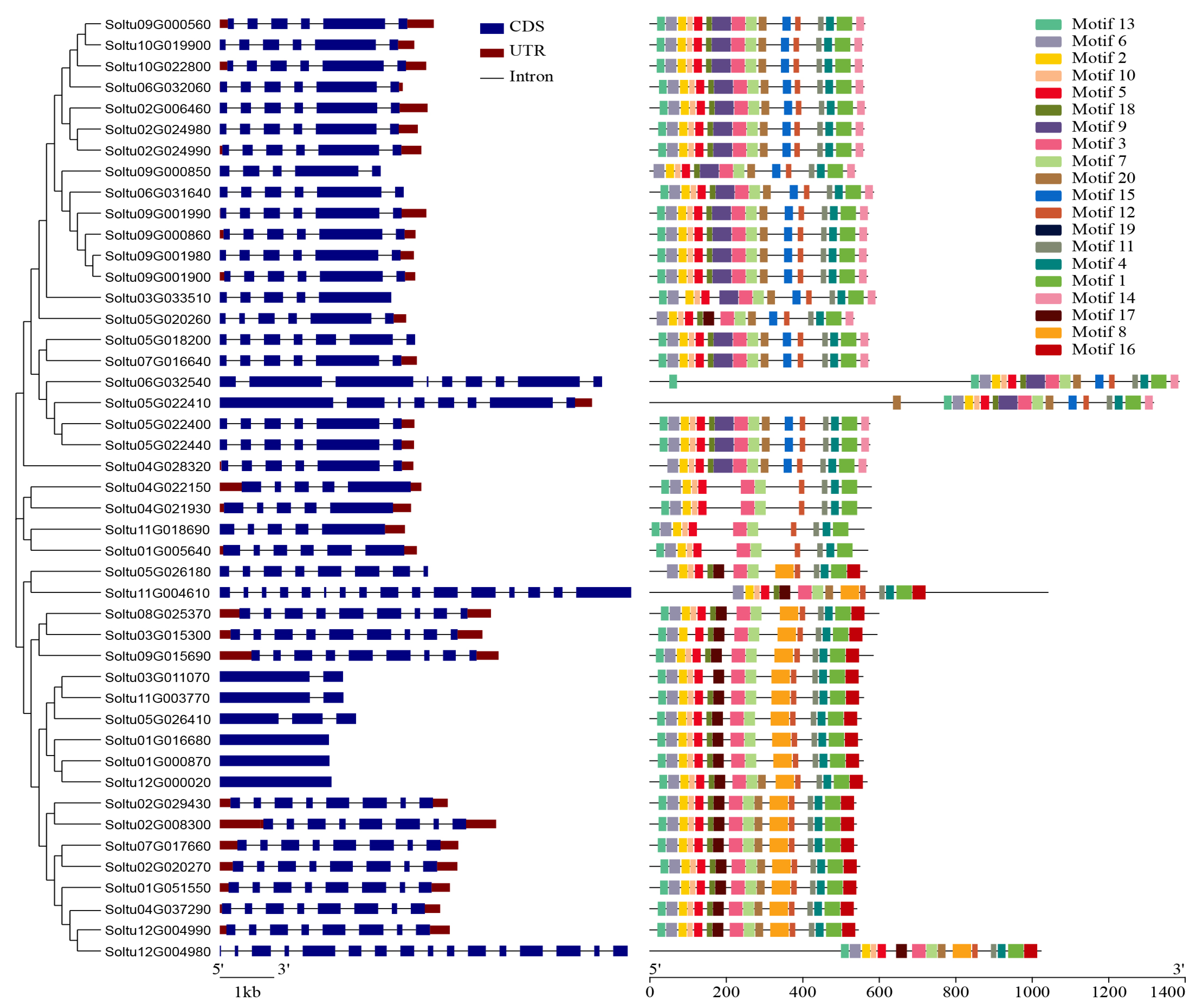
3.3. Gene Structure, Motif Composition, and Conserved Domain Analysis of StLACs
The phylogenetic tree was constructed for the phylogenetic relationship, gene structure, and conserved motif analysis of StLACs gene family. The structure of StLACs was mapped using GSDS. The exons of 45 StLACs range in number from 1 to 16. Nearly 44% of StLACs include six exons, among which three StLACs contain one exon, two genes contain two exons, five genes contain five exons, and eight genes contain eight exons. There are four genes containing nine exons and two genes containing sixteen exons (Figure 3). To further understand the conserved sequences of StLACs proteins, the MEME program was used to identify the conserved motifs of the 45 StLACs proteins (Figure 3 and Table S3). Overall, the members grouped in a cluster have a similar order of arrangement. Motif6, Motif2, Motif10, and Motif5 constitute the first Cu2+ oxidase domain. Motif2, Motif3, and Motif7 were distributed in the C-terminal of the second Cu2+ oxidase domain. Motif11, Motif4, and Motif1 are also located in the C-terminal of the third Cu2+ oxidase domain.
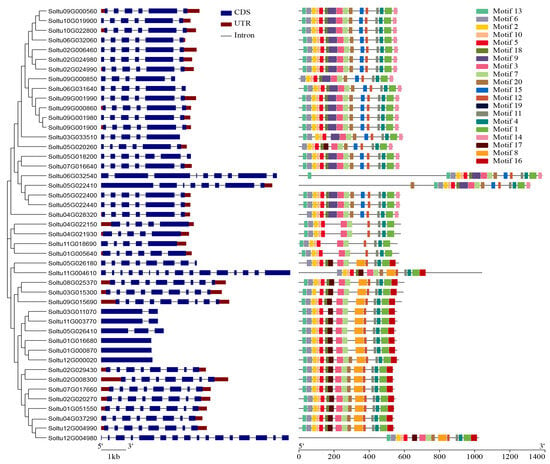
Figure 3.
Evolutionary relationships, gene structures, and conserved motif analyses of the StLACs gene family.
3.4. Gene Replication Events of StLACs and Collinearity Analysis with Other Species
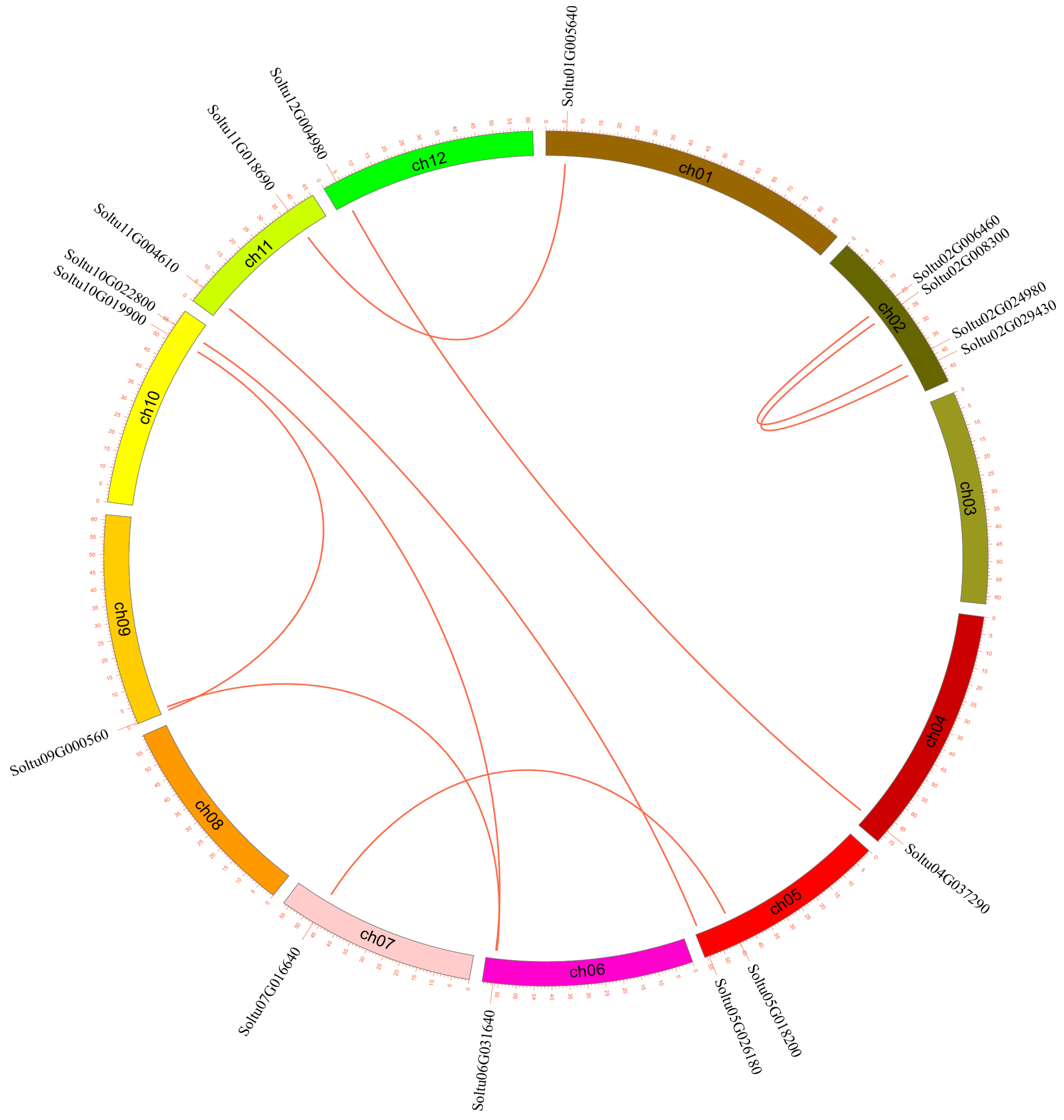
The 16 StLACs that underwent gene duplication were unevenly distributed across 10 chromosomes. Chromosome chr02 had the largest number of StLACs genes, with a total of four, while chromosomes chr01, chr012, chr07, chr06, and chr04 had the fewest, each containing only one StLAC gene (Figure 4).
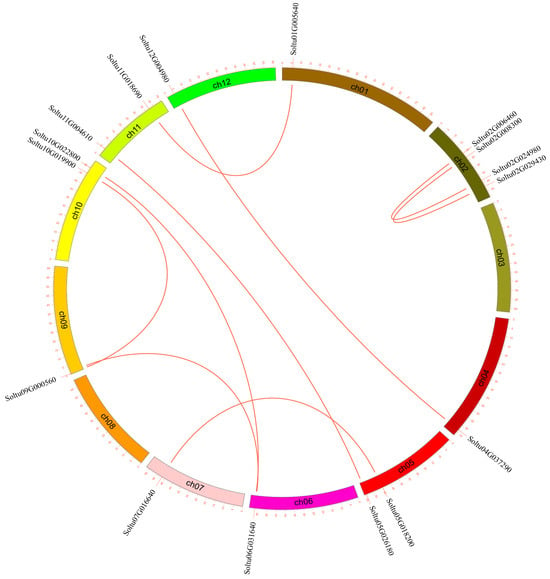
Figure 4.
Duplication events of the StLAC gene family. The red lines indicate the segmental duplication of StLAC genes.
In plant evolution, gene replication (including tandem and fragment replication) is a key driving factor. Gene replication events in the StLACs family were analyzed using MCScanX software. The analysis results showed nine pairs of replications in the StLACs. Soltu02G00640/Soltu02G024980, Soltu02G008300/Soltu02G09430, respectively, Soltu01G005640/Soltu11G018690, Soltu12G004980/Soltu04G037290, Soltu11G018690/Soltu01G005640, Soltu11G004610/Soltu05G026180, Soltu10G022800/Soltu06G031640, Soltu10G019900/Soltu09G000560, Soltu09G000560/Soltu06G031640 (Figure 4), and five for tandem repeat Soltu02G024980/Soltu02G024990, respectively, Soltu05G022400/Soltu05G022410, Soltu09G000850/Soltu09G000860, Soltu09G001980/Soltu09001990, Soltu12G004980/Soltu12G004990 (Figure 1). This demonstrates that gene replication and tandem duplication play an important role in expanding the StLAC gene family.
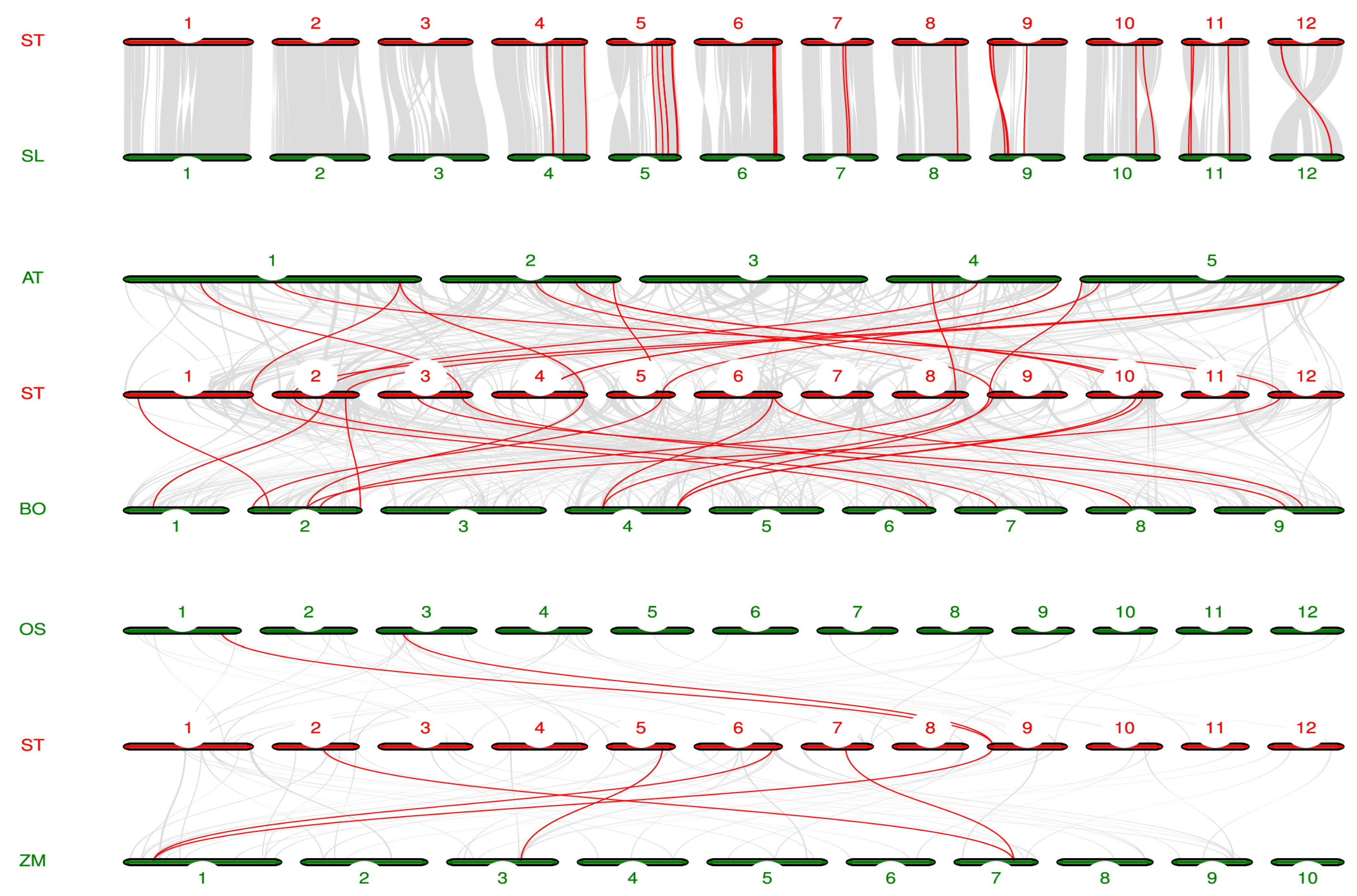
To further investigate the evolutionary relationship of LAC gene family, we analyzed the homology of potato and tomato (Solanum lycopersicum), Arabidopsis, cabbage (Brassica oleracea), rice, and maize genomes (Figure 5). Twenty homologous gene pairs were found in potato and tomato, and thirteen pairs of homologous genes were found in potato and Arabidopsis. There were 16 pairs of homologous genes in potato and cabbage. There were two pairs of homologous genes in potato and rice. Five pairs of homologous genes were identified in potato and maize. Notably, Bo4g065760 in cabbage, Os01g62490 in rice, and Zm00001d028599 in maize are directly homologous to Soltu09G001990 in potato.
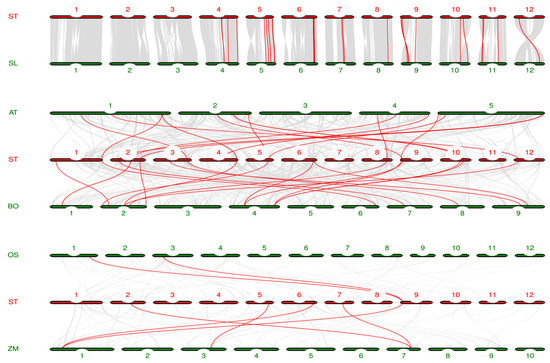
Figure 5.
Homology analysis of LAC genes in potato, tomato, Arabidopsis, cabbage, rice, and corn. The red lines represent the collinear relationships between potato and tomato, Arabidopsis, cabbage, rice, and corn, respectively.
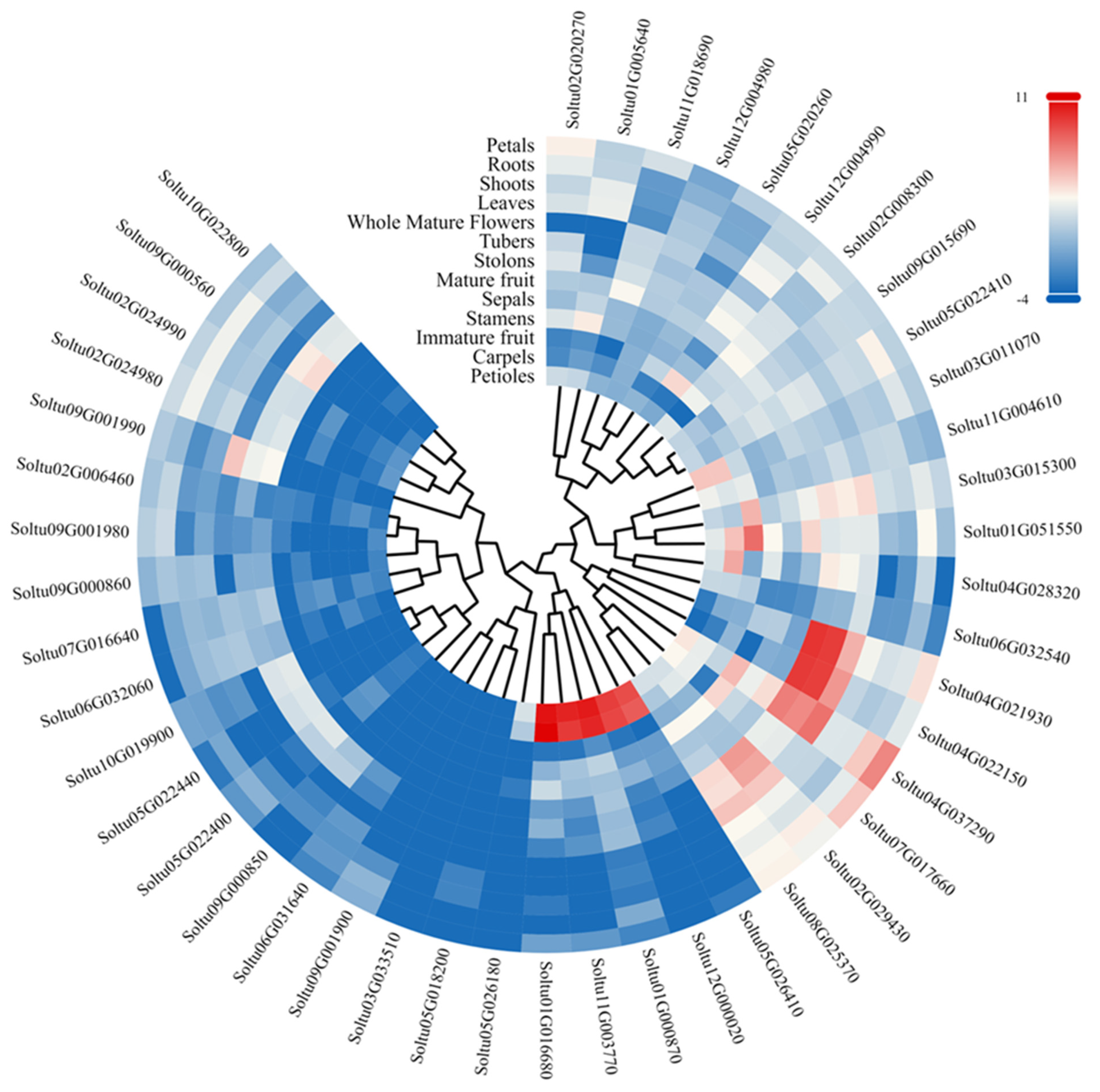
3.5. Analysis of StLAC Gene Expression in Different Tissues of DM Potato
The RNA-seq data were downloaded from PGSC to analyze the expression patterns of StLAC genes in thirteen tissue parts of DM potato, including roots, leaves, shoots, tubers, stolons, petioles, sepals, petals, flowers, carpels, stamens, immature fruits, and mature fruits (Figure 6 and Table S4). The results showed that one StLAC gene was not expressed in all tissues (FPKM = 0). Some StLAC genes were specifically expressed in certain tissues. Soltu03G015300, Soltu04G022150, and Soltu04G037290 were specifically expressed in stolons. Soltu02G029430 and Soltu04G022150 were specifically expressed in both stolons and tubers. Soltu01G005640 was specifically expressed in leaves. Soltu11G003770, Soltu05G026410, Soltu01G000870, Soltu12G000020, and Soltu01G016680 were specifically expressed in petioles and carpels. Soltu08G025370 and Soltu04G037290 were specifically expressed in whole flowers. The remaining StLACs (Soltu07G017660) were specifically expressed in petals and stamens, respectively (FPKM > 20).
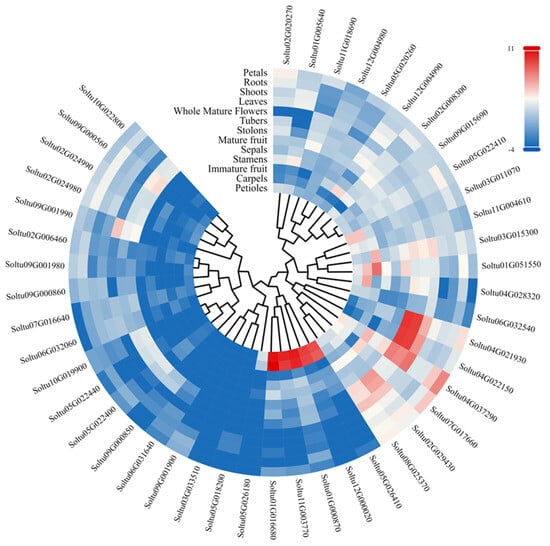
Figure 6.
Expression of StLACs in different tissue parts of DM potato. The logarithm of StLACs expression was taken as base 2, and the color code was drawn with log2FPKM.
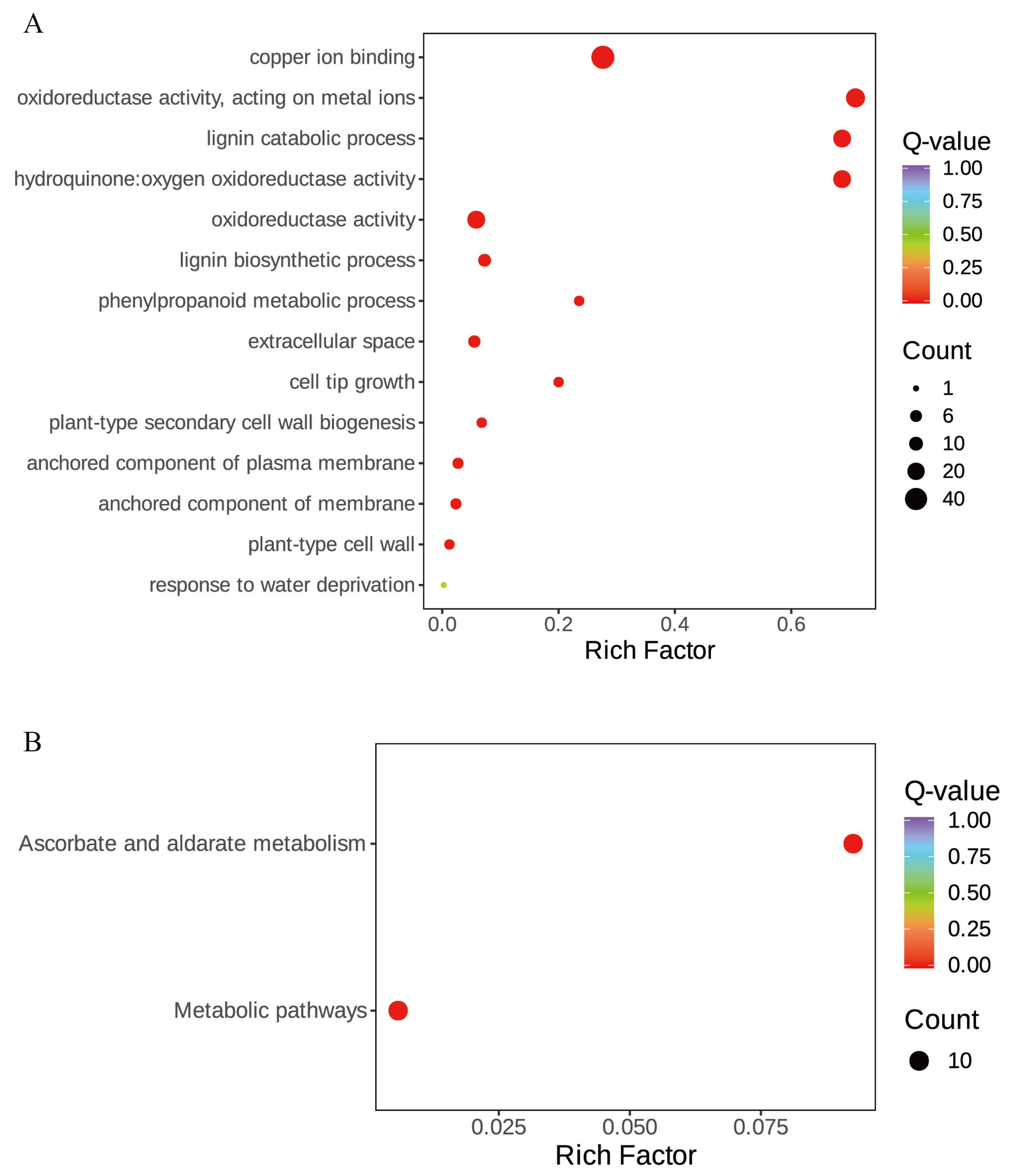
3.6. KEGG and GO Enrichment Analysis of StLACs
To further explore the biological function of StLAC genes, 45 StLAC genes were subjected to enrichment analysis (Figure 7 and Table S5). GO enrichment analysis revealed that the primary biological process involved lignin catabolism and biosynthetic pathways. The cellular components encompassed extracellular space, the anchored component of plasma membrane, the anchored component of membrane, and the plant-type cell wall. The molecular functions included copper ion binding, oxidoreductase activity, and hydroquinone/oxygen oxidoreductase activity. Ten genes are enriched in the Ascorbate and aldarate metabolism, belonging to the C1 (Soltu04G022150, Soltu11G018690, Soltu01G005640, and Soltu04G021930) and C2 (Soltu05G026410, Soltu03G011070, Soltu01G016680, Soltu01G000870, Soltu12G000020, and Soltu11G003770) groups in the evolutionary tree. Laccase is a copper-containing polyphenol oxidase, and it is speculated that laccase is highly likely to intervene in the redox process to perform oxidation or reduction reactions on ascorbic acid, aldaric acid, and their associated metabolites, thereby regulating the concentration and activity of the above metabolites in cells. According to the above findings, the StLACs gene family is likely to rely only on some specific genes to perform biological functions, implying a certain degree of functional redundancy within the gene family.
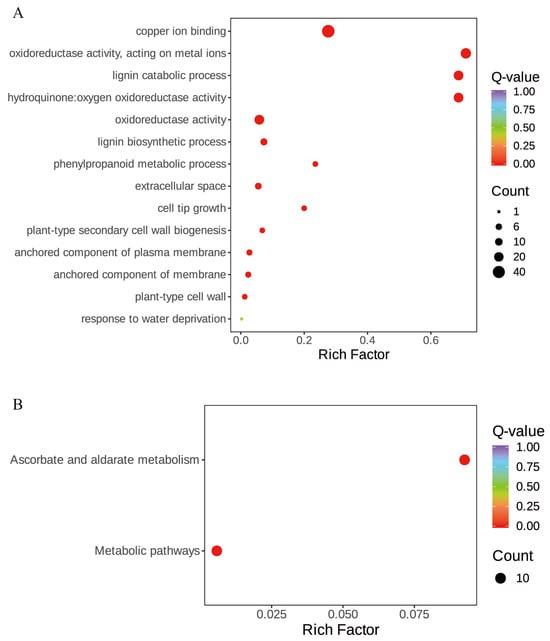
Figure 7.
KEGG and GO enrichment analysis of StLAC gene family. (A) Gene Ontology (GO) enrichment analysis; (B) KEGG enrichment analysis.
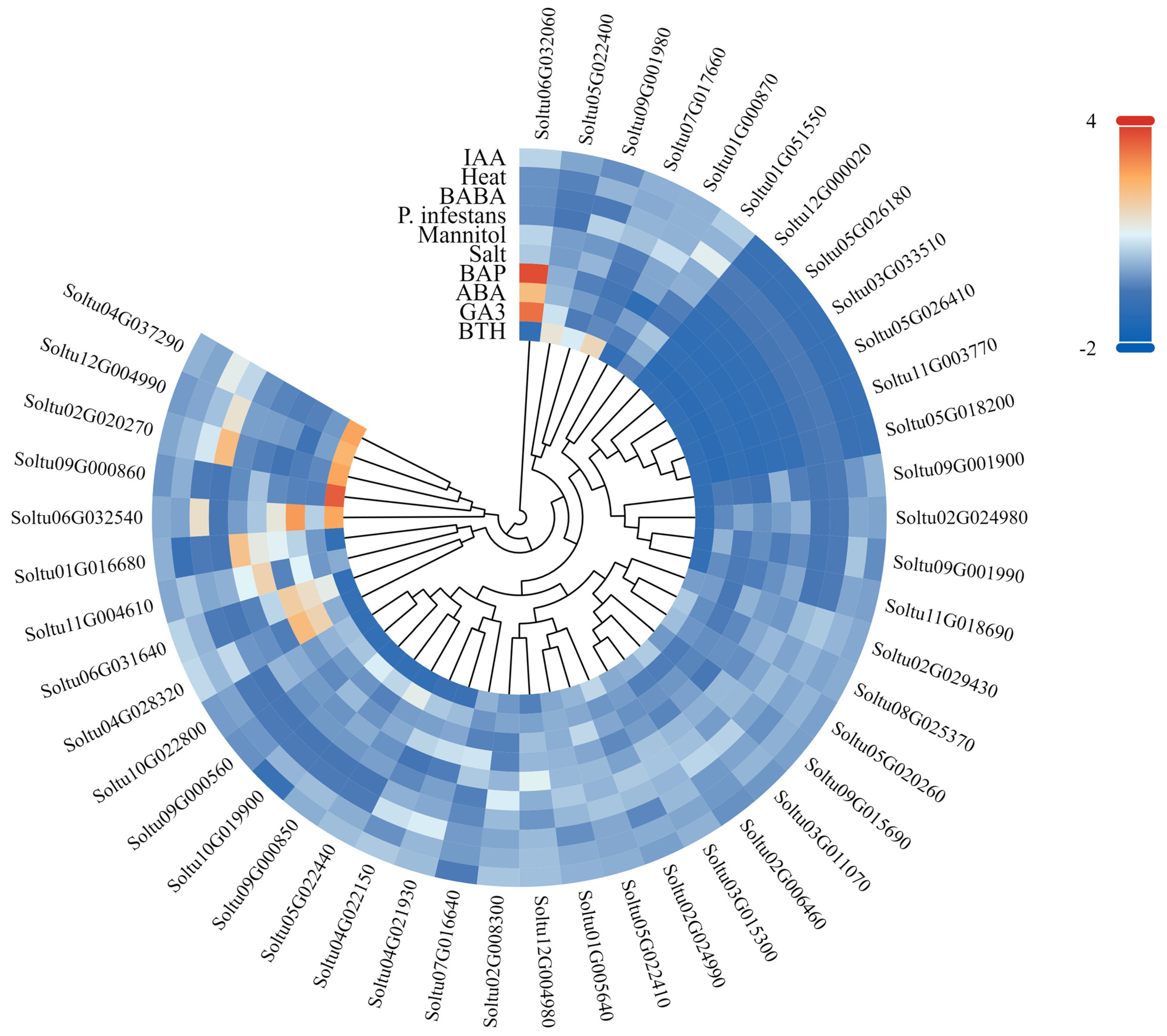
3.7. Expression Patterns of StLACs Under Different Treatments
In this study, the expression patterns of StLAC genes in DM potato under abiotic stress (salt, simulated drought, heat), biotic stress (P. infestans), biostimulant (β-aminobutyric acid and benzo-2,1,3-thiadiazole), and hormone treatment (ABA, IAA, GA, BAP) were analyzed using the RNA-Seq data downloaded by PGSC (Figure 8 and Table S6). The results showed that a total of seven genes were differentially expressed under three abiotic stresses (FPKM > 1 and |log2FC| > 1), among which two StLAC genes (Soltu07G017660 and Soltu04G022150) were differentially expressed in response to two different abiotic stresses (NaCl and mannitol). Six StLAC genes (Soltu09G001990, Soltu04G021930, Soltu04G028320, Soltu07G017660, Soltu02G020270, Soltu04G022150) were down-regulated only under NaCl treatment; Soltu02G008300 was upregulated in mannitol. Under biological stress, three StLAC genes (Soltu12G004980, Soltu04G028320, Soltu09G015690) were differently downregulated (FPKM > 1 and |log2FC| > 1). Under BTH treatment, four StLACs (Soltu12G004980, Soltu04G021930, Soltu04G028320, Soltu04G022150) were down-regulated and one StLAC gene (Soltu07G017660) was up-regulated. BABA treatment only down-regulated one StLAC (Soltu09G015690). Under hormone treatment, 22 StLAC genes were differentially expressed, among which three StLAC genes under ABA treatment were similar to those under NaCl treatment, which may play an important role in potato response to abiotic stress.
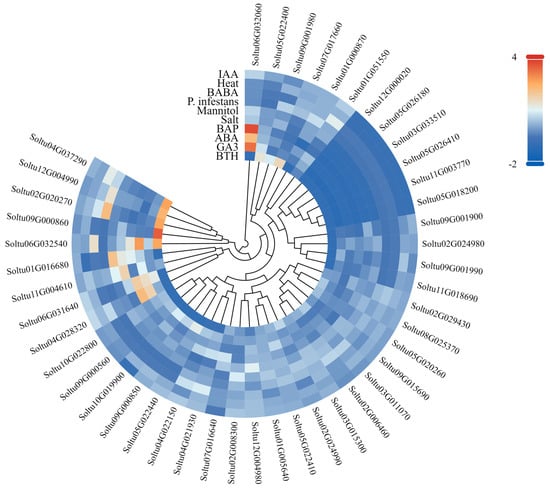
Figure 8.
Expression of StLAC gene in DM potato under stress conditions and hormonal treatments. The logarithm base 2 was taken as the basis, and the color code was drawn with log2FC.
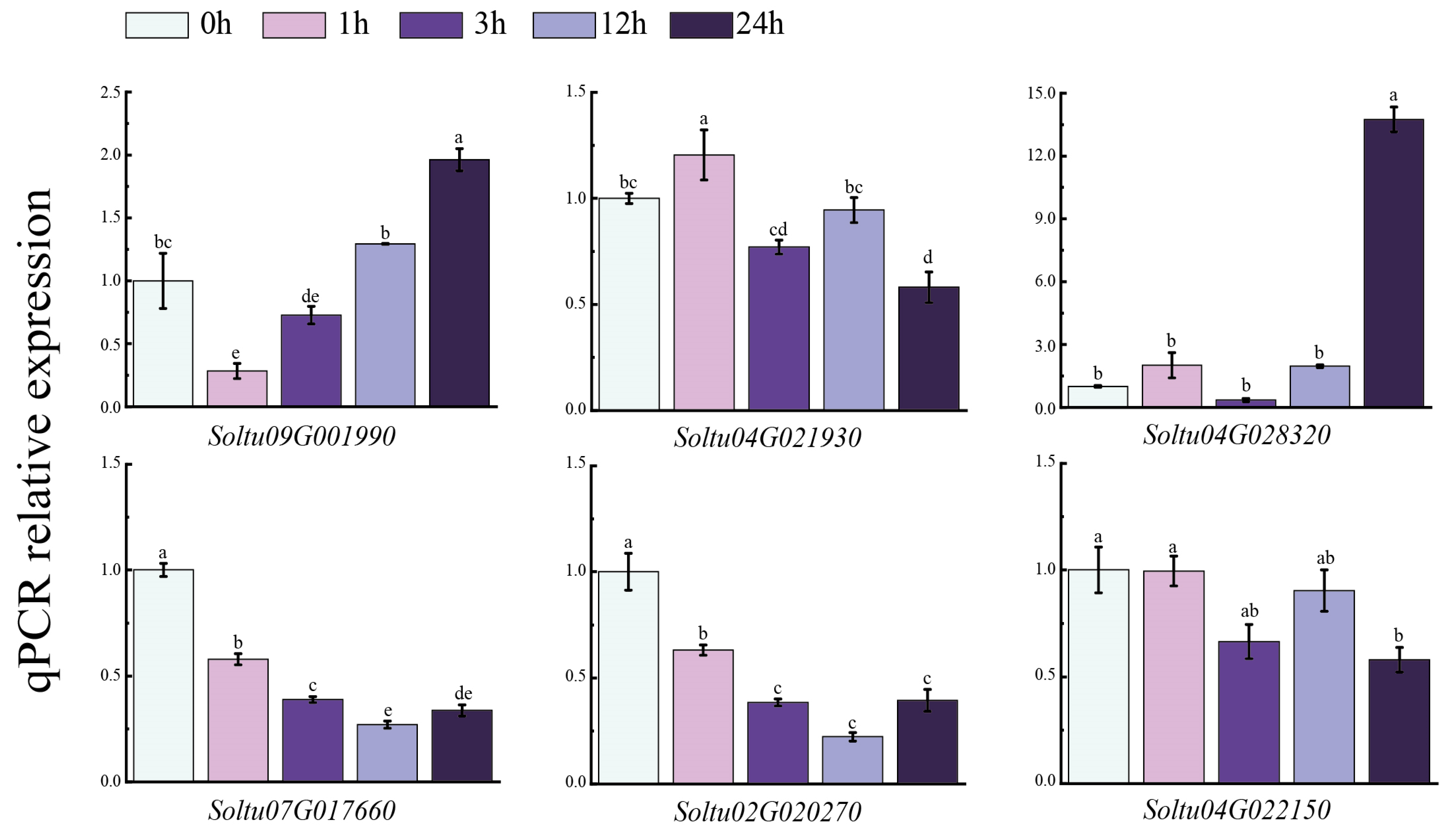
3.8. Expression of Six StLAC Genes in Potato with NaCl Treatment
To further verify the responses of the six StLAC genes (Soltu09G001990, Soltu04G021930, Soltu04G028320, Soltu07G017660, Soltu02G020270, Soltu04G022150) that were differentially expressed under NaCl treatment (Figure 9), we sampled the tissue-cultured seedlings of the tetraploid potato variety “Atlantic” at different time points (zero, one, three, twelve, twenty-four hours) during NaCl treatment. Then, the expression of StLACs was analyzed via qPCR. The results indicated that the expression levels of the two StLAC genes (Soltu07G017660 and Soltu02G020270) were significantly lower than the control at 1 h, and increased slightly over time until 24 h, but remained significantly lower than the control. The expression levels of Soltu04G021930 and Soltu04G022150 fluctuated in a pattern of increase–decrease–increase–decrease during the sampling period and were down-regulated compared to the control at 24 h. The expression level of Soltu09G001990 first decreased and then gradually increased over time, while the expression level of Soltu04G028320 was 13.74 times that of the control at 24 h. These genes may be involved in the material metabolism of potato plants in response to NaCl stress.
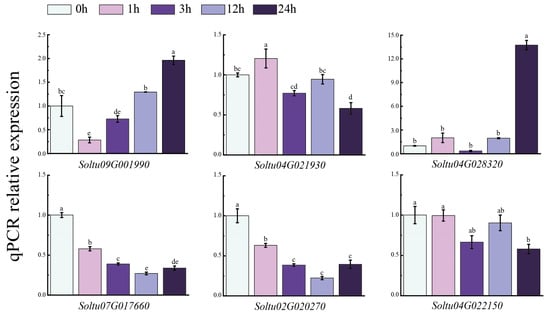
Figure 9.
Relative expression of StLAC gene in potato leaves treated with NaCl. The data were the means of three independent biological replicates (±SE). Error bars represent standard errors. Different letters above the bar chart indicate significant differences at p < 0.05.
4. Discussion
Laccases are multifunctional proteins. They have been demonstrated to contribute to cell morphology [30,31], pigment formation [11], and secondary cell wall biosynthesis [5,32], as well as plant resistance to biotic and abiotic stress [33,34]. In this study, 45 members of the laccase gene family were identified, and this number is higher than that of the LAC gene families in previously reported crops [35,36,37]. The StLAC gene family can be divided into five groups (C1, C2, C3, C4, and C5), among which the C1 and C2 groups only contain members of the potato. Interestingly, groups VII and VIII, which are not grouped with Arabidopsis, were also found in eggplants, suggesting that the LAC gene family may have evolved differently from Arabidopsis in Solanaceae crops. Nine pairs of segmental duplication genes (16 StLACs) and five pairs of tandem duplications (10 StLACs) were identified in the StLAC gene family. Of these 26 StLACs, 8 belong to the C2 group, 12 belong to the C5 group, 4 belong to the C4 group, and 2 belong to the C1 group. The above findings indicate that the StLAC gene family expands through segmental duplication, and the C5 group plays an important role in the expansion of the StLAC gene family, which is also the main reason why the C5 group has far more members than the other groups. Through collinearity analysis, it was found that Bo4g065760 in cabbage, Os01g62490 in rice, and Zm00001d028599 in corn are homologous to Soltu09G001990 in potato. These LAC genes may perform similar functions in different plants. The pI values of forty StLACs are higher than 7.0, and only five StLACs protein have pI values lower than 7.00, suggesting that they may function in an alkaline environment. Regarding the prediction analysis of subcellular locations, the vast majority of laccases (24 out of 45) are located extracellularly. Most potato laccases are secretory proteins, and this result is similar to that of the research on soybean (Glycine max) laccases [38].
Laccase is a poly-copper oxidase encoded by a multigene family [17]. In plant cells, it participates in the transport and deposition of lignin to the cell wall [30]. Lignin serves as a core structural element for cell construction and tissue differentiation during plant development [32]. Moreover, when plants face external environmental stresses such as salinity and drought, lignin plays an extremely critical and irreplaceable role in defense mechanisms like cell wall strengthening and reactive oxygen species removal. It is also of great significance for maintaining plant cell homeostasis and overall stress resistance [39]. In Arabidopsis, AtLAC4 and AtLAC17 are involved in the polymerization of lignin in the stems, facilitating the synthesis and deposition of lignin in the cell wall. This, in turn, enhances the development and structural support of vascular bundles, ensuring the effective transport of water, nutrients, and other substances in plants, which plays a crucial role in the growth, development, and abiotic stress responses of Arabidopsis thaliana [30,40,41]. Soltu09G001990, Soltu09G000860, Soltu09G001900, Soltu09G001980, and AtLAC17 are grouped together and specifically expressed in potato roots, leaves, shoots, petals, and mature flowers. Soltu09G000560, Soltu10G019900, Soltu10G022800, and AtLAC4 are grouped into one group, which is specifically expressed in roots, tubers, and stolons, and may participate in the development of vascular bundles and the deposition of lignin in tissues, thus affecting plant growth and development. Additionally, AtLAC15 is related to the oxidative polymerization of flavonoids in the Arabidopsis seed coat [4], while Soltu04G028320 and AtLAC15 are clustered together (Figure 2). This gene is highly expressed in the carpels (FPKM > 60), potentially related to the active substance metabolism at this site. KEGG enrichment analysis revealed that laccase, as a copper-containing polyphenol oxidase, is highly likely to intervene in redox reactions to precisely regulate the concentration and activity of related metabolites in the metabolic pathways of ascorbate and aldarate. This regulatory effect is of great significance, as it not only maintains the stability of plant basic metabolism but also closely links to the stress responses of plants when facing external stimuli, laying a solid foundation for their physiological defense systems. At the same time, GO enrichment analysis showed that StLAC genes were significantly related to lignin catabolism, with oxidoreductase activity and highly enriched copper ion binding functions. This indicates that StLAC genes are likely involved in constructing and remodeling plant cell structures, providing key physical and biochemical support for plant morphogenesis and material transport.
In soybeans, most GmLACs are strongly inhibited under drought conditions. However, GmLAC7/48/49 are highly induced after 6 to 24 h of drought treatment [42]. In salt-sensitive and salt-tolerant maize varieties, it was found that the expression level of ZmLAC17 (Zm00001d028599) in the tolerant variety shows a more significant upward trend under salt stress [32]. The rice OsChI1 gene, which targets the plasma membrane, is highly expressed under multiple abiotic stresses. Overexpressing this gene can enhance the tolerance of Arabidopsis to drought and salinity stresses [10]. In the DM materials, we identified that seven StLAC genes were differentially expressed under abiotic stresses. Among them, one StLAC was up-regulated upon mannitol treatment, while the other six were down-regulated under NaCl stress. Through further qPCR analysis, we discovered that the expression level of Soltu04G028320 gradually increased after 3 h of salt treatment, reaching 13.7 times that of the control at 24 h. Moreover, the expression level of Soltu09G001990 also significantly increased under NaCl treatment. The Soltu09G001990 and Soltu04G028320 genes may play positive roles in the plant’s response to NaCl stress, but their detailed biological functions remain to be further investigated.
5. Conclusions
In this study, 45 members of the potato LAC gene family (StLACs) were identified at the whole-genome level of the potato. These genes are unevenly distributed across 12 chromosomes. Based on their protein structures and phylogenetic characteristics, the 45 StLACs were classified into five sub-families. Segmental duplication and tandem duplication events play a vital role in the expansion of the potato LAC gene family. Through expression analysis, some candidate genes (Soltu09G001990 and Soltu04G028320) that might be involved in potato growth, development, and responses to biotic/abiotic stresses were preliminarily screened. This study provides a theoretical basis for further understanding the characteristics of the StLAC gene family and delving deeper into the biological functions of StLAC genes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy15030585/s1. Table S1: Primers utilized in PCR; Table S2: Characteristic features of 45 StLACs; Table S3: The conserved motifs identified by MEME among StLACs: Table S4: The FPKM value of StLACs in different tissues of DM potato. Table S5: GO and KEGG enrichment; Table S6: The FPKM values of StLOXs in DM potato under different treatments.
Author Contributions
Conceptualization, Y.L. and J.Z.; methodology, H.L. and Z.L. (Zhen Liu); software, H.L., Z.L. (Zhen Liu) and Z.L. (Zhitao Li); validation, H.L., Y.L. and Z.L. (Zhen Liu); formal analysis, J.Z. and Z.L. (Zhitao Li); investigation, H.L., J.Q. and J.Z.; resources, Y.L.; data curation, H.L., W.W. and X.Q.; writing—original draft preparation, H.L.; writing—review and editing, J.Z. and Z.L. (Zhen Liu); visualization, H.L., M.B. and C.G.; supervision, Y.L.; project administration, Y.L. and Z.L. (Zhen Liu); funding acquisition, Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
Central Government Guide Local Science and Technology Development Fund Project (23ZYQJ304), Gansu Science and Technology Program Project (24JRRA845), Gansu Provincial Colleges and Universities Industrial Support Program Project (2024CYZC-29), National Modern Agricultural Industrial Technology System (CARS-09-P14), and the “Fuxi Talent” Program project (Gaufx-02Y04) of Gansu Agricultural University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and the Supplementary Materials.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Yoshida, H.L. XIII.—Chemistry of lacquer (Urushi). Part I. Communication from the chemical Society of tokio. The United Kingdom of Great Britain and Northern Ireland. J. Chem. Soc. Trans. 1883, 43, 472–486. [Google Scholar] [CrossRef]
- Yao, W.; Liu, M.; Chen, M.; Liu, S.H.C. Genome-wide Identification and Expression Analysis of LAC Gene Family of Amaranthus tricolor L. Chin. Veg. 2024, 1, 48–56. [Google Scholar]
- Liu, N.; Jia, H.; Shen, K.; Cao, Z.; Dong, J. Fungal Laccase: Multi-biofunction and Complicated Natural Substrates. J. Agric. Biotechnol. 2020, 28, 333–341. (In Chinese) [Google Scholar]
- Turlapati, P.V.; Kim, K.W.; Davin, L.B.; Lewis, N.G. The laccase multigene family in Arabidopsis thaliana: Towards addressing the mystery of their gene function(s). Planta 2011, 233, 439–470. [Google Scholar] [CrossRef] [PubMed]
- Ranocha, P.; Chabannes, M.; Chamayou, S.; Danoun, S.; Jauneau, A.; Boudet, A.; Goffner, D. Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol. 2002, 129, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, C.L.; Zhu, M.L.; Yu, Y.; Zhang, Y.; Wei, Z. Generation and characterization of transgenic poplar plants overexpressing a cotton laccase gene. Plant Cell Tissue Organ Cult. 2008, 93, 303–310. [Google Scholar] [CrossRef]
- Liang, M.; Davis, E.; Gardner, D.; Cai, X.; Wu, Y. Involvement of AtLAC15 in lignin synthesis in seeds and root elongation of Arabidopsis. Planta 2006, 224, 1185–1196. [Google Scholar] [CrossRef]
- Cesarino, I.; Araújo, P.; Sampaio Mayer, J.L.; Vicentini, R.; Berthet, S.; Demedts, B.; Vanholme, B.; Boerjan, W.; Mazzafera, P. Expression of SofLAC, a new laccase in sugarcane, restores lignin content but not S: G ratio of Arabidopsis lac17 mutant. J. Exp. Bot. 2013, 64, 1769–1781. [Google Scholar] [CrossRef]
- Wang, Y.X.; Tao, J.L.; Zhu, H.S.; Xu, T.; Zhang, Y.F.; Cen, H.F. Heterologous expression of miR397-5p from Medicago sativa cv. ‘Pianguan’ improves the drought tolerance of tobacco. Acta Prataculturae Sin. 2024, 33, 123–134. [Google Scholar]
- Cho, H.Y.; Lee, C.; Hwang, S.G.; Park, Y.C.; Lim, H.L.; Jang, C.S. Overexpression of the OsChI1 gene, encoding a putative laccase precursor, increases tolerance to drought and salinity stress in transgenic Arabidopsis. Gene 2014, 552, 98–105. [Google Scholar] [CrossRef]
- Hu, Q.; Min, L.; Yang, X.Y.; Jin, S.; Zhang, L.; Li, Y.; Ma, Y.; Qi, X.; Li, D.; Liu, H.; et al. Laccase GhLac1 modulates broad-spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and jasmonic acid synthesis. Plant Physiol. 2018, 176, 1808–1823. [Google Scholar] [CrossRef]
- Stokstad, E. The new potato. Science 2019, 363, 574–577. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Liu, Z.H.; Zeng, Y.T.; Li, Z.T.; Chen, L.M.; Li, H.Y. Genome-wide identification of potato (Solanum tuberosum L.) PAL gene family and its expression analysis in abiotic stress and tuber anthocyanin synthesis. Acta Mater. Sin. 2023, 49, 2978–2990. [Google Scholar]
- Kong, J.J.; Xiong, R.; Qiu, K.L.; Lin, X.; Li, D.; Lu, L.; Zhou, J.; Zhu, S.; Liu, M.; Sun, Q. Genome-Wide Identification and Characterization of the Laccase Gene Family in Fragaria vesca and Its Potential Roles in Response to Salt and Drought Stresses. Plants 2024, 13, 3366. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Ren, C.; Xu, J.; Wang, H.; Wen, B.; Zhao, Q.; Zhang, W.; Yu, G.; Zhang, Y. Genome-wide analysis of the common bean (Phaseolus vulgaris) laccase gene family and its functions in response to abiotic stress. BMC Plant Biol. 2024, 24, 688. [Google Scholar] [CrossRef]
- Xu, X.Y.; Zhou, Y.P.; Wang, B.; Li, D.; Yue, W.; Li, L.; Yue, Z.H.; Wei, W.K. Genome-wide identification and characterization of the laccase gene family in Citrus sinensis. Gene 2019, 689, 114–123. [Google Scholar] [CrossRef]
- Wan, F.; Zhang, L.; Tan, M.; Wang, X.; Wang, G.-L.; Qi, M.; Liu, B.; Gao, J.; Pan, Y.; Wang, Y. Genome-wide identification and characterization of laccase family members in eggplant (Solanum melongena L.). PeerJ 2022, 10, e12922. [Google Scholar] [CrossRef] [PubMed]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, F.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Wang, J.K. Prediction of protein subcellular localization. Proteins: Structure. Funct. Bioinform. 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, Y.; Liang, M.; Kong, W.; Liu, J. The Citrus Laccase Gene CsLAC18 Contributes to Cold Tolerance. Int. J. Mol. Sci. 2022, 23, 14509. [Google Scholar] [CrossRef]
- Hall, B.G. Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- Bailey, T.L.; Mikael, B.; Buske, F.A.; Martin, F.; Grant, C.E.; Luca, C.; Ren, J.Y.; Li, W.F.; William, S.N. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.P.; Guo, A.Y. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Xu, X.; Pan, S.; Cheng, S.; Zhang, B.; Mu, D.; Ni, P.; Zhang, G.; Yang, S.; Li, R.; Wang, J. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. Tbtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, N.; Si, H.J.; Caideron, A. Selection and validation of reference genes for RT-qPCR analysis in potato under abiotic stress. Plant Methods 2017, 13, 85. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Berthet, S.; Demont-Caulet, N.; Pollet, B.; Bidzinski, P.; Cézard, L.; Le-Bris, P.; Borrega, N.; Hervé, J.; Blondet, E.; Balzergue, S.; et al. Disruption of Laccase4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 2011, 23, 1124–1137. [Google Scholar] [CrossRef]
- Sedbrook, J.C.; Carroll, K.L.; Hung, K.F.; Massonance, P.H.; Somerville, C.R. The arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 2002, 14, 1635–1648. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Nakashima, J.; Chen, F.; Yin, Y.; Fu, C.; Yun, J.; Shao, H.; Wang, X.; Wang, Z.Y.; Dixon, R.A. Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 2013, 25, 3976–3987. [Google Scholar] [CrossRef]
- Pourcel, L.; Routaboul, J.M.; Kerhoas, L.; Caboche, M.; Lepiniec, L.; Debeaujon, I. TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in the oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 2005, 17, 2966–2980. [Google Scholar] [CrossRef]
- Wang, T.H.; Liu, Y.; Zou, K.L. The Analysis, Description, and Examination of the Maize LAC Gene Family’s Reaction to Abiotic and Biotic Stress. Genes 2024, 15, 749. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Feng, J.J.; Jia, W.; Fan, P.; Bao, H.; Li, Y. Genome-wide Identification of Sorghum bicolor laccases reveals potential targets for lignin modification. Front. Plant Sci. 2017, 8, 714. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.P.; Lin, J.S.; Dong, F.; Ma, Q.; Ming, R. Genomic and allelic analyses of laccase genes in sugarcane (Saccharum spontaneum L.). Trop. Plant Biol. 2019, 12, 219–229. [Google Scholar] [CrossRef]
- Liu, Q.Q.; Luo, L.; Wang, X.X.; Shen, Z.; Zheng, L. Comprehensive Analysis of Rice Laccase Gene (OsLAC) Family and Ectopic Expression of OsLAC10 Enhances Tolerance to Copper Stress in Arabidopsis. Int. J. Mol. Sci. 2017, 18, 209. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, G.; Zheng, K.J.; Zhu, X.; Ma, J.; Wang, D.; Tang, K.; Feng, X.; Leng, J.; Yu, H.; et al. The soybean laccase gene family: Evolution and possible roles in plant defense and stem strength selection. Genes 2019, 10, 701. [Google Scholar] [CrossRef]
- Yadav, S.; Chattopadhyay, D. Lignin: The Building Block of Defense Responses to Stress in Plants. J. Plant Growth Regul. 2023, 42, 6652–6666. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Lise, J.; Serge, B.; Julien, M. Identification of laccases involved in lignin polymerization and strategies to deregulate their expression in order to modify lignin content in Arabidopsis and poplar. BMC Proc. 2011, 5, O39. [Google Scholar] [CrossRef]
- Liu, M.Y.; Dong, H.; Wang, M. Evolutionary divergence of function and expression of laccase genes in plants. J. Genet. 2020, 99, 23. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).