Genome-Wide Identification and Expression Analysis of the Pepper β-1,3-gucanase Gene Family in Response to Phytophthora capsici Stresses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Genome-Wide Characterization of the β-1,3-glucanase Gene Family in Peppers
2.3. Phylogenetic Analysis of β-1,3-glucanase Family Genes from Pepper, Arabidpsis, and Tomato
2.4. CaBGs Gene Motif and Structure Analysis
2.5. Cis-Element and Synteny Analysis of CaBGs
2.6. Analysis of Expression Patterns in Roots and Stems of Pepper Plants Under P. capsici Biostress
2.7. RNA Extraction and cDNA Reverse Transcription
2.8. RT-qPCR Analysis
2.9. Data Collection and Statistical Analysis
3. Results
3.1. Identification and Physicochemical Characteristics of the β-1,3-glucanase (BG) Family Members in Pepper
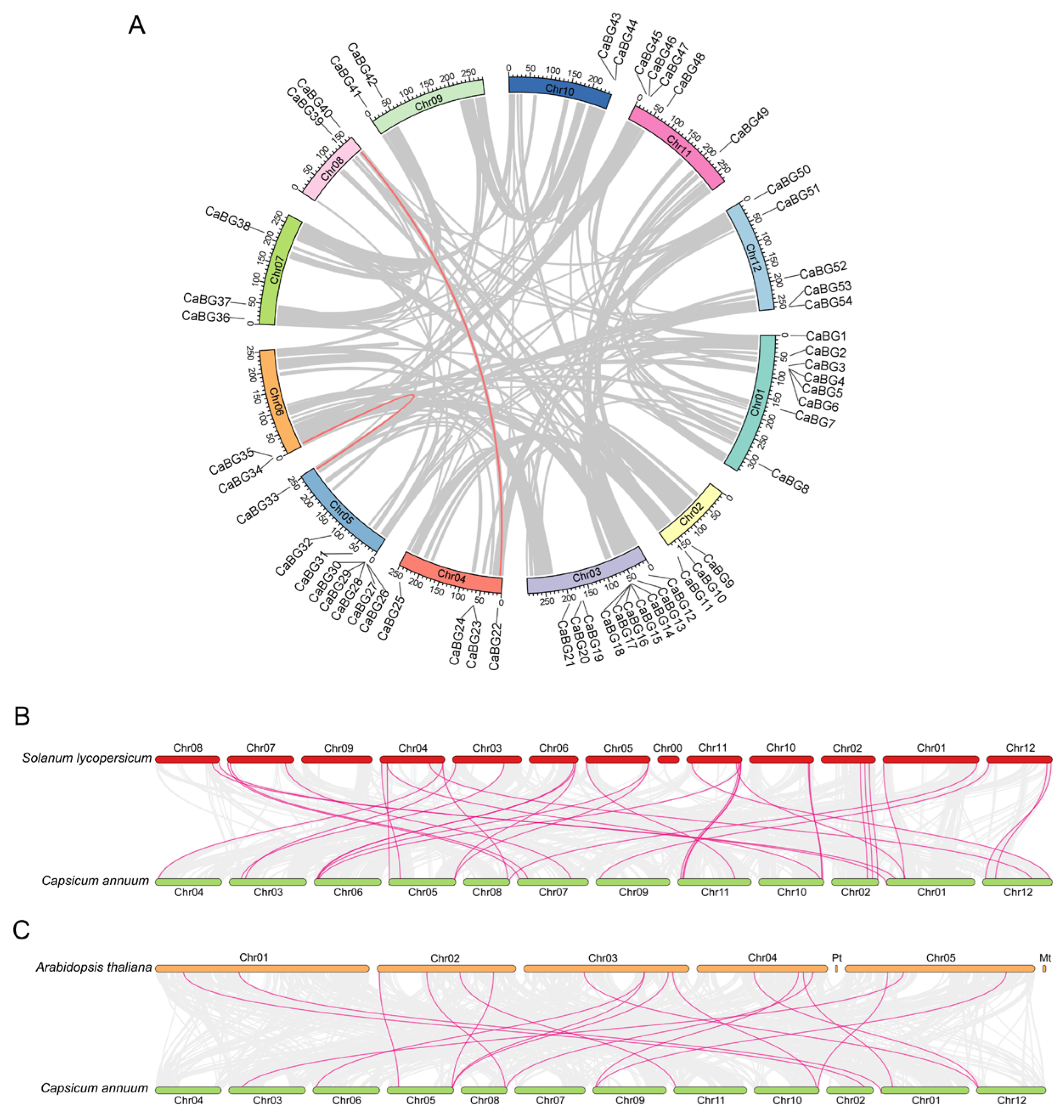
3.2. Chromosome Localization of Pepper BG Genes
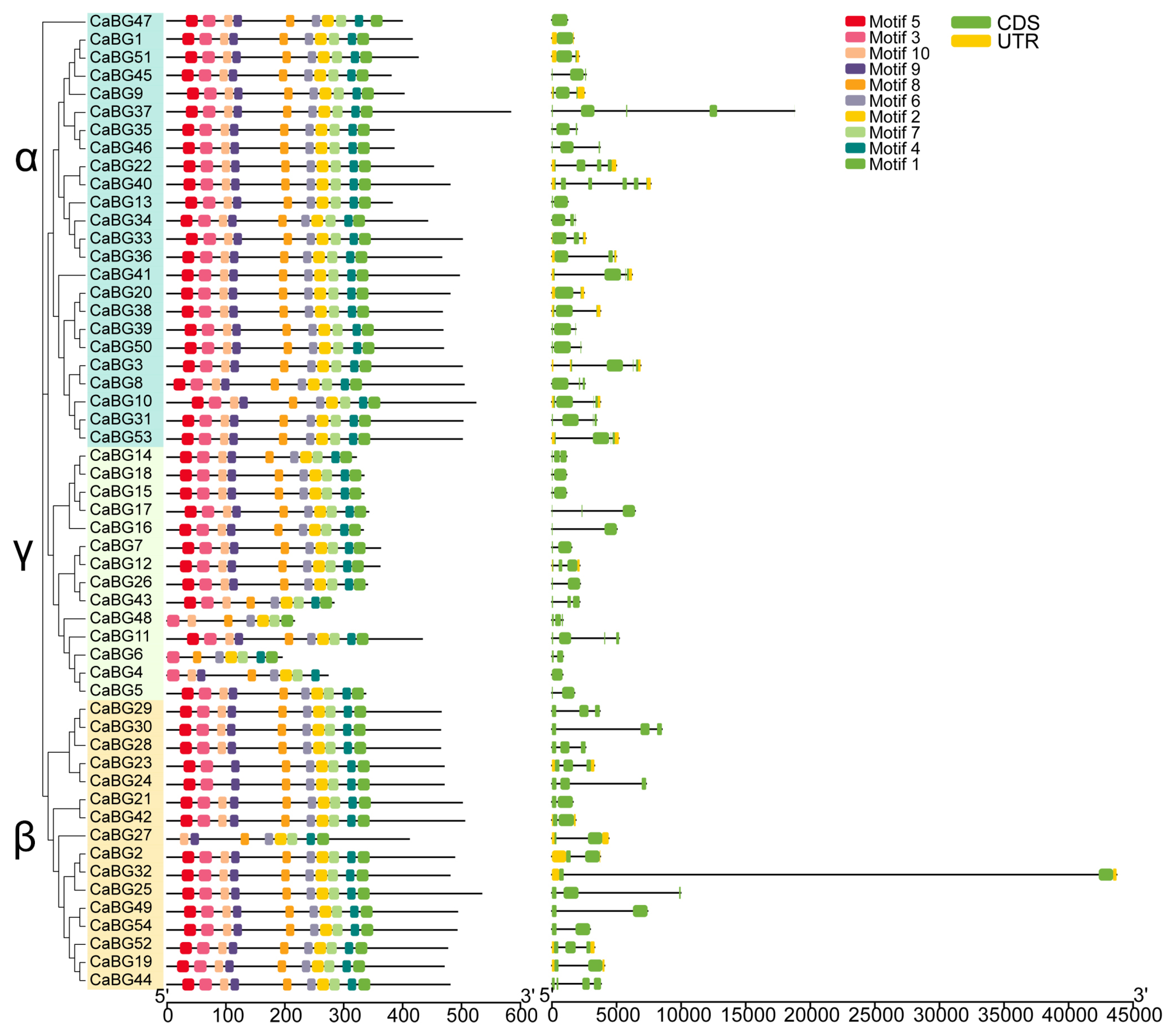
3.3. Phylogenetic Tree Analysis of CaBG Family Genes
3.4. Gene Structure and Conserved Motif Analysis of Pepper BG Genes
3.5. Synteny Analysis of BG Genes Among Pepper, Arabidopsis, and Tomato
3.6. Analysis of CaBG Promoter Cis-Elements
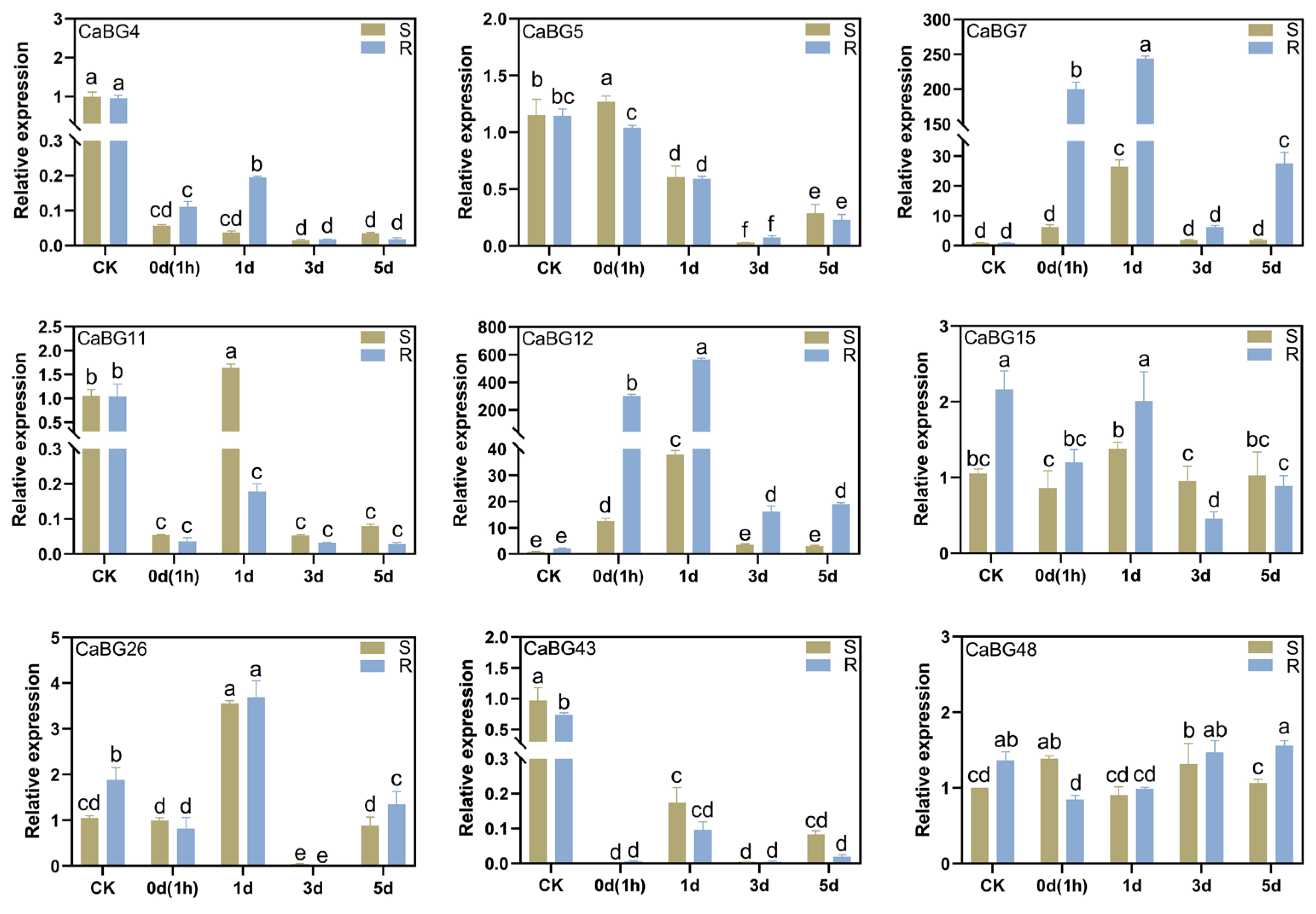
3.7. Expression Pattern Analysis of CaBG Genes Under P. capsici Infestans Stress
3.8. RT-qPCR Analysis of the Pepper BG Gene Family
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| BG | β-1,3-glucanase |
| P. capsici | Phytophthora capsici |
| R | resistant |
| S | susceptible |
| D | day |
| hpi | hour post-infection |
References
- Li, Q.; Zhu, H.; Ai, G.; Yu, J.; Dou, D. Plant genes related to Phytophthora pathogens resistance. Phytopathol. Res. 2024, 6, 15. [Google Scholar] [CrossRef]
- Li, Y.; Yu, T.; Wu, T.; Wang, R.; Wang, H.; Du, H.; Xu, X.; Xie, D.; Xu, X. The dynamic transcriptome of pepper (Capsicum annuum) whole roots reveals an important role for the phenylpropanoid biosynthesis pathway in root resistance to Phytophthora capsici. Gene 2020, 728, 144288. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Li, X.; Lu, X.; Hu, J.; Zhang, C.; Shi, L.; Zhu, C.; Guo, Y.; Wang, X.; Huang, Z.; et al. Identification and Functional Analysis of the Ph-2 Gene Conferring Resistance to Late Blight (Phytophthora infestans) in Tomato. Plants 2024, 13, 3572. [Google Scholar] [CrossRef] [PubMed]
- Lei, G.; Zhou, K.-H.; Chen, X.-J.; Huang, Y.-Q.; Yuan, X.-J.; Li, G.-G.; Xie, Y.-Y.; Fang, R. Transcriptome and metabolome analyses revealed the response mechanism of pepper roots to Phytophthora capsici infection. BMC Genom. 2023, 24, 626. [Google Scholar] [CrossRef]
- De Andrade, L.D.; Branco, I.; Choupina, A. Phytopathogenic oomycetes: A review focusing on Phytophthora cinnamomi and biotechnological approaches. Mol. Biol. Rep. 2020, 47, 9179–9188. [Google Scholar] [CrossRef]
- Volynchikova, E.; Kim, K.D. Biological Control of Oomycete Soilborne Diseases Caused by Phytophthora capsici, Phytophthora infestans, and Phytophthora nicotianae in Solanaceous Crops. Mycobiology 2022, 50, 269–293. [Google Scholar] [CrossRef]
- Gow, N.A.R.; Latge, J.P.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5, 28513415. [Google Scholar] [CrossRef]
- Edo, G.I.; Ndudi, W.; Makia, R.S.; Ainyanbhor, I.E.; Yousif, E.; Gaaz, T.S.; Zainulabdeen, K.; Jikah, A.N.; Opiti, R.A.; Akpoghelie, P.O.; et al. Beta-glucan: An overview in biological activities, derivatives, properties, modifications and current advancements in food, health and industrial applications. Process Biochem. 2024, 147, 347–370. [Google Scholar] [CrossRef]
- Islam, M.M.; El-Sappah, A.H.; Ali, H.M.; Zandi, P.; Huang, Q.; Soaud, S.A.; Alazizi, E.M.Y.; Wafa, H.A.; Hossain, M.A.; Liang, Y. Pathogenesis-related proteins (PRs) countering environmental stress in plants: A review. S. Afr. J. Bot. 2023, 160, 414–427. [Google Scholar] [CrossRef]
- Zhang, X.; Zheng, P.; Wen, X.; Bei, Z. Comprehensive Analysis of β-1,3-Glucanase Genes in Wolfberry and Their Implications in Pollen Development. Plants 2025, 14, 52. [Google Scholar]
- Hao, X.; Zhang, Y.; Zhang, H.; Yang, G.; Liu, Z.; Lv, H.; Zhou, X. Genome-Wide Identification, Expression and Interaction Analysis of GLN Gene Family in Soybean. Curr. Issues Mol. Biol. 2024, 46, 14154–14167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Bei, Z.; Li, J.; Ma, H.; Wang, C.; Xu, W.; Ren, Y.; Zhou, J.; Yan, X. Regulatory Mechanisms of Pollen Development: Transcriptomic and Bioinformatic Insights into the Role of β-1,3 Glucanase Gene (LbGlu1) in Lycium barbarum. Horticulturae 2024, 10, 512. [Google Scholar] [CrossRef]
- Liu, H.; Lu, X.; Li, M.; Lun, Z.; Yan, X.; Yin, C.; Yuan, G.; Wang, X.; Liu, N.; Liu, D.; et al. Plant immunity suppression by an exo-β-1,3-glucanase and an elongation factor 1α of the rice blast fungus. Nat. Commun. 2023, 14, 5491. [Google Scholar] [CrossRef]
- Balasubramanian, V.; Vashisht, D.; Cletus, J.; Sakthivel, N. Plant β-1,3-glucanases: Their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 2012, 34, 1983–1990. [Google Scholar] [CrossRef]
- Aimanianda, V.; Simenel, C.; Garnaud, C.; Clavaud, C.; Tada, R.; Barbin, L.; Mouyna, I.; Heddergott, C.; Popolo, L.; Ohya, Y.; et al. The Dual Activity Responsible for the Elongation and Branching of β-(1,3)-Glucan in the Fungal Cell Wall. mBio 2017, 8, e00619-17. [Google Scholar] [CrossRef]
- Doxey, A.C.; Yaish, M.W.; Moffatt, B.A.; Griffith, M.; McConkey, B.J. Functional divergence in the Arabidopsis beta-1,3-glucanase gene family inferred by phylogenetic reconstruction of expression states. Mol. Biol. Evol. 2007, 24, 1045–1055. [Google Scholar] [CrossRef]
- Paniagua, C.; Perry, L.; Benitez-Alfonso, Y. A phylogenetic and transcriptomic study of the β-1, 3-glucanase family in tomato identifies candidate targets for fruit improvement. bioRxiv 2021. [Google Scholar] [CrossRef]
- Cheong, Y.H.; Kim, C.Y.; Chun, H.J.; Moon, B.C.; Park, H.C.; Kim, J.K.; Lee, S.; Han, C.; Lee, S.Y.; Cho, M.J. Molecular cloning of a soybean class III beta-1,3-glucanase gene that is regulated both developmentally and in response to pathogen infection. Plant Sci. Int. J. Exp. Plant Biol. 2000, 154, 71–81. [Google Scholar] [CrossRef]
- Su, Y.; Wang, Z.; Liu, F.; Li, Z.; Peng, Q.; Guo, J.; Xu, L.; Que, Y. Isolation and Characterization of ScGluD2, a New Sugarcane beta-1,3-Glucanase D Family Gene Induced by Sporisorium scitamineum, ABA, H2O2, NaCl, and CdCl2 Stresses. Front. Plant Sci. 2016, 7, 1348. [Google Scholar] [CrossRef]
- Zhang, S.B.; Zhang, W.J.; Zhai, H.C.; Lv, Y.Y.; Cai, J.P.; Jia, F.; Wang, J.S.; Hu, Y.S. Expression of a wheat β-1,3-glucanase in Pichia pastoris and its inhibitory effect on fungi commonly associated with wheat kernel. Protein Exp. Purif. 2019, 154, 134–139. [Google Scholar] [CrossRef]
- Taif, S.; Zhao, Q.; Pu, L.; Li, X.; Liu, D.; Cui, X. A β-1,3-glucanase gene from Panax notoginseng confers resistance in tobacco to Fusarium solani. Ind. Crops Prod. 2020, 143, 111947. [Google Scholar] [CrossRef]
- Shen, Y.; Mao, L.; Zhou, Y.; Sun, Y.; Liu, Z.; Liang, C. Integrated Transcriptome and Metabolome Analysis Revealed the Molecular Mechanism of Anthocyanin Synthesis in Purple Leaf Pepper (Capsicum annuum L.) under Different Light Intensities. Agronomy 2023, 9, 814. [Google Scholar]
- Quesada-Ocampo, L.M.; Parada-Rojas, C.H.; Hansen, Z.; Vogel, G.; Smart, C.; Hausbeck, M.K.; Carmo, R.M.; Huitema, E.; Naegele, R.P.; Kousik, C.S.; et al. Phytophthora capsici: Recent Progress on Fundamental Biology and Disease Management 100 Years After Its Description. Annu. Rev. Phytopathol. 2023, 61, 185–208. [Google Scholar] [CrossRef]
- Guo, Y.; Qiu, X.; Lai, B.; Ou, C.; Wang, H.; Guo, H.; Li, L.; Lin, L.; Yu, D.; Liu, W.; et al. TBCC Domain-Containing Protein Regulates Sporulation and Virulence of Phytophthora capsici via Nutrient-Responsive Signaling. Int. J. Mol. Sci. 2024, 25, 12301. [Google Scholar] [CrossRef]
- López-Reyes, P.K.; De la Torre-Zavala, S.; Cortés-González, M.M.; Galán-Wong, L.J.; Avilés-Arnaut, H. Biological Control of Streptomyces sp. PR69 Against Phytophthora capsici and Its Growth-Promoting Effects on Plants. Horticulturae 2024, 10, 1365. [Google Scholar] [CrossRef]
- Zhou, J.; Liang, J.; Zhang, X.; Wang, F.; Qu, Z.; Gao, T.; Yao, Y.; Luo, Y. Trichoderma brevicompactum 6311: Prevention and Control of Phytophthora capsici and Its Growth-Promoting Effect. J. Fungi 2025, 11, 105. [Google Scholar] [CrossRef]
- Candole, B.L.; Conner, P.J.; Ji, P.J.H. Screening Capsicum annuum Accessions for Resistance to Six Isolates of Phytophthora capsici. HortScience 2010, 45, 254–259. [Google Scholar]
- Zhang, K.; Wang, X.; Chen, S.; Liu, Y.; Zhang, L.; Yang, X.; Yu, H.; Cao, Y.; Zhang, L.; Cai, C.; et al. The gap-free assembly of pepper genome reveals transposable-element-driven expansion and rapid evolution of pericentromeres. Plant Commun. 2025, 6, 101177. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Cackett, L.; Luginbuehl, L.H.; Schreier, T.B.; Lopez-Juez, E.; Hibberd, J.M. Chloroplast development in green plant tissues: The interplay between light, hormone, and transcriptional regulation. New Phytol. 2022, 233, 2000–2016. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wan, H.; Yuan, W.; Ruan, M.; Ye, Q.; Wang, R.; Li, Z.; Zhou, G.; Yao, Z.; Zhao, J.; Liu, S.; et al. Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem. Biophys. Res. Commun. 2011, 416, 24–30. [Google Scholar] [CrossRef]
- Benitez-Alfonso, Y.; Faulkner, C.; Pendle, A.; Miyashima, S.; Helariutta, Y.; Maule, A. Symplastic intercellular connectivity regulates lateral root patterning. Dev. Cell 2013, 26, 136–147. [Google Scholar] [CrossRef]
- Vaddepalli, P.; Fulton, L.; Schneitz, K. Asymmetric Redundancy of ZERZAUST and ZERZAUST HOMOLOG in Different Accessions of Arabidopsis thaliana. G3 2019, 9, 2245–2252. [Google Scholar] [CrossRef]
- Pei, Y.; Xue, Q.; Zhang, Z.; Shu, P.; Deng, H.; Bouzayen, M.; Hong, Y.; Liu, M. β-1,3-GLUCANASE10 regulates tomato development and disease resistance by modulating callose deposition. Plant Physiol. 2023, 192, 2785–2802. [Google Scholar] [CrossRef]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef]
- Zhang, X.; Mo, Y.; Zhou, H.; Li, M.; Cheng, H.; Li, P.; Zhang, R.; Huang, Y.; Wang, Y.; Xu, J.; et al. CaHY5 Mediates UV-B Induced Anthocyanin Biosynthesis in Purple Pepper. Agronomy 2025, 15, 28. [Google Scholar]
- Liu, J.; Cheng, Y.; Ruan, M.; Ye, Q.; Wang, R.; Yao, Z.; Zhou, G.; Liu, C.; Wan, H. Phylogenetic, Structural, and Evolutionary Insights into Pepper NBS-LRR Resistance Genes. Int. J. Mol. Sci. 2025, 26, 1828. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Weng, Y.; Chen, Y.; Liu, K.; Liu, Y.; Zhang, K.; Shi, L.; He, S.; Liu, Z. CaARP1/CaSGT1 Module Regulates Vegetative Growth and Defense Response of Pepper Plants against Phytophthora capsici. Plants 2024, 13, 2849. [Google Scholar] [CrossRef]
- Foster, J.M.; Hausbeck, M.K. Resistance of Pepper to Phytophthora Crown, Root, and Fruit Rot Is Affected by Isolate Virulence. Plant Dis. 2010, 94, 24–30. [Google Scholar] [CrossRef]
- McGrath, M.T.; Strauss, J.; Dillard, H.R. First Report of Phytophthora Blight Caused by Phytophthora capsici on Snap Bean in New York. Plant Dis. 2011, 95, 1028. [Google Scholar] [CrossRef]
- Sanogo, S.; Lamour, K.; Kousik, C.S.; Lozada, D.N.; Parada-Rojas, C.H.; Quesada-Ocampo, L.M.; Wyenandt, C.A.; Babadoost, M.; Hausbeck, M.K.; Hansen, Z.; et al. Phytophthora capsici, 100 Years Later: Research Mile Markers from 1922 to 2022. Phytopathology 2023, 113, 921–930. [Google Scholar] [CrossRef]
- Enoki, S.; Fujimori, N.; Yamaguchi, C.; Hattori, T.; Suzuki, S. High Constitutive Overexpression of Glycosyl Hydrolase Family 17 Delays Floral Transition by Enhancing FLC Expression in Transgenic Arabidopsis. Plants 2017, 6, 31. [Google Scholar] [CrossRef]
- Adams, D.J. Fungal cell wall chitinases and glucanases. Microbiology 2004, 150, 2029–2035. [Google Scholar] [CrossRef]
- Roncero, C.; Vázquez de Aldana, C.R. Glucanases and Chitinases. Curr. Top. Microbiol. Immunol. 2020, 425, 131–166. [Google Scholar] [CrossRef]
- Linthorst, H.J.; Melchers, L.S.; Mayer, A.; van Roekel, J.S.; Cornelissen, B.J.; Bol, J.F. Analysis of gene families encoding acidic and basic beta-1,3-glucanases of tobacco. Proc. Natl. Acad. Sci. USA 1990, 87, 8756–8760. [Google Scholar] [CrossRef]
- Jin, W.; Horner, H.T.; Palmer, R.G.; Shoemaker, R.C. Analysis and mapping of gene families encoding beta-1,3-glucanases of soybean. Genetics 1999, 153, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Zha, W.; Cheng, X.; Liu, C.; Lv, L.; Liu, C.; Wang, Z.; Du, B.; Chen, R.; Zhu, L.; et al. A rice β-1,3-glucanase gene Osg1 is required for callose degradation in pollen development. Planta 2011, 233, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Feng, Y.; Fang, S.; Xu, J.; Wang, X.; Guo, W. Genome-wide characterization of the β-1,3-glucanase gene family in Gossypium by comparative analysis. Sci. Rep. 2016, 6, 29044. [Google Scholar] [CrossRef]
- Zhan, C.; Shen, S.; Yang, C.; Liu, Z.; Fernie, A.R.; Graham, I.A.; Luo, J. Plant metabolic gene clusters in the multi-omics era. Trends Plant Sci. 2022, 27, 981–1001. [Google Scholar] [CrossRef]
- Shoji, T.; Yuan, L. ERF Gene Clusters: Working Together to Regulate Metabolism. Trends Plant Sci. 2021, 26, 23–32. [Google Scholar] [CrossRef]
- Polturak, G.; Osbourn, A. Defense-related phenylpropanoid biosynthetic gene clusters in rice. Sci. Bull. 2022, 67, 13–16. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Maere, S.; Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 2009, 10, 725–732. [Google Scholar] [CrossRef]
- Cheng, F.; Wu, J.; Cai, X.; Liang, J.; Freeling, M.; Wang, X. Gene retention, fractionation and subgenome differences in polyploid plants. Nat. Plants 2018, 4, 258–268. [Google Scholar] [CrossRef]
- Feng, X.; Chen, Q.; Wu, W.; Wang, J.; Li, G.; Xu, S.; Shao, S.; Liu, M.; Zhong, C.; Wu, C.I.; et al. Genomic evidence for rediploidization and adaptive evolution following the whole-genome triplication. Nat. Commun. 2024, 15, 1635. [Google Scholar] [CrossRef]
- Van de Rhee, M.D.; Lemmers, R.; Bol, J.F. Analysis of regulatory elements involved in stress-induced and organ-specific expression of tobacco acidic and basic beta-1,3-glucanase genes. Plant Mol. Biol. 1993, 21, 451–461. [Google Scholar] [CrossRef]
- Ali, S.; Chandrashekar, N.; Rawat, S.; Nayanakantha, N.M.C.; Mir, Z.A.; Manoharan, A.; Sultana, M.; Grover, A. Isolation and molecular characterization of pathogenesis related PR2 gene and its promoter from Brassica juncea. Biol. Plant. 2017, 61, 763–773. [Google Scholar] [CrossRef]
- Parveen, S.; Mazumder, M.; Bhattacharya, A.; Mukhopadhyay, S.; Saha, U.; Mukherjee, A.; Mondal, B.; Debnath, A.J.; Das, S.; Sikdar, S.; et al. Identification of Anther-Specific Genes from Sesame and Functional Assessment of the Upstream Region of a Tapetum-Specific β-1,3-glucanase Gene. Plant Mol. Biol. Report. 2018, 36, 149–161. [Google Scholar] [CrossRef]
- Wang, L.; Li, R.; Li, K.; Qu, Z.; Zhou, R.; Lu, G.; Li, P.; Li, G. Genome-wide identification of the grapevine β-1,3-glucanase gene (VviBG) family and expression analysis under different stresses. BMC Plant Biol. 2024, 24, 911. [Google Scholar] [CrossRef] [PubMed]
- Mohammadizadeh-Heydari, N.; Tohidfar, M.; Maleki Zanjani, B.; Mohsenpour, M.; Ghanbari Moheb Seraj, R.; Esmaeilzadeh-Salestani, K. Co-overexpression of chitinase and β-1,3-glucanase significantly enhanced the resistance of Iranian wheat cultivars to Fusarium. BMC Biotechnol. 2024, 24, 35. [Google Scholar] [CrossRef]
- Shi, Y.; Zhang, Y.; Shih, D.S. Cloning and expression analysis of two β-1,3-glucanase genes from Strawberry. J. Plant Physiol. 2006, 163, 956–967. [Google Scholar] [CrossRef]
- Liu, B.; Xue, X.; Cui, S.; Zhang, X.; Han, Q.; Zhu, L.; Liang, X.; Wang, X.; Huang, L.; Chen, X.; et al. Cloning and characterization of a wheat beta-1,3-glucanase gene induced by the stripe rust pathogen Puccinia striiformis f. sp. tritici. Mol. Biol. Rep. 2010, 37, 1045–1052. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Li, D.; Zhang, Y.; Wang, Y.; Song, T.; Yan, C.; Jia, L.; Jiang, H. Genome-Wide Identification and Expression Analysis of the Pepper β-1,3-gucanase Gene Family in Response to Phytophthora capsici Stresses. Agronomy 2025, 15, 802. https://doi.org/10.3390/agronomy15040802
Wang H, Li D, Zhang Y, Wang Y, Song T, Yan C, Jia L, Jiang H. Genome-Wide Identification and Expression Analysis of the Pepper β-1,3-gucanase Gene Family in Response to Phytophthora capsici Stresses. Agronomy. 2025; 15(4):802. https://doi.org/10.3390/agronomy15040802
Chicago/Turabian StyleWang, Han, Dongchen Li, Yu Zhang, Yanping Wang, Tingting Song, Congsheng Yan, Li Jia, and Haikun Jiang. 2025. "Genome-Wide Identification and Expression Analysis of the Pepper β-1,3-gucanase Gene Family in Response to Phytophthora capsici Stresses" Agronomy 15, no. 4: 802. https://doi.org/10.3390/agronomy15040802
APA StyleWang, H., Li, D., Zhang, Y., Wang, Y., Song, T., Yan, C., Jia, L., & Jiang, H. (2025). Genome-Wide Identification and Expression Analysis of the Pepper β-1,3-gucanase Gene Family in Response to Phytophthora capsici Stresses. Agronomy, 15(4), 802. https://doi.org/10.3390/agronomy15040802




