Abstract
Wheat is one of the most essential food crops globally, but diseases significantly threaten its yield and quality, resulting in considerable economic losses. The identification of wheat diseases faces challenges, such as interference from complex environments in the field, the inefficiency of traditional machine learning methods, and difficulty in deploying the existing deep learning models. To address these challenges, this study proposes a multi-scale feature fusion shuffle network model (MFFSNet) for wheat disease identification from complex environments in the field. MFFSNet incorporates a multi-scale feature extraction and fusion module (MFEF), utilizing inflated convolution to efficiently capture diverse features, and its main constituent units are improved by ShuffleNetV2 units. A dual-branch shuffle attention mechanism (DSA) is also integrated to enhance the model’s focus on critical features, reducing interference from complex backgrounds. The model is characterized by its smaller size and fast operation speed. The experimental results demonstrate that the proposed DSA attention mechanism outperforms the best-performing Squeeze-and-Excitation (SE) block by approximately 1% in accuracy, with the final model achieving 97.38% accuracy and 97.96% recall on the test set, which are higher than classical models such as GoogleNet, MobileNetV3, and Swin Transformer. In addition, the number of parameters of this model is only 0.45 M, one-third that of MobileNetV3 Small, which is very suitable for deploying on devices with limited memory resources, demonstrating great potential for practical applications in agricultural production.
1. Introduction
Wheat is one of the most widely cultivated cereals worldwide, ranking as the third most consumed grain after maize and rice. Wheat is also a vital food crop in China, where its planting area and total production hold a significant position on the global scale. According to data from the National Bureau of Statistics, in 2024, China’s wheat planting area was 23,587.4 thousand hectares, with a total production of 140.10 million tons. However, wheat diseases, particularly fungal pathogens such as rusts, crown and root rot, and powdery mildew, severely impact both yield and quality across China’s major wheat belts. According to the National Agro-Tech Extension and Service Center (2022), wheat rusts cause annual production losses of 1.5–3.2 million metric tons in China, with severe epidemics reducing yields by over 40% in affected regions. Accurate wheat disease identification depends on plant pathologists, but farmers in remote areas have limited contact with them due to geographical and economic factors. To address these issues, machine learning offers an efficient alternative for wheat disease identification. Tian et al. [1] analyzed the color, shape, and texture features of wheat leaves affected by fungal diseases like powdery mildew and rust, then applied support vector machines to distinguish and identify these diseases based on the extracted features. Azadbakht et al. [2] used four machine learning techniques for wheat disease classification, achieving the best results with support vector regression. Traditional machine learning for wheat disease identification depends on manual feature extraction, a process that is not only time- and labor-intensive but also constrained by the subjectivity of the extracted features, leading to limited accuracy. Additionally, this approach requires specific models to be designed for different diseases, which lacks generalization.
In recent years, deep learning techniques, such as convolutional neural networks (CNNs) and other artificial neural network (ANN) architectures [3,4], have been extensively applied to agricultural disease identification [5,6,7], with the primary advantage of automatically learning features from image data. This method not only enhances the efficiency of disease diagnosis but also, by leveraging deep learning models, can process vast amounts of data and extract more abstract and higher-level features, leading to improved disease identification accuracy. Additionally, deep learning models offer strong generalization capabilities, enabling them to adapt to diverse datasets and practical application scenarios. Lu et al. [8] designed a real-time automatic wheat disease diagnosis system, utilizing image processing and machine learning to identify wheat diseases in field environments, offering technical support for early detection and precise management of wheat diseases. Nema and Dixit utilized support vector machines (SVMs) to detect and prevent wheat leaf diseases in their study [9]. Their approach involved k-means clustering for classifying wheat leaf samples and SVMs for identifying healthy and diseased leaves. Abade et al. [10] reviewed the application of CNNs for plant disease image recognition, analyzing 121 research papers published over the past decade. The review highlighted trends in CNN applications for plant disease identification, offering insights to guide future studies in the field. In the study by Picon [11], the use of deep convolutional neural networks successfully enabled the classification of crop disease images captured under field conditions using mobile devices. Their experiments, specifically targeting wheat diseases, improved the balanced accuracy to 0.87. Mi et al. proposed a deep-learning-based grading method for wheat stripe rust [12]. The proposed C-DenseNet model significantly improved the classification accuracy of different severity levels of wheat stripe rust.
In addition, CNNs have achieved remarkable results in the disease identification of other agricultural products. Albayrak’s team assessed 20 CNN architectures for mushroom disease classification using a 3195-image dataset [13]. Some of these CNN architectures produced good results. Pan’s team proposed DualTransAttNet, a rapid corn seed classification method using multi-source images [14]. The model combines hyperspectral and RGB data, leveraging CNNs and transformers to capture features, achieving 90.01% accuracy. Ou’s team used hyperspectral imaging and multi-dimensional features for early strawberry disease detection [15]. They extracted and selected key features, then built models with deep learning methods. The CNN model achieved the highest accuracy of 96.6%.
Although the aforementioned methods have shown remarkable results in plant disease identification, they depend heavily on large amounts of training data and powerful computational resources. This poses limitations for applying these methods in resource-constrained settings, particularly in real-time applications on mobile devices. In contrast, lightweight convolutional neural networks (LWCNNs) have shown considerable potential for real-time disease detection on mobile devices thanks to their lower resource requirements, efficient computational performance, and faster processing speeds. Thakur et al. presented an LWCNN-based method for crop leaf disease identification, with the model comprising about 6 million parameters, fewer than traditional CNNs and many of their improved models, allowing the model to maintain high recognition accuracy while running more efficiently on resource-limited devices [16]. Zhang et al. proposed a novel method to achieve rapid and accurate identification of greenhouse cucumber diseases in naturally complex environments [17]. The method reduces the number of parameters and computational cost by optimizing the network’s width, depth, and resolution while maintaining high accuracy. Barman and his team developed a real-time citrus leaf disease classification system based on smartphone images using their own CNN model and MobileNet to enable fast identification of citrus leaf diseases on mobile devices [18].
Lightweight neural networks have demonstrated strong performance in recognizing plant disease images with simple backgrounds. However, they often face difficulties in extracting enough feature details from images with complex backgrounds, primarily due to parameter constraints and insufficient utilization of multi-scale feature information. To overcome this challenge, researchers have turned to attention mechanisms, which are an effective feature extraction technique. Attention mechanisms enhance the model’s ability to focus on important regions within an image, improving its feature recognition ability. Bao et al. introduced an LWCNN model named SimpleNet for the automatic identification of wheat spike diseases from images captured in field conditions via mobile devices [19]. This model employs inverted residual blocks and integrates the convolutional block attention module (CBAM) to enhance attention to critical disease features. Li et al. proposed a transformer module based on spatial convolutional self-attention (SCSA) for the recognition of strawberry diseases in complex backgrounds [20]. The SCSA-Transformer module integrates multi-head self-attention (MSA) with spatial convolutional self-attention, greatly enhancing recognition accuracy and efficiency.
Based on the preceding discussion, this study proposes a lightweight network model (MFFSNet) for wheat disease recognition in complex environments. The model first captures features at multiple scales through a multi-scale feature extraction and fusion module (MFEF) and then further refines these features using lightweight modules. On this basis, an attention mechanism is introduced to enhance the model’s recognition of key features while suppressing interference from complex backgrounds, significantly improving the network’s performance in disease feature representation. The model is characterized by its small size and fast operation speed, meeting the requirements for real-time applications. The experimental results show that this method outperforms other networks in terms of performance. The main contributions of this study can be summarized as follows:
- A novel lightweight network model (MFFSNet) was developed for wheat disease recognition in complex field backgrounds. MFFSNet employs a lightweight module to reduce parameters and enhance operational speed, making it suitable for real-time disease recognition.
- A multi-scale feature extraction (MFE) module was proposed, using dilated convolutions and the inception module to extract multi-scale feature information. This approach captures detailed and contextual information across multiple scales, improving disease recognition accuracy.
- A multi-scale feature fusion (MFF) module was introduced, which integrates the extracted multi-scale features. This module overcomes the challenge of fragmented feature representations by combining information from different scales, thereby enhancing the network’s ability to comprehensively recognize wheat diseases.
- An efficient channel attention mechanism was proposed to improve key feature recognition in complex backgrounds, enhancing model performance and reliability in real-world applications.
2. Materials and Methods
2.1. Datasets
The dataset used in this study was derived from the LWDCD wheat disease dataset [21], with 40% of the images collected in the field, while the remaining images were sourced from previously published datasets. The LWDCD2020 dataset comprises approximately 12,000 images, covering nine categories of wheat diseases with single-disease annotations per image and one additional healthy category introduced to explicitly distinguish healthy plants from diseased cases. The images have been preprocessed for dimensional uniformity. These images feature complex backgrounds, varying capture conditions, different characteristics at various stages of disease progression, and similar features among different wheat diseases. We selected three common wheat diseases in China: crown and root rot, leaf rust, and wheat loose smut, along with healthy wheat as a control category. This selection enables us to focus on specific disease identification challenges while working with limited data resources. The dataset of 3430 images was randomly split into training and testing sets at a 9:1 ratio. This division ensures sufficient samples for robust model training while preserving an adequate testing set for reliable performance evaluation. The test set’s sample distribution is detailed in Table 1. This is crucial for practical applications where data collection may be limited by resources or conditions.

Table 1.
The sample distribution of the test set.
2.2. Image Preprocessing
Before model training, the preprocessing steps for all wheat disease images were as follows: training set images were randomly cropped to 224 × 224 pixels and randomly horizontally flipped to enhance feature diversity while preventing dataset expansion. Considering that the disease distribution is not limited to the center of the leaf, a random cropping method was used to retain more lesion information. The test set images were first resized to 256 × 256 pixels and then center-cropped to a fixed size of 224 × 224 pixels. All images were standardized to reduce data variability, which in turn enhances the model’s convergence speed and improves its generalization ability.
2.3. Experiment Details
To evaluate the effectiveness of the MFFSNet model in wheat disease image recognition and classification, we conducted a series of experiments. The experimental hardware setup in this study includes a Linux operating system and an Intel(R) Xeon(R) E5-2686 v4 processor, while model training and testing are accelerated using an NVIDIA GeForce RTX 3060 GPU. During the model training process, hyperparameters are critical in determining the model’s overall performance. Therefore, after referring to related literature on model design, we set the hyperparameters as shown in Table 2.

Table 2.
Experiment details.
2.4. Evaluation Indexes
Regarding evaluation metrics, relying solely on accuracy to assess the model’s performance on wheat disease images is inadequate. When the data are insufficient or imbalanced, the model’s accuracy may appear high, but its overall performance could still be suboptimal. In such cases, the model may perform well in minority classes, but it is also prone to misclassification, which negatively impacts its practical effectiveness. To address this, we employ four evaluation metrics: accuracy, precision, recall, and F1-score—to comprehensively evaluate the performance of the proposed method. The calculation formulas for these evaluation metrics are presented below:
Here, TP (true positive) represents the number of samples that are correctly predicted as positive, FP (false positive) represents the number of samples that are incorrectly predicted as positive, FN (false negative) represents the number of samples incorrectly predicted as negative, and TN (true negative) represents the number of samples correctly predicted as negative.
2.5. Model Structure
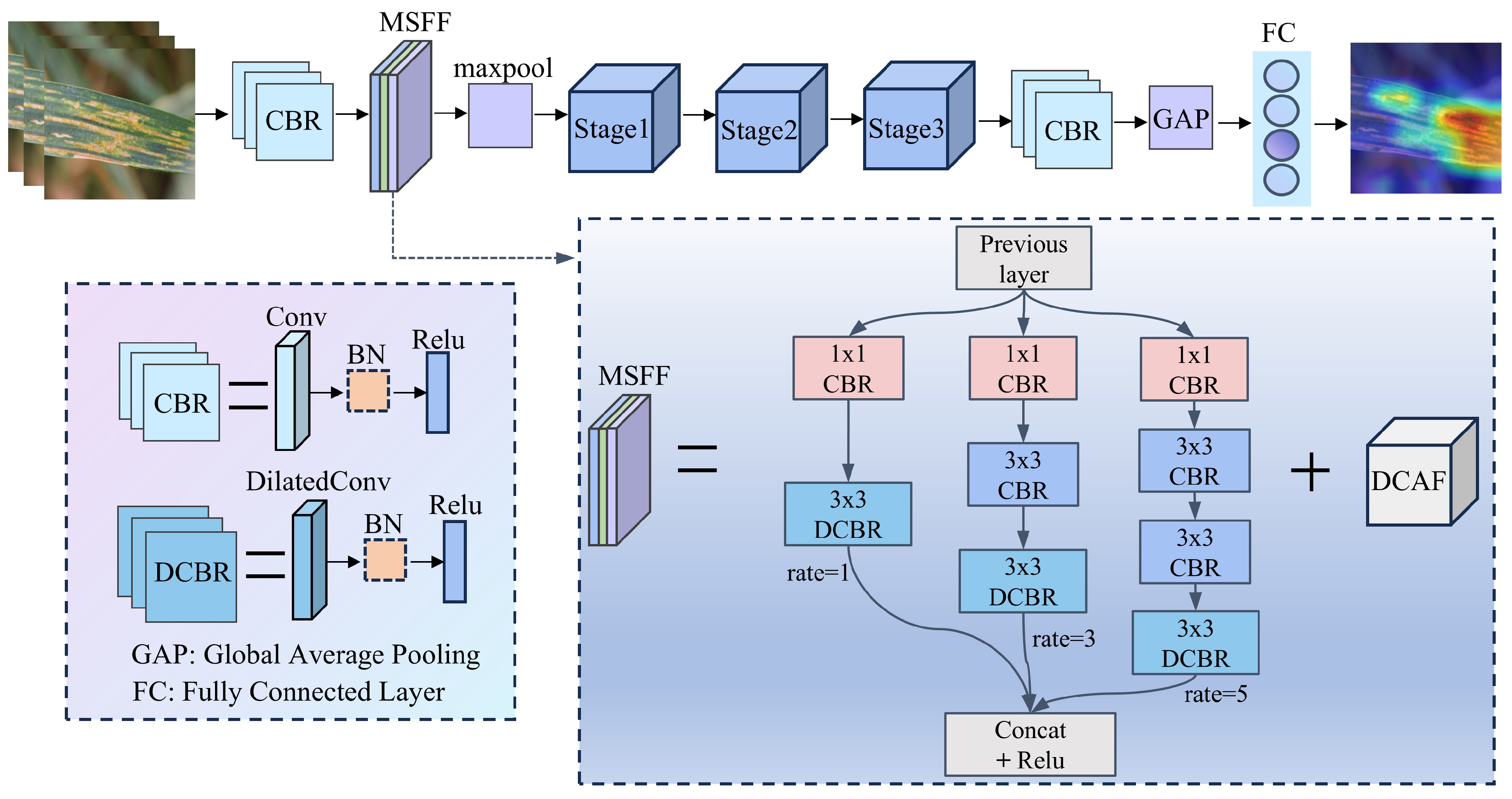
The proposed MFFSNet mainly consists of the MFEF module and three key stages, achieving efficient extraction and recognition of wheat disease features. The MFEF module serves as the core of MFFSNet, effectively capturing fine textures and macrostructures in images through a multi-scale feature fusion strategy, providing rich feature support for disease recognition. The structure of MFFSNet is shown in Figure 1. MFFSNet first employs a CBR layer to perform preliminary downsampling on the input image, extracting important features and reducing computational load. Subsequently, the MFEF module is applied to further extract multi-scale features, and, through the feature fusion strategy, it reduces information loss and enhances the network’s ability to express disease features. Following this, MaxPool is used for downsampling, reducing feature dimensions and laying the foundation for deep feature extraction. The network then proceeds with three stages of deep feature extraction, with each stage consisting of a different number of units to accommodate varying levels of feature learning requirements.

Figure 1.
Structure of MFFSNet.
The architecture of MFFSNet is shown in Table 3. Referring to classic network models, for multi-channel input images (each sized ), an output feature map (sized ) is obtained through a convolution layer (Conv1 with a kernel size of and ), batch normalization (BN) [22], and ReLU6 activation function. Subsequently, the feature map passes through the MFEF module, which includes multi-scale feature extraction and deep attention feature fusion, maintaining an output channel count of 24 with the feature map size unchanged. Next, a max pooling layer (MaxPool) with a stride of 2 halves the spatial size of the feature map to while retaining 24 channels. Then, the process enters three stage-wise deep feature extraction modules, which progressively increase the number of output channels (from 48 to 96, and then to 192) to deepen feature representation. Finally, through Conv5 and a global average pooling layer (GAP), a fixed-length feature vector is obtained. The final classification is achieved through a fully connected layer (FC), resulting in the final category output. In MFFSNet, the specific output channel count and feature map size at each stage are designed to help the model capture image features at different levels, enhancing the accuracy of wheat disease identification.

Table 3.
Layered description of the proposed MFFSNet.
2.6. Component Units
In the process of designing the MFFSNet network architecture, we faced the challenge of identifying wheat disease features at different scales. To effectively capture and recognize these disease features, we needed a network structure capable of densely capturing features at different scales. This structure should be able to maintain computational efficiency while enhancing the network’s ability to capture detailed contextual information. The ShuffleNetV2 network architecture [23], with its unique design advantages, served as an inspiration for our approach.
ShuffleNetV2 is a lightweight convolutional neural network. It effectively improves network information flow and parameter efficiency by introducing channel shuffle operations and pointwise grouped convolutions. The core advantage of ShuffleNetV2 lies in its channel shuffle layer, which rearranges the channel dimensions of feature maps to enhance information exchange between different groups, thereby improving the network’s feature representation capability. Moreover, ShuffleNetV2’s pointwise grouped convolutions not only reduce computational cost but also maintain the network’s depth and complexity by utilizing grouped operations. This allows the network to learn rich feature representations despite having fewer parameters.
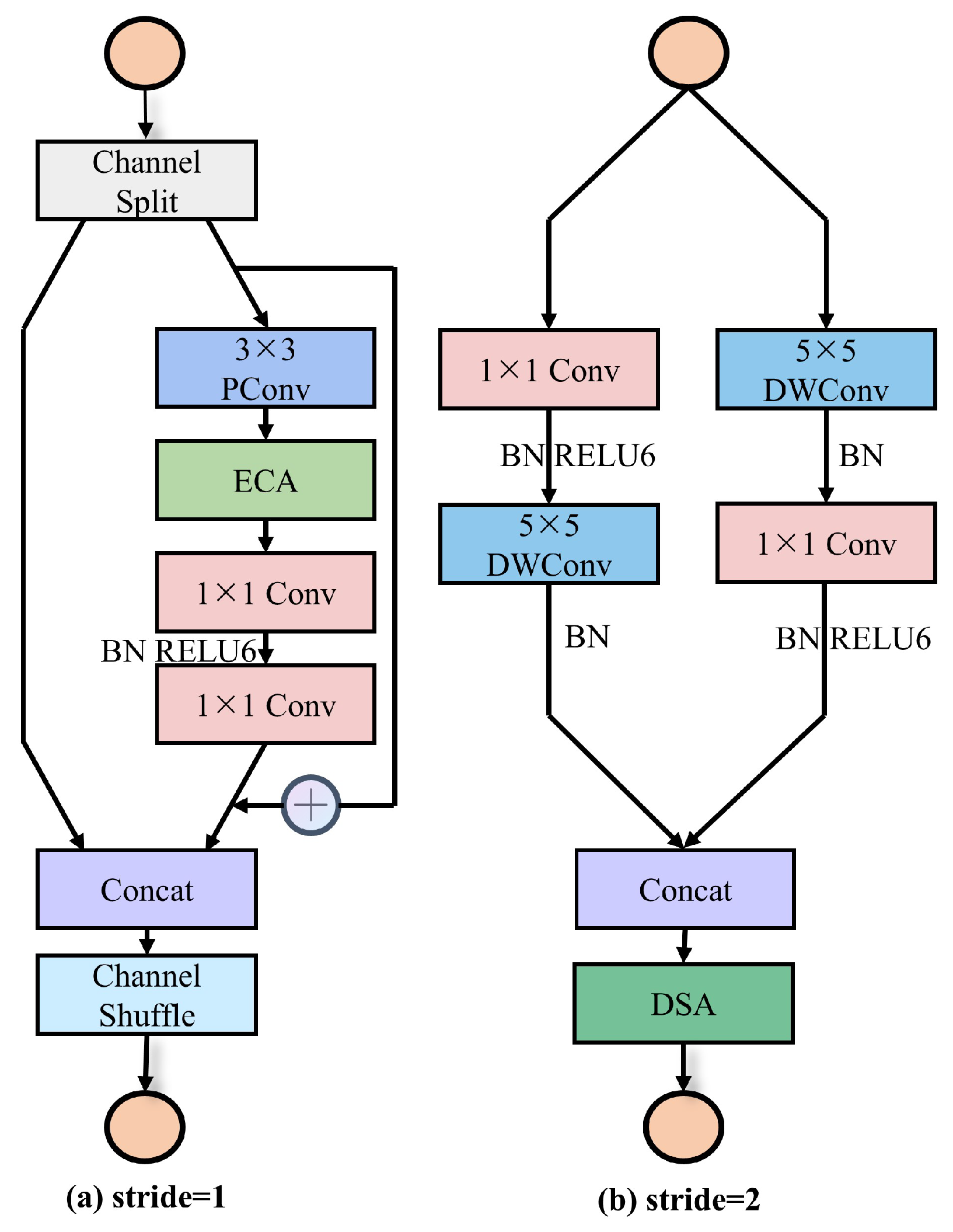
However, the basic unit of the ShuffleNet network suffers from partial structural redundancy, as well as poor network operation speed. To solve the problem of the ShuffleNetV2, we designed the basic unit of MFFSNet, as shown in Figure 2. Its basic units will be reused in the three stages of MFFSNet for progressive extraction of deep features. To improve the model’s performance and computational efficiency, we introduced key optimizations in the units, including replacing the original depthwise separable convolution (DWConv) [24] with a DWConv to capture a broader spatial context. Additionally, one pointwise convolution layer was removed to reduce the computational burden while preserving feature diversity. Moreover, DSA mechanism was introduced after concat operation, replacing the conventional channel shuffle to improve the model’s capability in recognizing disease features. In the stride = 1 unit, we introduced the Fasternet Block module [25], combined with the efficient channel attention (ECA) mechanism [26]. This module consists of partial convolution (PConv) and two pointwise convolutions (PWConv), reducing redundant computation and memory access while accelerating the network’s processing speed. The receptive field of the PConv + PWConv combination resembles a T-shaped convolution, focusing primarily on the central region, which is critical for the precise localization of disease features. The ECA mechanism is both computationally efficient and parameter-light. When incorporated into PConv, it selectively enhances the extracted features, reinforcing important ones while suppressing irrelevant ones, thus reducing the learning of these redundant features by the subsequent layers and improving the overall computational efficiency.

Figure 2.
MFFSNet component unit. (a) Stride = 1 unit; (b) stride = 2 units.
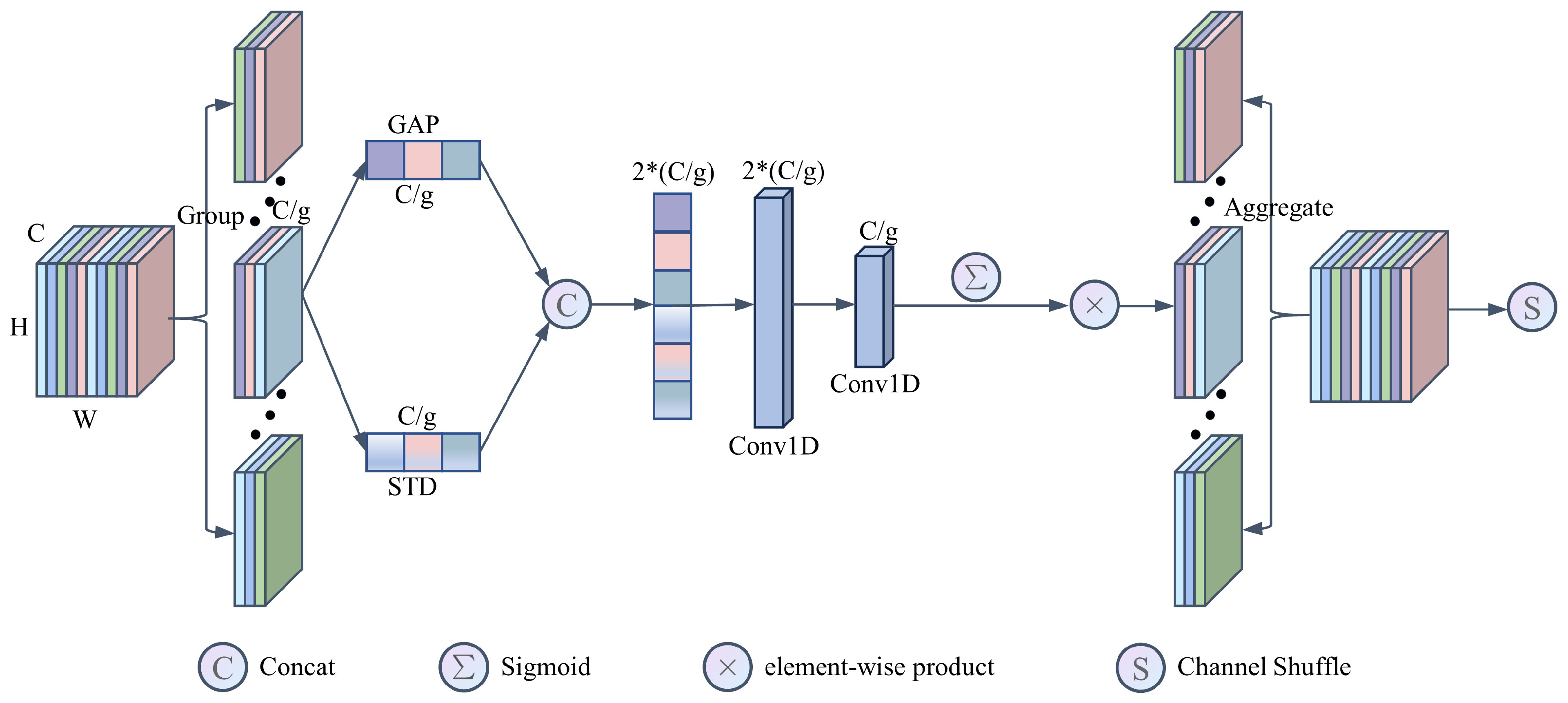
2.7. Dual-Branch Shuffle Attention (DSA)
Wheat disease images often display substantial inter-class differences and subtle intra-class variations, making the recognition process vulnerable to interference from redundant information within the images. To suppress background noise and address image quality issues that can affect classification performance, we propose an improved shuffle attention mechanism [27] (DSA) to help the network focus more effectively on critical disease features in the images. The DSA mechanism incorporates group convolutions and channel shuffle operations, which are tightly aligned with the core design principles of MFFSNet, and they will replace channel shuffle operations and be applied to basic units with a stride of 2. Given an intermediate feature map into G groups along the channel dimension, denoted as , where . Each sub-feature progressively captures specific semantic information during training. Subsequently, the channel attention module assigns importance coefficients to each sub-feature. As depicted in Figure 3, one branch employs global average pooling (GAP) to capture the statistical properties of each channel globally, denoted as , by compressing across the spatial dimensions :

Figure 3.
Construction of the DSA module.
Another branch adopts standard deviation pooling to capture intra-channel distribution characteristics, offering a more comprehensive feature representation for the model, as calculated below:
where the mean for each channel c is given by
The result of the standard deviation pooling reduces the dimensions from to :
The features are then concatenated along the channel dimension, resulting in the aggregated feature t. To capture local cross-channel interactions and enhance channel attention learning, we employ Conv1D convolutions inspired by ECA. The first 1D convolution facilitates the integration of standard deviation pooling and global average pooling results along the channel dimension, as described below:
The second 1D convolution is used for dimensionality reduction, filtering out the most representative information from the enhanced features, thus boosting the channel attention mechanism’s effectiveness, as shown below:
Finally, all sub-features are aggregated, and the “channel shuffle” operator is applied to enable cross-group information flow along the channel dimension.
2.8. Multi-Scale Feature Extraction and Fusion Module (MFEF)
The multi-scale feature extraction and fusion module comprises an extraction module for capturing wheat disease features at different scales and a fusion module for integrating these features to improve disease recognition.
2.8.1. Multi-Scale Feature Extraction Module (MFE)
In investigating the complex visual features of wheat diseases, a key challenge lies in capturing their multi-scale details. To comprehensively extract disease features ranging from subtle to prominent, we reference and improve the Receptive Field Block (RFB) [28] module to serve as a multi-scale feature extraction module, as depicted in Figure 1. This module employs dilated convolutions [29] to expand the network’s receptive field, thereby capturing disease features at varying scales. The introduction of dilated convolutions allows the module to significantly enhance its perception of image details while maintaining a low computational cost. Specifically, the module adopts a parallel processing structure, consisting of multiple branches, each with convolutional kernels at varying dilation rates, paired with convolutions for dimensionality reduction. The outputs from these branches are subsequently merged to enable multi-scale feature fusion. This approach enhances the network’s capacity to identify disease features without compromising computational efficiency.
2.8.2. Multi-Scale Feature Fusion Module (MFF)
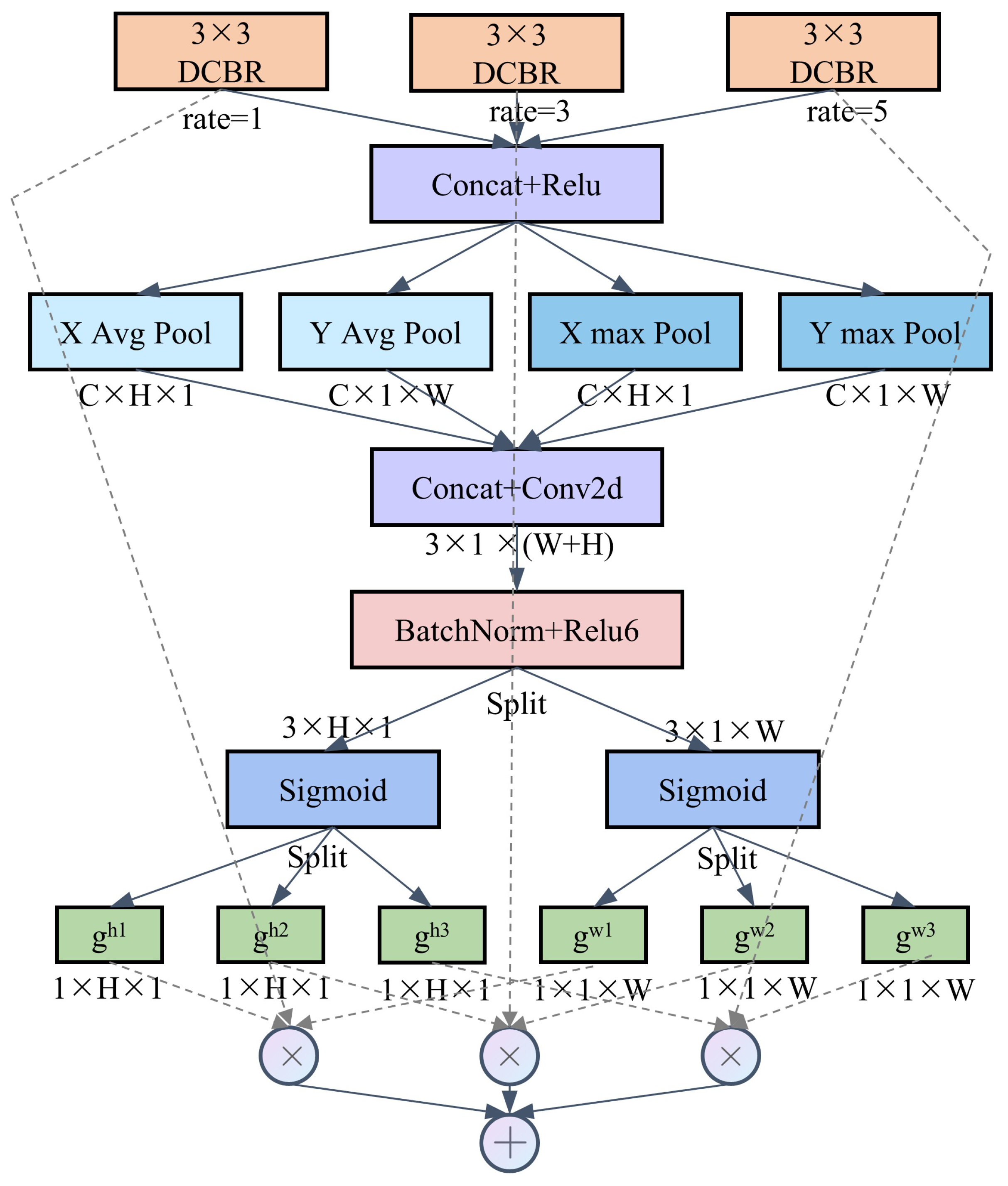
Based on multi-scale feature extraction, to fully utilize the advantages of information at different scales, it is necessary to fuse feature maps with varying receptive fields, thereby achieving multi-scale feature integration and enhancing the model’s representation capability. In this study, the adopted feature fusion strategy is inspired by the kernel selection technique in LSKNet [30]. This technique enables each neuron in the network to adaptively adjust its receptive field size based on the content of the input features, allowing for more effective capture of multi-scale information. In terms of selection mechanism, we referred to Coordinate Attention [31] and proposed an alternative spatial selection mechanism. The spatial selection mechanism in LSKNet encodes spatial information globally, but it compresses the global spatial information into a single channel descriptor, making it difficult to preserve the positional information necessary for capturing spatial structures in vision tasks. Therefore, we use Coordinate Attention to capture long-range spatial interactions while retaining precise positional information.
As illustrated in Figure 4, the information extracted by the MFE module is first processed using max pooling and average pooling kernels with spatial ranges of (H, 1) or (1, W), encoding each channel along the horizontal and vertical axes, respectively. The output of the channel at a given height h can be expressed using the following two formulas:

Figure 4.
Construction of the MFF module.
Similarly, the output of the channel at width w can be written as
To fully utilize the captured positional information, accurately locate regions of interest, and facilitate information interaction between different spatial descriptors, we first concatenate them and then use a convolutional layer to transform the pooled features (with channels) into N spatial attention maps.
Then, f is divided into two separate tensors and along the spatial dimension, and a sigmoid activation function is applied to obtain the individual spatial selection mask for each of the branch inputs:
where is the sigmoid function, and and are the attention weights in the H and W directions.
Then, different branch inputs are weighted using their corresponding spatial selection masks to obtain the final output.
3. Experiments and Results
3.1. Impact of the Position of MFEF Module
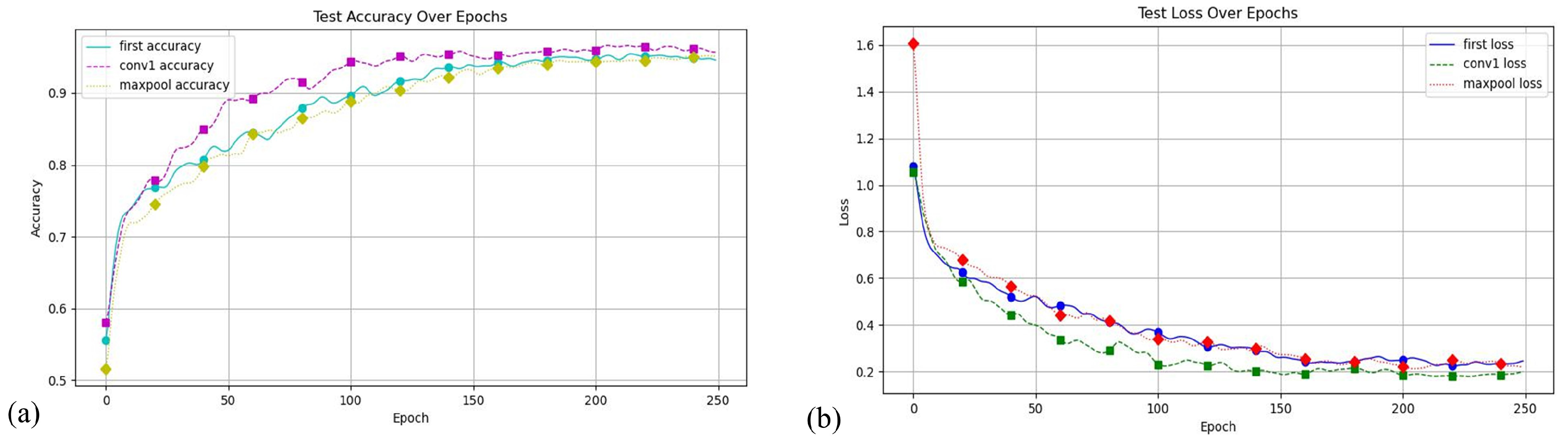
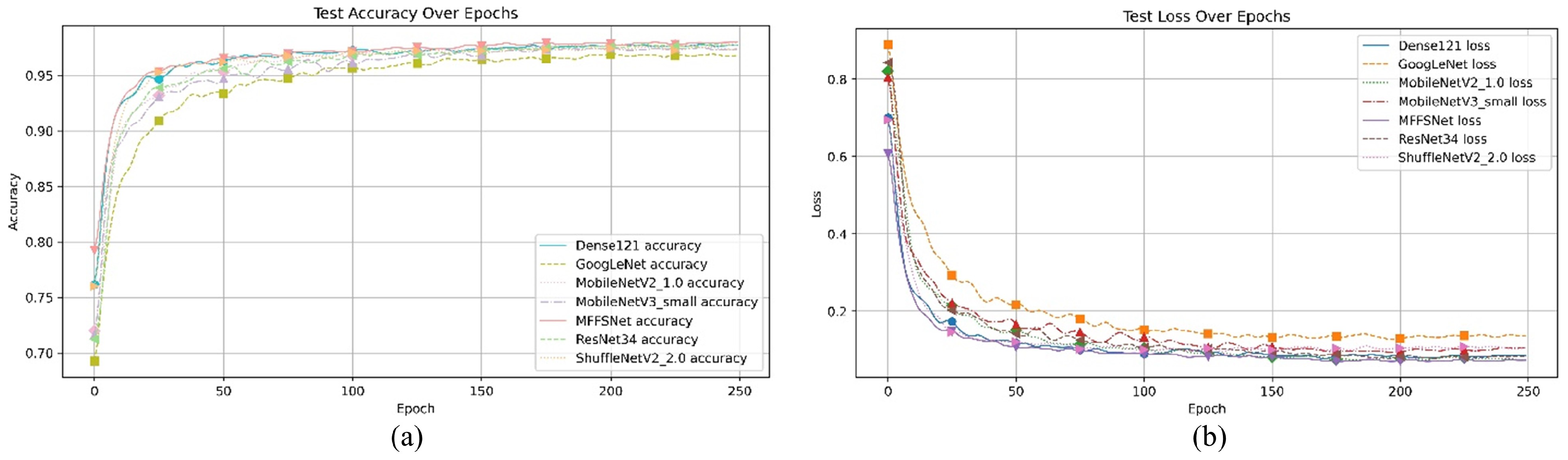
The core advantage of the MFEF module is its ability to process and integrate features at different scales, which is crucial for understanding disease progression and identifying subtle differences in disease symptoms. Its feature extraction primarily targets details such as texture and color in wheat diseases, which are typically captured in the shallow layers of convolutional neural networks. Consequently, placing the MFEF module in the initial layers of the network introduces multi-scale information early in the feature extraction process, which aids the network in better understanding disease traits and improving performance in later stages. To identify the optimal placement of the MFEF module, we conducted comparative experiments by inserting it at different network positions: the first layer, after the Conv1 layer, and after the MaxPool layer. We tested the models with these different MFEF positions, and their loss and accuracy curves are shown in Figure 5. The MFEF module’s performance varies with its placement within the network. When positioned as the network’s first layer, it achieves 96.50% accuracy and 96.13% precision. Placing the module after the MaxPool () layer results in 94.47% accuracy and 94.84% precision. However, when positioned after the Conv1 layer, the module attains its highest accuracy of 97.38% alongside 96.79% precision. Therefore, subsequent experiments in this study were conducted by placing the MFEF module after the Conv1 layer.

Figure 5.
The effect of different positions of the MFEF module in the model. (a) Accuracy value curve; (b) loss value curve.
3.2. Comparative Experiment on Different Network Models
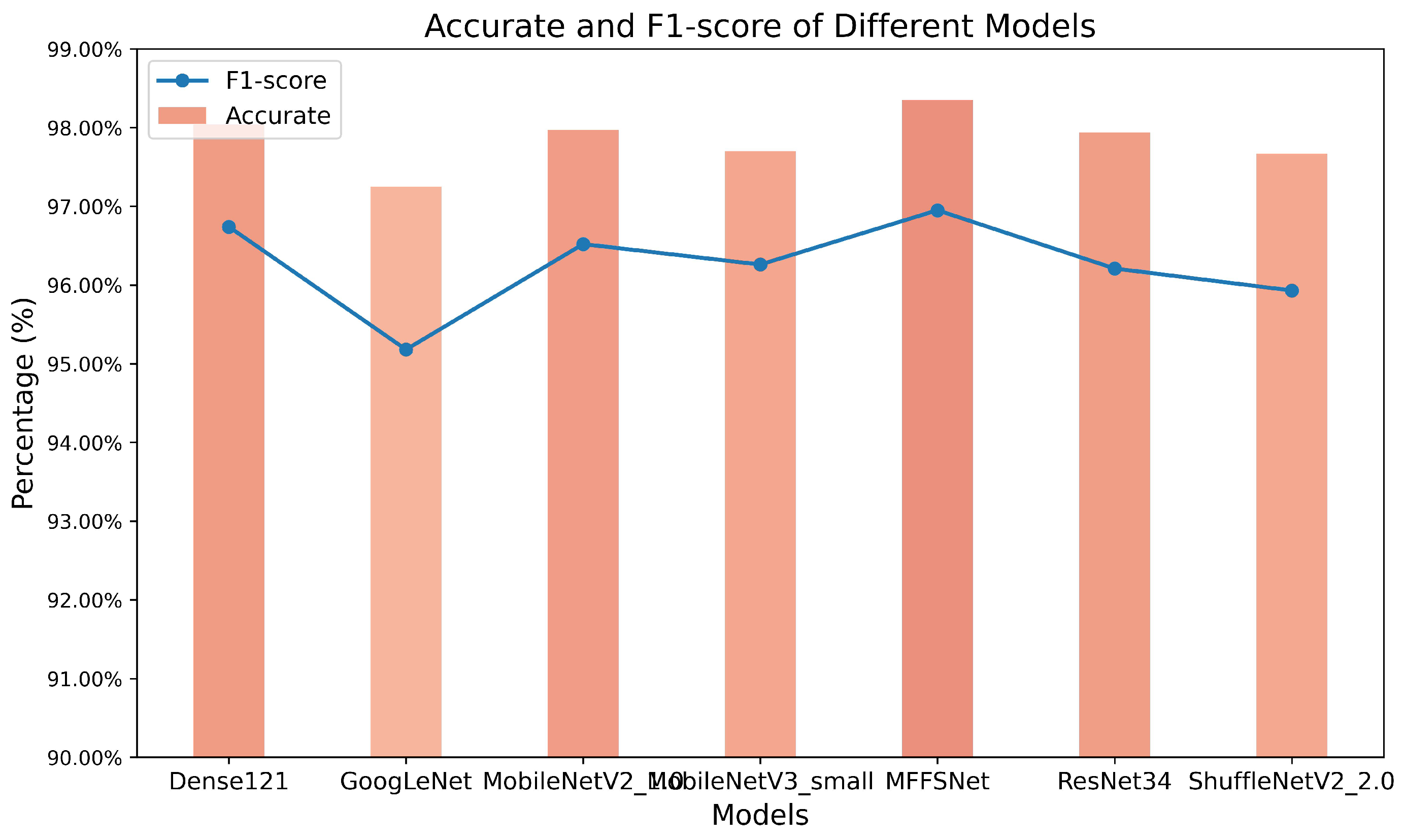
Extensive experiments were conducted on the wheat disease dataset, comparing the proposed MFFSNet with several classical models, with the highest accuracy achieved over 250 epochs taken as the optimal recognition result for each model. As shown in Table 4, the proposed network model is tens of times better in terms of the number of parameters and floating point operations, and its accuracy, precision, recall, and F1-score are better than those of ResNet50 when compared to the heavyweight network (ResNet50 [32]). When compared to several classical lightweight networks (GoogleNet [33], Densenet-121 [34], MobileNetV2 1.0x [35], MobileNetV3 Small [36], ShuffleNetV2 2.0x, and EfficientNetV2-S [37]), the proposed model has the fewest parameters—approximately one-third of the smallest among the other models. Additionally, its accuracy, precision, recall, and F1-score are the highest among the compared models, reaching 97.38% (approximately 3% higher than GoogleNet), 96.79% (around 2% higher than MobileNetV3 Small), 97.96%, and 97.34% (roughly 1.5% higher than EfficientNetV2-S), respectively. Although the proposed model does not exhibit significant superiority over Densenet-121 and ShuffleNetV2 2.0x in terms of performance metrics, its parameter count is less than one-tenth of these two models, and its floating point operations are substantially lower. When compared to the state-of-the-art transformer-based Swin Transformer module [38], the proposed method also exhibits superior performance.

Table 4.
Comparison of the identification results of different models.
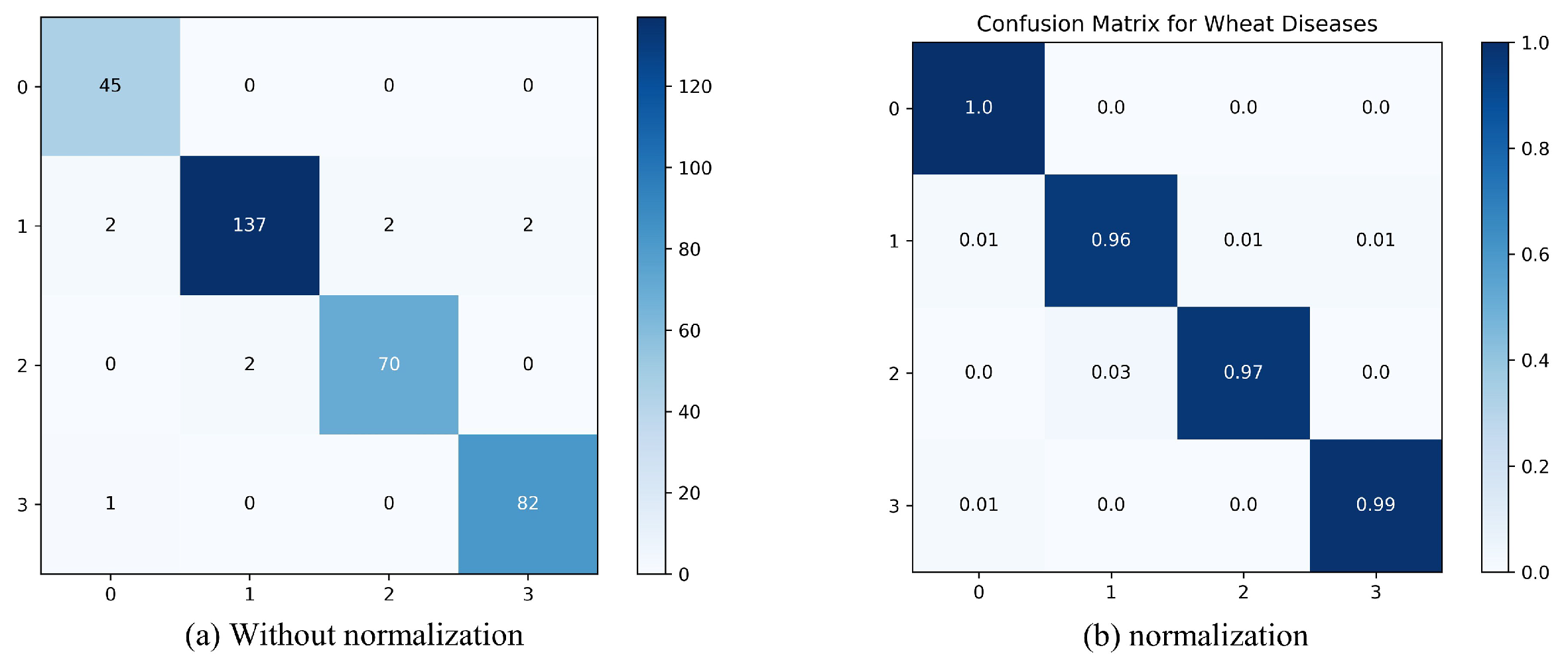
To visually present the classification performance of the proposed method, we used a confusion matrix to analyze the classification results of each disease category in the MFFSNet model, as illustrated in Figure 6. Categories A, B, C, and D represent the four different types, and the values on the main diagonal of the confusion matrix indicate the classification accuracy for each category. As shown in the figure, the darkest colors on the main diagonal are the maximum values for each row and column, indicating that the model demonstrates better classification performance for each category. Since crown and root rot has a relatively simple background, its classification accuracy reaches 100%. In the healthy wheat category, 137 images were correctly classified, with two images each misclassified into other categories. For leaf rust, two images were misclassified as healthy wheat. As for wheat loose smut, one image was misclassified as crown and root rot. These categories are susceptible to interference from complex backgrounds, and the presence of similar features among different diseases causes some degree of misclassification.

Figure 6.
Confusion matrix analysis. (a) Without normalization; (b) with normalization.
3.3. Results of Experiments for the Attention Mechanism
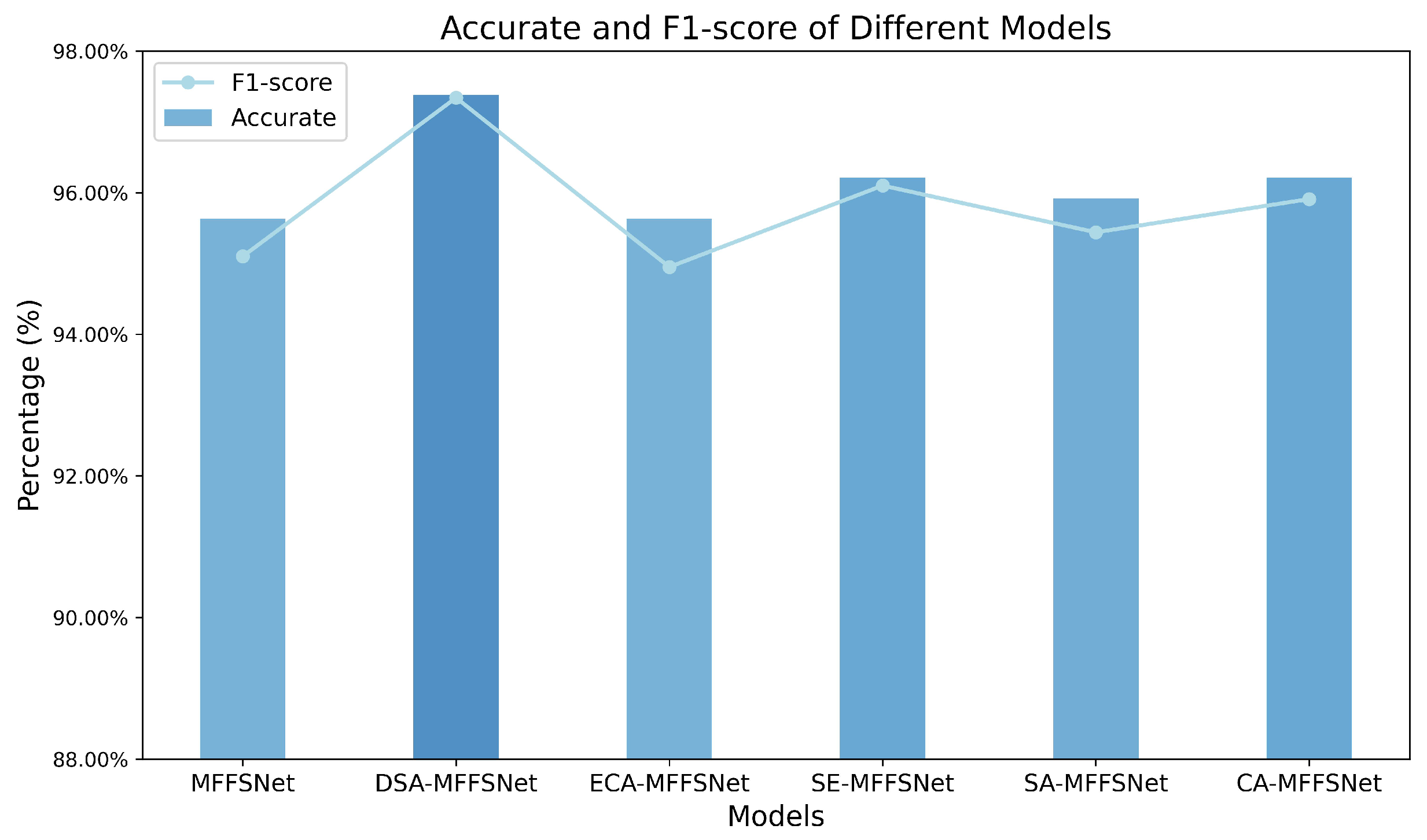
To evaluate the impact of the proposed DSA module on model performance, we designed a series of experiments based on different attention mechanisms. We replaced the DSA module with various other attention mechanisms for comparative analysis, and the results of these experiments are depicted in Figure 7. As shown in the figure, incorporating the DSA module led to a noticeable performance enhancement. When compared to networks using ECA, SE [39], SA, and CA attention mechanisms, it outperformed the best-performing SE-MFFSNet by approximately 1% in accuracy, with a corresponding increase in F1-score.
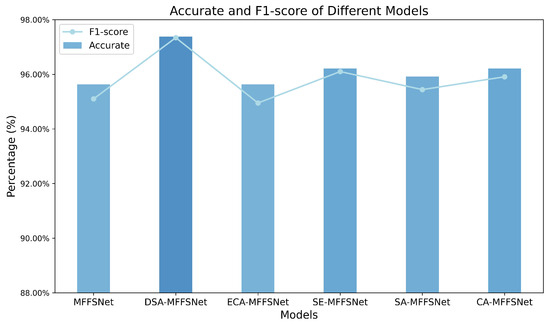
Figure 7.
Comparison of accuracy and F1-scores of different attention mechanism models.
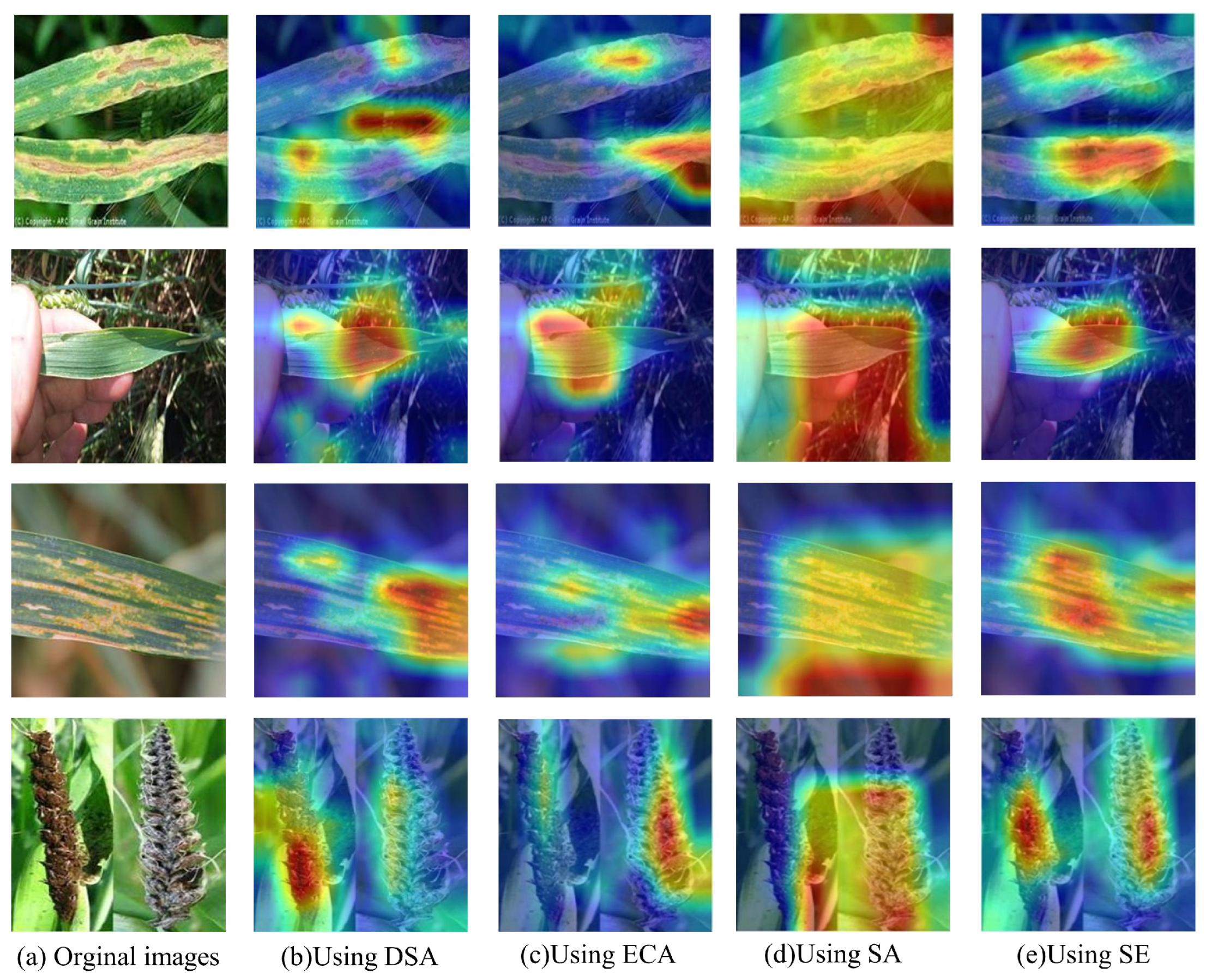
To further validate the effectiveness of the DSA module, we utilized Class Activation Mapping (CAM) [40] to visualize the disease recognition process of the model, as illustrated in Figure 8. The heatmap indicates that, the darker the red regions in the image, the more effectively the model extracts features from these areas. Conversely, if the heatmap appears blue, it suggests that the region may be prone to redundancy. As illustrated in the figure, the model incorporating DSA can better focus on diseased regions and extract critical features, thereby enhancing its recognition accuracy.

Figure 8.
CAM visualization results for different models of attention mechanisms.
3.4. Comparison of Experiments Performed on Different Datasets
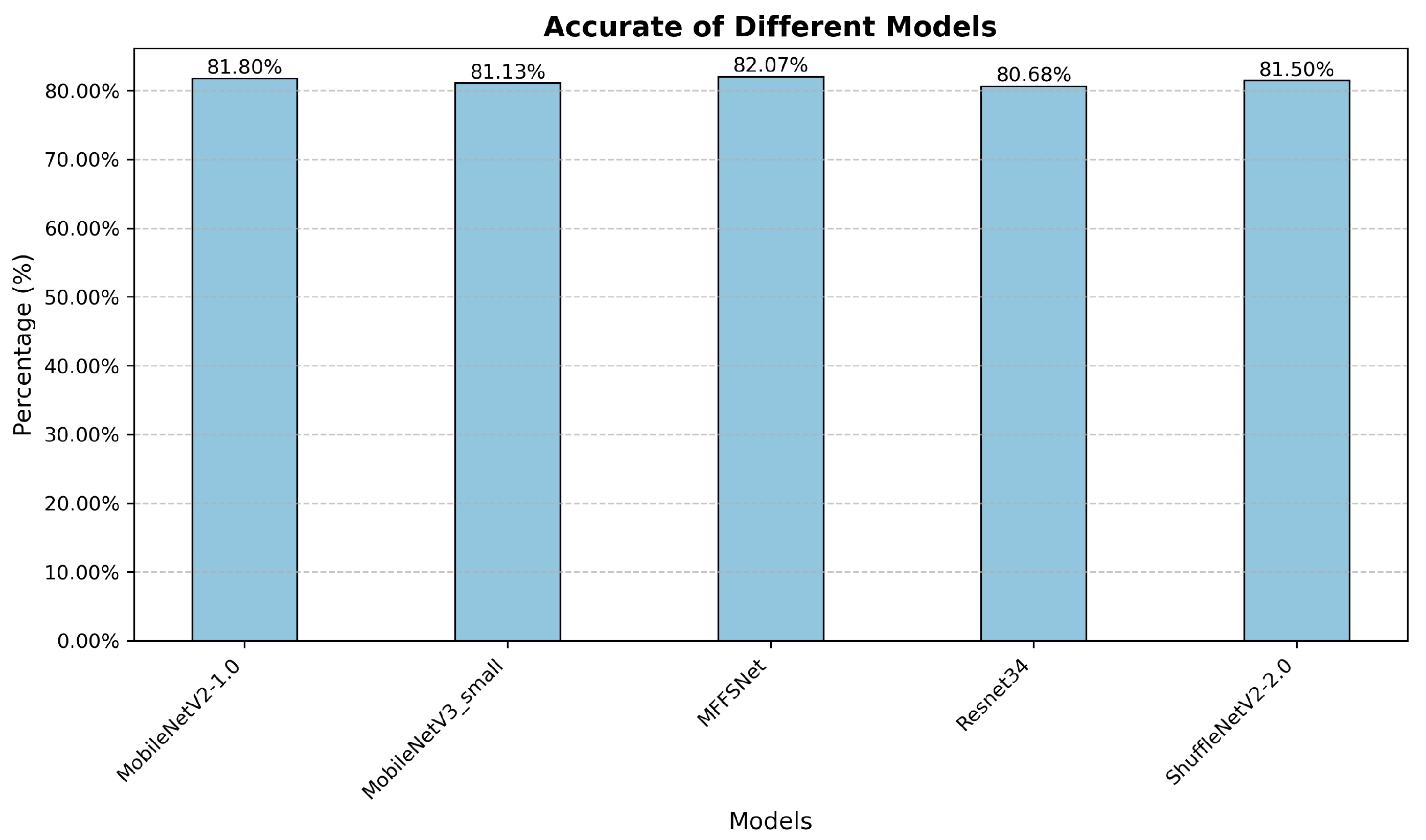
To verify the generalizability of our method across different datasets, we conducted comparative experiments on the AppleLeaf9 dataset [41], which consists of nine classes of apple leaf diseases, with a total of 14,582 images, 94% of which were captured in natural outdoor environments. The experimental results are illustrated in Figure 9. Compared with other models, our proposed method demonstrated superior performance. Although its accuracy is slightly higher than that of DenseNet121, the parameter count is substantially reduced compared to DenseNet121. We also displayed the loss curve of the proposed module along with those of other excellent recognition methods, as shown in Figure 10b. From the loss curves, it is evident that the convergence rate of our model is on par with other models. As observed from the accuracy curves in Figure 10a, our method outperforms the other classifiers in terms of accuracy.

Figure 9.
Recognition results of different models on AppleLeaf9 dataset.

Figure 10.
Comparative evaluation of experiments performed on AppleLeaf9 dataset. (a) Accuracy value curve; (b) loss value curve.
3.5. Comparison of Experiments on the Wheat Plant Diseases Dataset
To verify the applicability of our method on wheat datasets with more disease categories, we conducted experiments on the Wheat Plant Diseases dataset [42]. This dataset provides high-resolution images of real-world wheat diseases without artificial augmentation, ensuring natural disease variations and improved model generalizability. It contains 13,104 non-repetitive images across 15 categories. We divided the dataset into training, validation, and test sets in a 1/2, 1/6, 1/3 ratio. As shown in Figure 11, our proposed method outperforms the others in accuracy. However, our method’s accuracy still has room for improvement, and we plan to optimize it further in the future.
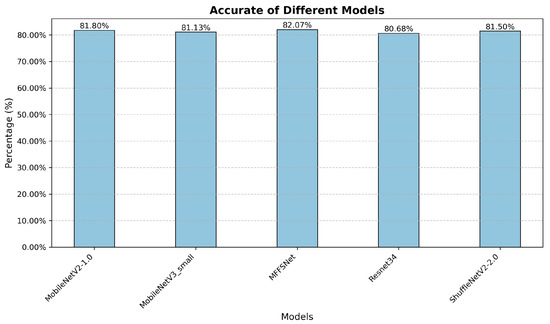
Figure 11.
Recognition results of different models on Wheat Plant Diseases dataset.
3.6. Ablation Experiments
We performed a series of ablation experiments to further investigate the practicality of the ECA mechanism, MFF fusion method, MFEF module, and the distinctions among different fusion techniques, with the results summarized in Table 5. First, when the ECA mechanism is not used in the proposed method, the accuracy decreases by about 1%. Secondly, without the DSA module, the accuracy and other metrics also decrease. Finally, in comparison to models without the MFEF module and those utilizing either MFF or LSK fusion methods, the MFF fusion technique yielded the best recognition performance. These findings highlight the effectiveness of both the ECA mechanism and the MFEF module.

Table 5.
Ablation experiments.
3.7. Visualization of Recognition Models and Results
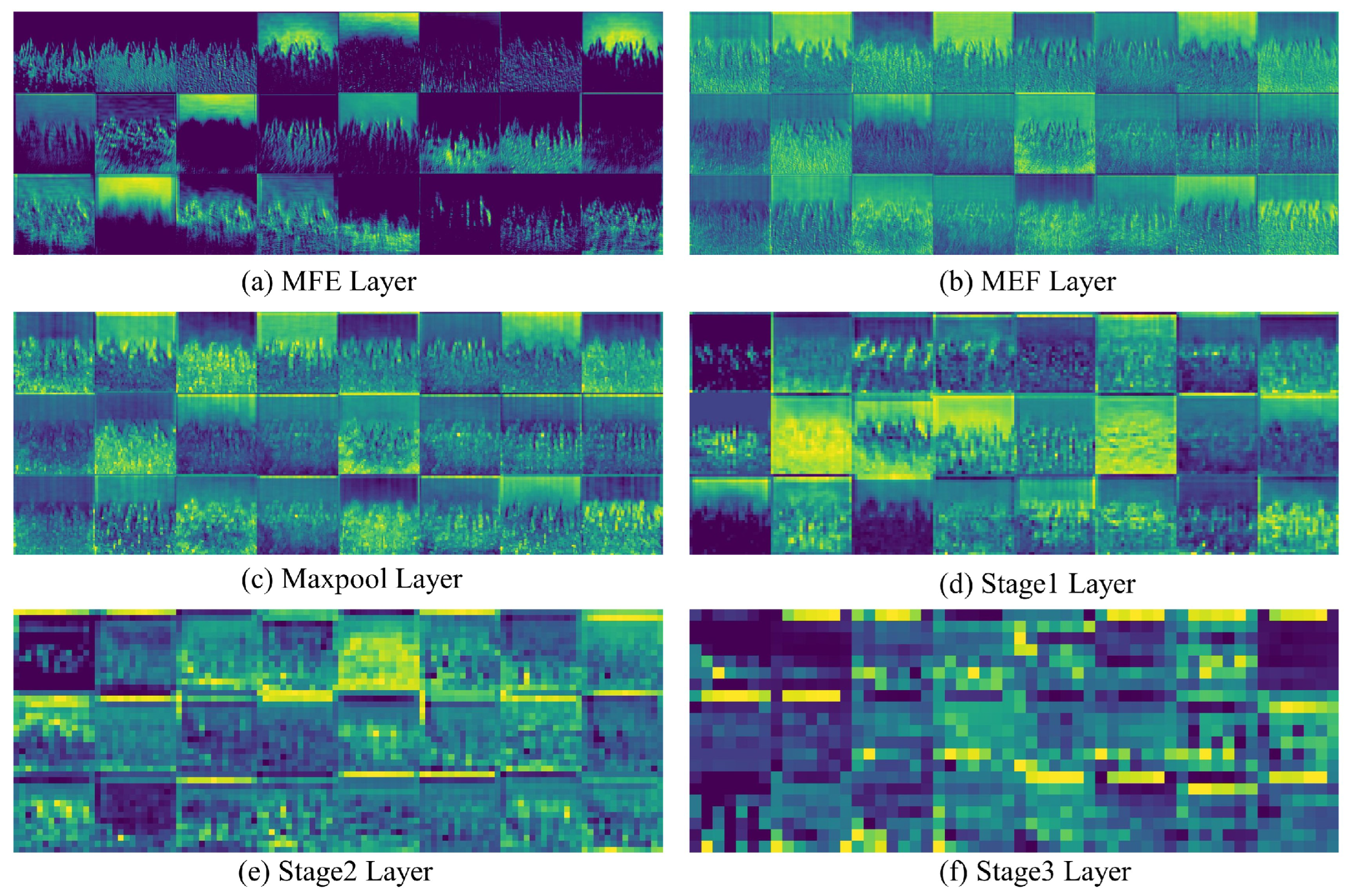
In this study, Figure 12 shows the visual analysis of the feature heatmaps output by specific layers in the model. From the figure, it is evident that, after being processed by the MFEF module, the model successfully captures rich multi-scale information from the images, encompassing, but not limited to, textures of varying scales, edge features, and color distributions. As the image progresses through the three key stages, the amount of detailed visual information gradually decreases, while more abstract feature information begins to emerge. This indicates that the model, during the feature extraction process, is progressively shifting from low-level pixel features to higher-level semantic features.

Figure 12.
Visualization results of the output feature maps of the convolution layer.
4. Discussion
In this study, we proposed MFFSNet, a novel model for accurately identifying wheat diseases in complex environments. It uses an MFEF module with inflated convolution to capture diverse features across scales. MFFSNet’s primary units are enhanced ShuffleNetV2 units for better efficiency and performance. A DSA mechanism is also integrated to help the model focus on key features and reduce background interference. MFFSNet demonstrates superior performance compared to several classical models. It not only has fewer parameters and float operations but also better performance metrics thanks to its unique architecture. The effectiveness of the DSA module, which enhances the model’s focus on diseased areas and improves recognition accuracy, is verified by the comparative analysis in Figure 7 and the visualization in Figure 8. Furthermore, experiments on the AppleLeaf9 dataset, containing apple leaf disease images, show that MFFSNet can generalize well to different crops, indicating its potential for broader agricultural applications. However, this study also has certain limitations. The research mainly focuses on wheat disease identification in field environments with complex backgrounds, but it does not consider many factors such as plant age at infection, plant organ infected, wheat variety, stage of pathogen life cycle, etc. Future research should be conducted in collaboration with plant pathologists to more comprehensively evaluate wheat disease identification performance.
5. Conclusions
This study successfully designed a lightweight convolutional neural network model, MFFSNet, for wheat disease recognition in complex backgrounds. The model efficiently captures subtle features of wheat diseases using multi-scale feature extraction and fusion modules, refines these features through lightweight components, and employs attention mechanisms to strengthen the identification of crucial features while mitigating interference from complex backgrounds. MFFSNet offers advantages such as a compact model size and fast computational speed. The experimental results demonstrate that the model achieves an accuracy of 97.38%, approximately 3% higher than the classical model GoogleNet, and a precision about 2% higher than the mobile network MobileNetV3 Small. Furthermore, with a parameter count of just 0.45 M, the model is well suited for deployment on resource-constrained devices. The limitation of this study is that it does not consider the stage of the pathogen life cycle. For future work, improvements can be made in the following areas: (1) collaboration with plant pathologists to conduct a more comprehensive evaluation of wheat disease identification performance; (2) we intend to deploy MFFSNet on mobile devices or field-programmable gate arrays (FPGAs) for real-time rapid wheat disease identification in agricultural fields, offering farmers a convenient tool for effective disease management.
Author Contributions
Conceptualization, M.X., R.Y., S.D. and L.W.; Methodology, M.X., R.Y., S.D. and L.W.; Investigation, M.X., J.S., L.X. and Z.L.; Writing—original draft preparation, M.X.; Project administration, M.X., R.Y. and S.D.; Supervision, M.X., R.Y., S.D. and L.W.; Validation, M.X., J.W., J.S., L.X., Z.L., R.Y., S.D. and L.W.; Formal analysis, J.W., J.S., L.X. and Z.L.; Data curation, J.S., L.X. and Z.L.; Writing—review and editing, J.W. and L.W. All authors have read and agreed to the published version of the manuscript.
Funding
Project supported by the National Natural Science Foundation of China (Grant Nos. U20A20227, 62076208, 62076207, 62306246, and 62406260), Chongqing Talent Plan Project (Grant No. CQYC20210302257), Fundamental Research Funds for the Central Universities (Grant Nos. SWU-XDZD22009, SWU-XDJH202319, and SWU-ZLPY07), Chongqing Higher Education Teaching Reform Research Project (Grant No. 211005), and Open Fund Project of State Key Laboratory of Intelligent Vehicle Safety Technology (Grant No. IVSTSKL-202309).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Acknowledgments
Special thanks to the reviewers for their valuable comments.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Tian, Y.; Zhao, C.J.; Lu, S.G.; Guo, X.Y. Multiple Classifier Combination for Recognition of Wheat Leaf Diseases. Intell. Autom. Soft Comput. 2011, 17, 519–529. [Google Scholar]
- Azadbakht, M.; Ashourloo, D.; Aghighi, H.; Radiom, S.; Alimohammadi, A. Wheat leaf rust detection at canopy scale under different LAI levels using machine learning techniques. Comput. Electron. Agric. 2019, 156, 119–128. [Google Scholar] [CrossRef]
- Dong, Z.; Ji, X.; Lai, C.S.; Qi, D. Design and Implementation of a Flexible Neuromorphic Computing System for Affective Communication via Memristive Circuits. IEEE Commun. Mag. 2023, 61, 74–80. [Google Scholar] [CrossRef]
- Ji, X.; Dong, Z.; Han, Y.; Lai, C.S.; Qi, D. A Brain-Inspired Hierarchical Interactive In-Memory Computing System and Its Application in Video Sentiment Analysis. IEEE Trans. Circuits Syst. Video Technol. 2023, 33, 7928–7942. [Google Scholar]
- Zhu, X.; Li, J.; Jia, R.; Liu, B.; Yao, Z.; Yuan, A.; Huo, Y.; Zhang, H. LAD-Net: A Novel Light Weight Model for Early Apple Leaf Pests and Diseases Classification. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 20, 1156–1169. [Google Scholar] [CrossRef]
- Chen, J.; Chen, W.; Zeb, A.; Yang, S.; Zhang, D. Lightweight Inception Networks for the Recognition and Detection of Rice Plant Diseases. IEEE Sens. J. 2022, 22, 14628–14638. [Google Scholar]
- Bidarakundi, P.M.; Kumar, B.M. Coffee-Net: Deep Mobile Patch Generation Network for Coffee Leaf Disease Classification. IEEE Sens. J. 2025, 25, 7355–7362. [Google Scholar]
- Lu, J.; Hu, J.; Zhao, G.; Mei, F.; Zhang, C. An in-field automatic wheat disease diagnosis system. Comput. Electron. Agric. 2017, 142, 369–379. [Google Scholar] [CrossRef]
- Nema, S.; Dixit, A. Wheat leaf detection and prevention using support vector machine. In Proceedings of the 2018 International Conference on Circuits and Systems in Digital Enterprise Technology (ICCSDET), Kottayam, India, 21–22 December 2018; pp. 1–5. [Google Scholar]
- Abade, A.; Ferreira, P.A.; de Barros Vidal, F. Plant diseases recognition on images using convolutional neural networks: A systematic review. Comput. Electron. Agric. 2021, 185, 106125. [Google Scholar]
- Picon, A.; Alvarez-Gila, A.; Seitz, M.; Ortiz-Barredo, A.; Echazarra, J.; Johannes, A. Deep convolutional neural networks for mobile capture device-based crop disease classification in the wild. Comput. Electron. Agric. 2019, 161, 280–290. [Google Scholar] [CrossRef]
- Mi, Z.; Zhang, X.; Su, J.; Han, D.; Su, B. Wheat stripe rust grading by deep learning with attention mechanism and images from mobile devices. Front. Plant Sci. 2020, 11, 558126. [Google Scholar] [CrossRef] [PubMed]
- Albayrak, U.; Golcuk, A.; Aktas, S.; Coruh, U.; Tasdemir, S.; Baykan, O.K. Classification and Analysis of Agaricus bisporus Diseases with Pre-Trained Deep Learning Models. Agronomy 2025, 15, 226. [Google Scholar] [CrossRef]
- Pan, F.; He, D.; Xiang, P.; Hu, M.; Yang, D.; Huang, F.; Peng, C. DualTransAttNet: A Hybrid Model with a Dual Attention Mechanism for Corn Seed Classification. Agronomy 2025, 15, 200. [Google Scholar] [CrossRef]
- Ou, Y.; Yan, J.; Liang, Z.; Zhang, B. Hyperspectral Imaging Combined with Deep Learning for the Early Detection of Strawberry Leaf Gray Mold Disease. Agronomy 2024, 14, 2694. [Google Scholar] [CrossRef]
- Thakur, P.S.; Sheorey, T.; Ojha, A. VGG-ICNN: A lightweight CNN model for crop disease identification. Multimed. Tools Appl. 2023, 82, 497–520. [Google Scholar] [CrossRef]
- Zhang, P.; Yang, L.; Li, D. EfficientNet-B4-Ranger: A novel method for greenhouse cucumber disease recognition under natural complex environment. Comput. Electron. Agric. 2020, 176, 105652. [Google Scholar] [CrossRef]
- Barman, U.; Choudhury, R.D.; Sahu, D.; Barman, G.G. Comparison of convolution neural networks for smartphone image based real time classification of citrus leaf disease. Comput. Electron. Agric. 2020, 177, 105661. [Google Scholar] [CrossRef]
- Bao, W.; Yang, X.; Liang, D.; Hu, G.; Yang, X. Lightweight convolutional neural network model for field wheat ear disease identification. Comput. Electron. Agric. 2021, 189, 106367. [Google Scholar] [CrossRef]
- Li, G.; Jiao, L.; Chen, P.; Liu, K.; Wang, R.; Dong, S.; Kang, C. Spatial convolutional self-attention-based transformer module for strawberry disease identification under complex background. Comput. Electron. Agric. 2023, 212, 108121. [Google Scholar] [CrossRef]
- Goyal, L.; Sharma, C.M.; Singh, A.; Singh, P.K. Leaf and spike wheat disease detection & classification using an improved deep convolutional architecture. Inform. Med. Unlocked 2021, 25, 100642. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. Int. Conf. Mach. Learn. (ICML) 2015, 37, 448–456. [Google Scholar]
- Ma, N.; Zhang, X.; Zheng, H.-T.; Sun, J. ShuffleNet v2: Practical guidelines for efficient CNN architecture design. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 116–131. [Google Scholar]
- Howard, A.G. MobileNets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Chen, J.; Kao, S.-H.; He, H.; Zhuo, W.; Wen, S.; Chan, C.-H.G. Run, don’t walk: Chasing higher FLOPS for faster neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 18–22 June 2023; pp. 12021–12031. [Google Scholar]
- Wang, Q.; Wu, B.; Zhuo, P.; Li, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient channel attention for deep convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 11534–11542. [Google Scholar]
- Zhang, Q.-L.; Yang, Y.-B. SA-Net: Shuffle attention for deep convolutional neural networks. In Proceedings of the ICASSP 2021—2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Virtual, 6–12 June 2021; pp. 2235–2239. [Google Scholar]
- Liu, S.; Huang, D. Receptive field block net for accurate and fast object detection. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 385–400. [Google Scholar]
- Yu, F.; Koltun, V. Multi-scale context aggregation by dilated convolutions. arXiv 2015, arXiv:1511.07122. [Google Scholar]
- Li, Y.; Hou, Q.; Zheng, Z.; Cheng, M.-M.; Yang, J.; Li, X. Large selective kernel network for remote sensing object detection. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 4–6 October 2023; pp. 16794–16805. [Google Scholar]
- Hou, Q.; Zhou, D.; Feng, J. Coordinate attention for efficient mobile network design. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 19–25 June 2021; pp. 13713–13722. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.-C.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V.; et al. MobileNetV3. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 1314–1324. [Google Scholar]
- Tan, M.; Le, Q. EfficientNet: Rethinking model scaling for convolutional neural networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Yang, Q.; Duan, S.; Wang, L. Efficient identification of apple leaf diseases in the wild using convolutional neural networks. Agronomy 2022, 12, 2784. [Google Scholar] [CrossRef]
- kushagra3204. Wheat Plant Diseases [Dataset]. Kaggle. 2024. Available online: https://www.kaggle.com/datasets/kushagra3204/wheat-plant-diseases/data (accessed on 1 March 2025).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).