Use of Wild Relatives and Closely Related Species to Adapt Common Bean to Climate Change
Abstract
:1. Introduction
2. Results and Discussion
2.1. Climate Change
2.2. Genetic Resources and Conservation

| Sections | Species | Total |
|---|---|---|
| Clade A (8 sections) |  | |
Not assigned to any section, nor forming any section with each other:  | 2 | |
| Bracteati Freytag | macrolepis, talamancensis | 2 |
| Brevilegumeni Freytag | 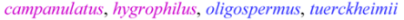 | 4 |
| Chiapasana Delgado | chiapasanus | 1 |
| Digitati Freytag | albiflorus, albiviolaceus, altimontanus, neglectus | 4 |
| Minkelersia (Mart. & Gal.) Maréchal, Mascherpa, Stainier |  | 9 |
| Pedicellati (Benth.) Freytag |  | 9 |
| Revoluti Freytag | leptophyllus | 1 |
| Xanthotricha Delgado |  | 6 |
| Clade B (6 sections) |  | |
| Acutifolii Freytag | acutifolius, parvifolius | 2 |
| Coriacei Freytag |  | 5 |
| Falcati Freytag |  | 3 |
| Paniculati Freytag |  | 20 |
| Phaseoli DC |  | 6 |
| Rugosi Freytag |  | 3 |
| Total (no. sections): 14. Total (no. species): 77 | ||

| Species (wild), country | Known populations | Possible No. of populations | Accessions in genebanks | Gap as % | Populations in protected areas |
|---|---|---|---|---|---|
| P. albescens | |||||
| Mexico | 13 | 18 | 1 | 85 | 2 |
| P. coccineus | |||||
| Mexico | 342 | 462 | 106 | 77 | 12 |
| Guatemala | 64 | 66 | 59 | 10 | 4 |
| Honduras | 4 | 12 | 3 | 50 | 0 |
| P. costaricensis | |||||
| Costa Rica | 47 | 49 | 8 | 83 | 2 |
| Panama | 10 | 12 | 0 | 100 | 2 |
| P. dumosus | |||||
| Guatemala | 11 | 19 | 9 | 53 | 2 |
| Mexico | 1 | 1 | 0 | 100 | 0 |
| P. persistentus | |||||
| Guatemala | 1 | 2 | 0 | 100 | 0 |
| P. vulgaris | |||||
| Mexico | 110 | 420 | 395 | 6 | 6 |
| Guatemala | 16 | 45 | 39 | 14 | 3 |
| Honduras | 5 | 18 | 6 | 65 | 0 |
| El Salvador | 2 | 4 | 1 | 50 | 1 |
| Nicaragua | 7 | 8 | 4 | 50 | 0 |
| Costa Rica | 24 | 25 | 30 | 1 | 1 |
| Panama | 0 | 2 | 0 | 100 | 0 |
| Colombia | 74 | 79 | 74 | 6 | 0 |
| Venezuela | 2 | 18 | 0 | 100 | 0 |
| Ecuador | 10 | 12 | 10 | 4 | 1 |
| Peru | 32 | 38 | 31 | 12 | 2 |
| Bolivia | 11 | 14 | 11 | 4 | 0 |
| Argentina | 87 | 96 | 66 | 31 | 4 |
2.3. Use of Wild Relatives and Closely Related Species in Common Bean Breeding
2.4. Future Breeding and Genomic Strategies for Use of CWR

3. Conclusions
Acknowledgements
References
- Crops and Plants [Online]. USDA-NASS: Washington, DC, USA, 2012. Available online: http://www.nass.usda.gov/ (accessed on 6 May 2013).
- Palomino, V.R. Bayesian Analysis of a Linear Mixed Model to Measure the Impact of Climate Change on Yield of Common Bean for the Year 2030 Worldwide. Master’s Thesis, University of Puerto Rico, Mayaguez, Puerto Rico, 2012. [Google Scholar]
- Acosta-Gallegos, J.A.; Kelly, J.D.; Gepts, P. Prebreeding in common bean and use of genetic diversity from wild germplasm. Crop Sci. 2007, 47, S44–S49. [Google Scholar]
- Rowlands, D.; Frame, D.J.; Ackerley, D.; Aina, T.; Booth, B.B.B.; Christensen, C.; Collins, M.; Faull, N.; Forest, C.E.; Grandey, B.S.; et al. Broad range of 2050 warming from an observationally constrained large climate model ensemble. Nat. Geosci. 2012, 5, 256–260. [Google Scholar] [CrossRef]
- Intergovernmental Panel on Climate Change (IPCC), Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change; Solomon, S.; Qin, D.; Manning, M.; Chen, Z.; Marquis, M.; Averyt, K.B.; Tignor, M.; Miller, H.L. (Eds.) Cambridge University Press: Cambridge, UK, 2007.
- Beebe, S.; Ramirez, J.; Jarvis, A.; Rao, I.M.; Mosquera, G.; Bueno, J.M.; Blair, M.W. Genetic Improvement of Common beans and the Challenges of Climate Change. In Crop Adaptation to Climate Change; Yadav, S.S., Redden, R.J., Hatfield, J.L., Lotze-Campen, H., Hall, A.E., Eds.; Wiley-Blackwell: Chichester, UK, 2012; pp. 356–369. [Google Scholar]
- Singh, S.P.; Terán, H.; Lema, M.; Webster, D.M.; Strausbaugh, C.A.; Miklas, P.N.; Schwartz, H.F.; Brick, M.A. Seventy-five years of breeding dry bean of the Western USA. Crop Sci. 2007, 47, 1–9. [Google Scholar] [CrossRef]
- Shisanya, C.A. Improvement of drought adapted tepary bean (Phaseolus acutifolius A. Gray var. latifolius) yield through biological nitrogen fixation in semi-arid SE-Kenya. Eur. J. Agron. 2002, 16, 13–24. [Google Scholar] [CrossRef]
- Blair, M.W.; Iriarte, G.; Beebe, S. QTL analysis of yield traits in an advanced backcross population derived from a cultivated Andean × wild common bean (Phaseolus vulgaris L.) cross. Theor. Appl. Genet. 2006, 112, 1149–1163. [Google Scholar] [CrossRef]
- Koinange, E.M.K.; Singh, S.P.; Gepts, P. Genetic control of the domestication syndrome in common bean. Crop Sci. 1996, 36, 1037–1045. [Google Scholar] [CrossRef]
- Isely, D. Phaseolus. In The Jepson Manual—Higher Plants of California; Hickman, J.C., Ed.; University of California Press: Berkeley, CA, USA, 1993; p. 641. [Google Scholar]
- Westphal, E. Pulses in Ethiopia: Their Taxonomy and Agricultural Significance; Centre for Agricultural Publishing and Documentation: Wageningen, The Netherlands, 1974; Volume 815, pp. 140–151. [Google Scholar]
- Gepts, P. Phaseolin as an Evolutionary Marker. In Genetic Resources of Phaseolus Beans; Gepts, P., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1988; pp. 215–241. [Google Scholar]
- Toro-Chica, O.; Ocampo, C.H.; Debouck, D.G. Phaseolin: Variability and reference materials in wild and cultivated common bean. Annu. Rep. Bean Improv. Coop. (USA) 2007, 50, 69–70. [Google Scholar]
- Khairallah, M.M.; Sears, B.B.; Adams, M.W. Mitochondrial restriction fragment length polymorphisms in wild Phaseolus vulgaris L.: Insights on the domestication of the common bean. Theor. Appl. Genet. 1992, 84, 915–922. [Google Scholar]
- Becerra-Velásquez, V.L.; Gepts, P. RFLP diversity of common bean (Phaseolus vulgaris) in its centres of origin. Genome 1994, 37, 256–263. [Google Scholar] [CrossRef]
- Bitocchi, E.; Nanni, L.; Bellucci, E.; Rossi, M.; Giardini, A.; Spagnoletti-Zeuli, P.; Logozzo, G.; Stougaard, J.; McClean, P.; Attene, G.; Papa, R. Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proc. Natl. Acad. Sci. USA 2012, 109, 788–796. [Google Scholar] [CrossRef]
- Nabhan, G.P. Native crop diversity in Aridoamerica: Conservation of regional gene pools. Econ. Bot. 1985, 39, 387–399. [Google Scholar] [CrossRef]
- Drewes, S.I. Prospección y colecta de germoplasma silvestre de Phaseolus vulgaris en la zona central de Argentina. Plant Genet. Resour. Newsl. 2008, 155, 9–14. [Google Scholar]
- Becerra-Velásquez, V.; Paredes-Cárcomo, M.; Debouck, D.G. Genetic relationships of common bean (Phaseolus vulgaris L.) race Chile with wild Andean and Mesoamerican germplasm. Chilean J. Agric. Res. 2011, 71, 3–15. [Google Scholar] [CrossRef]
- Toro-Chica, O.; Tohme, J.; Debouck, D.G. Wild Bean (Phaseolus vulgaris L.): Description and Distribution; International Board for Plant Genetic Resources and International Center for Tropical Agriculture: Cali, Colombia, 1990; p. 106. [Google Scholar]
- Debouck, D.G. Cahiers de Phaséologie: Section PHASEOLI; International Center for Tropical Agriculture (CIAT): Cali, Colombia, 2012; p. 172. Available online: http://www.ciat.cgiar.org/urg (accessed on 2 October 2012).
- Debouck, D.G.; Toro, O.; Paredes, O.M.; Johnson, W.C.; Gepts, P. Genetic diversity and ecological distribution of Phaseolus vulgaris (Fabaceae) in northwestern South America. Econ. Bot. 1993, 47, 408–423. [Google Scholar] [CrossRef]
- Gepts, P.; Osborn, T.C.; Rashka, K.; Bliss, F.A. Phaseolin protein variability in wild forms and landraces of the common bean (Phaseolus vulgaris L.): Evidence for multiple centers of domestication. Econ. Bot. 1986, 40, 451–468. [Google Scholar] [CrossRef]
- Koenig, R.L.; Singh, S.P.; Gepts, P. Novel phaseolin types in wild and cultivated common bean (Phaseolus vulgaris, Fabaceae). Econ. Bot. 1990, 44, 50–60. [Google Scholar] [CrossRef]
- Freyre, R.; Ríos, R.; Guzmán, L.; Debouck, D.G.; Gepts, P. Ecogeographic distribution of Phaseolus spp. (Fabaceae) in Bolivia. Econ. Bot. 1996, 50, 195–215. [Google Scholar] [CrossRef]
- Tohme, J.; González, D.O.; Beebe, S.; Duque, M.C. AFLP analysis of gene pools of a wild bean core collection. Crop Sci. 1996, 36, 1375–1384. [Google Scholar] [CrossRef]
- Chacón, S.M.I.; Pickersgill, B.; Debouck, D.G.; Arias, J.S. Phylogeographic analysis of the chloroplast DNA variation in wild common bean (Phaseolus vulgaris L.) in the Americas. Plant Syst. Evol. 2007, 266, 175–195. [Google Scholar] [CrossRef]
- Chacón, S.M.I.; Pickersgill, B.; Debouck, D.G. Domestication patterns in common bean (Phaseolus vulgaris L.) and the origin of the Mesoamerican and Andean cultivated races. Theor. Appl. Genet. 2005, 110, 432–444. [Google Scholar] [CrossRef]
- Kami, J.; Becerra-Velásquez, V.; Debouck, D.G.; Gepts, P. Identification of presumed ancestral DNA sequences of phaseolin in Phaseolus vulgaris. Proc. Natl. Acad. Sci. USA 1995, 92, 1101–1004. [Google Scholar]
- Graham, A. Late Cretaceous and Cenozoic history of Latin American vegetation and terrestrial environments; Missouri Botanical Garden Press: St. Louis, MI, USA, 2010; p. 617. [Google Scholar]
- Romero-Andreas, J.; Yandell, B.S.; Bliss, F.A. Bean arcelin. 1. Inheritance of a novel seed protein of Phaseolus vulgaris L. and its effect on seed composition. Theor. Appl. Genet. 1986, 72, 123–128. [Google Scholar]
- Acosta-Gallegos, J.A.; Quintero, C.; Vargas, J.; Toro, O.; Tohme, J.; Cardona, C. A new variant of arcelin in wild common bean, Phaseolus vulgaris L., from southern Mexico. Genet. Resour. Crop Evol. 1998, 45, 235–242. [Google Scholar] [CrossRef]
- Osborn, T.C.; Blake, T.; Gepts, P.; Bliss, F.A. Bean arcelin. 2. Genetic variation, inheritance and linkage relationships of a novel seed protein of Phaseolus vulgaris L. Theor. Appl. Genet. 1986, 71, 847–855. [Google Scholar]
- Lynch, J.; González, A.; Tohme, J.M.; Garcia, J.A. Variation in characters related to leaf photosynthesis in wild bean populations. Crop Sci. 1992, 32, 633–640. [Google Scholar] [CrossRef]
- González, A.; Lynch, J.; Tohme, J.M.; Beebe, S.E.; Macchiavelli, R.E. Characters related to leaf photosynthesis in wild populations and landraces of common bean. Crop Sci. 1995, 35, 1468–1476. [Google Scholar] [CrossRef]
- Acosta-Gallegos, J.A.; Aguilar Garzón, B.; Rodríguez-Guerra, R.; Mendoza Hernández, M.; Guzman-Maldonado, H.; Kelly, J.D. Seed yield of black seeded lines introgressed with wild Phaseolus vulgaris. Annu. Rep. Bean Improv. Coop. (USA) 2007, 50, 23–24. [Google Scholar]
- Tanksley, S.D.; Grandillo, S.; Fulton, T.M.; Zamir, D.; Eshed, Y.; Petiard, V.; López, J.; Beck-Bunn, T. Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor. Appl. Genet. 1996, 92, 213–224. [Google Scholar] [CrossRef]
- Xiao, J.; Li, J.; Grandillo, S.; Nag-Ahn, S.; Yuan, L.; Tanksley, S.D.; McCouch, S.R. Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics 1998, 150, 899–909. [Google Scholar]
- Berglund-Brücher, O.; Brücher, H. The South American wild bean (Phaseolus aborigineus Burk.) as ancestor of the common bean. Econ. Bot. 1976, 30, 257–272. [Google Scholar] [CrossRef]
- Gentry, H.S. Origin of the common bean, Phaseolus vulgaris L. Econ. Bot. 1969, 23, 55–69. [Google Scholar] [CrossRef]
- Miranda-Colín, S. Origen de Phaseolus vulgaris L. (Frijol común). Agrociencia 1967, 1, 99–109. [Google Scholar]
- Heiser, C.B. Cultivated plants and cultural diffusion in nuclear America. Am. Anthropol. 1965, 67, 930–949. [Google Scholar]
- Kaplan, L. Phaseolus: Diffusion and Centers of Origin. In Man across the Sea: Problems in Pre-Columbian Contacts; Riley, C.L., Kelley, J.C., Pennington, C.W., Randa, R.L., Eds.; University of Texas Press: Austin, TX, USA, 1971; pp. 416–427. [Google Scholar]
- Kwak, M.; Kami, J.A.; Gepts, P. The putative Mesoamerican domestication center of Phaseolus vulgaris is located in the Lerma-Santiago basin of Mexico. Crop Sci. 2009, 49, 554–563. [Google Scholar] [CrossRef]
- Maréchal, R.; Mascherpa, J.; Stainier, F. Etude taxonomique d’un groupe complexe d’espèces des genres Phaseolus et Vigna. (Papilionaceae) sur la base de données morphologiques et polliniques, traitées par l’analyse informatique. Boissiera 1978, 28, 1–273. [Google Scholar]
- Schmit, V.; du Jardin, P.; Baudoin, J.P.; Debouck, D.G. Use of chloroplast DNA polymorphisms for the phylogenetic study of seven Phaseolus taxa including P. vulgaris and P. coccineus. Theor. Appl. Genet. 1993, 87, 506–516. [Google Scholar]
- Delgado-Salinas, A.O.; Turley, T.; Richman, A.; Lavin, M. Phylogenetic analysis of the cultivated and wild species of Phaseolus (Fabaceae). Syst. Bot. 1999, 24, 438–460. [Google Scholar] [CrossRef]
- Delgado-Salinas, A.; Bibler, R.; Lavin, M. Phylogeny of the genus Phaseolus (Leguminosae): A recent diversification in an ancient landscape. Syst. Bot. 2006, 31, 779–791. [Google Scholar] [CrossRef]
- Freytag, G.F.; Debouck, D.G. Taxonomy, distribution, and ecology of the genus Phaseolus (Leguminosae-Papilionoideae) in North America, Mexico and Central America. SIDA Bot. Misc. 2002, 23, 1–300. [Google Scholar]
- Schmit, V.; Debouck, D.G. Observations on the origin of Phaseolus polyanthus Greenman. Econ. Bot. 1991, 45, 345–364. [Google Scholar] [CrossRef]
- Neill, D.A.; Klitgaard, B.B.; Lewis, G.P. Fabaceae. In Catalogue of the Vascular Plants of Ecuador; Jorgensen, P.M., León-Yánez, S., Eds.; Missouri Botanical Garden Press: St. Louis, MI, USA, 1999; pp. 468–484. [Google Scholar]
- Debouck, D.G.; Smartt, J. Beans, Phaseolus spp. (Leguminosae-Papilionoideae). In Evolution of Crop Plants, 2nd; Smartt, J., Simmonds, N.W., Eds.; Longman Scientific & Technical: London, UK, 1995; pp. 287–294. [Google Scholar]
- Araya-Villalobos, R.; González-Ugalde, W.G.; Camacho-Chacón, F.; Sánchez-Trejos, P.; Debouck, D.G. Observations on the geographic distribution, ecology and conservation status of several Phaseolus bean species in Costa Rica. Genet. Resour. Crop Evol. 2001, 48, 221–232. [Google Scholar] [CrossRef]
- Freytag, G.F.; Debouck, D.G. Phaseolus costaricensis, a new wild bean species (Phaseolinae, Leguminosae) from Costa Rica and Panama, Central America. Novon 1996, 6, 157–163. [Google Scholar] [CrossRef]
- Ramírez-Delgadillo, R.; Delgado-Salinas, A. A new species of Phaseolus (Fabaceae) from west-central Mexico. SIDA 1999, 18, 637–646. [Google Scholar]
- Delgado-Salinas, A.; Thulin, M.; Pasquet, R.; Weeden, N.; Lavin, M. Vigna (Leguminosae) sensu lato: The names and identities of the American segregate genera. Am. J. Bot. 2011, 98, 1694–1715. [Google Scholar] [CrossRef]
- Lackey, J.A. A review of generic concepts in American Phaseolinae (Fabaceae, Faboideae). Iselya 1983, 2, 21–64. [Google Scholar]
- Lavin, M.; Herendeen, P.S.; Wojciechowski, M.F. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst. Biol. 2005, 54, 575–594. [Google Scholar] [CrossRef]
- Gepts, P.; Papa, R.; Coulibaly, S.; González-Mejía, A.; Pasquet, R. Wild Legume Diversity and DomesticationߞInsights from Molecular Methods. In Wild Legumes; Oono, K., Ed.; National Institute of Biological Resources: Tsukuba, Japan, 2000; pp. 19–31. [Google Scholar]
- Serrano-Serrano, M.L.; Hernández-Torres, J.; Castillo-Villamizar, G.; Debouck, D.; Chacón, M.I. Gene pools in wild Lima bean (Phaseolus lunatus L.) from the Americas: Evidences for an Andean origin and past migrations. Mol. Phylogenet. Evol. 2010, 54, 76–87. [Google Scholar] [CrossRef]
- Kuboyama, T.; Shintaku, Y.; Takeda, G. Hybrid plant of Phaseolus vulgaris L. and P. lunatus L. obtained by means of embryo rescue and confirmed by restriction endonuclease analysis of rDNA. Euphytica 1991, 54, 177–182. [Google Scholar]
- Leonard, M.F.; Stephens, L.C.; Summers, W.L. Effect of maternal genotype on development of Phaseolus vulgaris L. x P. lunatus L. interspecific hybrid embryos. Euphytica 1987, 36, 327–332. [Google Scholar] [CrossRef]
- Kaplan, L.; Lynch, T. Phaseolus (Fabaceae) in archaeology: AMS radiocarbon dates and their significance for pre-Colombian agriculture. Econ. Bot. 1999, 53, 261–272. [Google Scholar] [CrossRef]
- Piperno, D.L. New Archaeobotanical Information on Early Cultivation and Plant Domestication Involving Microplant (Phytolith and Starch Grain) Remains. In Biodiversity in AgricultureߞDomestication, Evolution, and Sustainability; Gepts, P., Famula, Th.R., Bettinger, R.L., Brush, S.B., Damania, A.B., McGuire, P.E., Qualset, C.O., Eds.; Cambridge University Press: Cambridge, UK, 2012; pp. 136–159. [Google Scholar]
- Piperno, D.L.; Dillehay, T.D. Starch grains on human teeth reveal early broad crop diet in northern Peru. Proc. Natl. Acad. Sci. USA 2008, 105, 19622–19627. [Google Scholar] [CrossRef]
- Ishimoto, M.; Suzuki, K.; Iwanaga, M.; Kikuchi, F.; Kitamura, K. Variation of seedߞAmylase inhibitors in the common bean. Theor. Appl. Genet. 1995, 90, 425–429. [Google Scholar]
- Seigler, D.S.; Maslin, B.R.; Conn, E.E. Cyanogenesis in the Leguminosae. In Advances in Legume Biology, Monographs in Systematic Botany from the Missouri Botanical Garden; Stirton, C.H., Zarucchi, J.L., Eds.; Missouri Botanical Garden: St. Louis, MO, USA, 1989; Volume 29, pp. 645–672. [Google Scholar]
- Shreve, F.; Wiggins, I.L. Vegetation and Flora of the Sonoran Desert; Stanford University Press: Stanford, CA, USA, 1964; Volume 1, p. 840. [Google Scholar]
- Balasubramanian, P.; Ahmad, F.; Vandenberg, A.; Hucl, P.J. Barriers to interspecific hybridization of common bean with Phaseolus angustissimus A. Gray and P. filiformis Bentham. J. Genet. Breed. 2005, 59, 321–328. [Google Scholar]
- Maréchal, R.; Baudoin, J.P. Observations sur quelques hybrids dans le genre Phaseolus IV. L’hybride Phaseolus vulgaris × Phaseolus filiformis. Bull. Rech. Agron. Gembloux. 1978, 13, 233–240. [Google Scholar]
- Bayuelo-Jiménez, J.; Debouck, D.G.; Lynch, J. Salinity tolerance in Phaseolus species during early vegetative growth. Crop Sci. 2002, 42, 2184–2192. [Google Scholar] [CrossRef]
- Buhrow, R. The wild beans of southwestern North America. Desert Plants 1983, 5, 67–88. [Google Scholar]
- Muñoz, L.C.; Blair, M.W.; Duque, M.C.; Tohme, J.; Roca, W. Introgression in common bean × tepary bean interspecific congruity-backcross lines as measured by AFLP markers. Crop Sci. 2004, 44, 637–645. [Google Scholar] [CrossRef]
- Lin, T.Y.; Markhart, A.H. Phaseolus acutifolius A. Gray is more heat tolerant than P. vulgaris L. in the absence of water stress. Crop Sci. 1996, 36, 110–114. [Google Scholar] [CrossRef]
- Miklas, P.N.; Rosas, J.C.; Beaver, J.S.; Telek, L.; Freytag, G.F. Field performance of select tepary bean germplasm in the Tropics. Crop Sci. 1994, 34, 1639–1644. [Google Scholar] [CrossRef]
- Brücher, H. The Wild Ancestor of Phaseolus vulgaris in South America. In Genetic Resources of Phaseolus Beans; Gepts, P., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1988; pp. 185–214. [Google Scholar]
- Singh, S.P.; Terán, H.; Schwartz, H.; Otto, K.; Lema, M. White mold-resistant interspecific common bean germplasm lines VCW 54 and VCW 55. J. Plant Regist. 2009, 3, 191–197. [Google Scholar] [CrossRef]
- Ramírez-Villegas, J.; Khoury, C.; Jarvis, A.; Debouck, D.G.; Guarino, L. A gap analysis methodology for collecting crop genepools: A case study with Phaseolus beans. PLoS One 2010, 5, 1–18. [Google Scholar]
- Vázquez-García, J.A.; Cuevas-Guzmán, R.; Cochrane, T.S.; Iltis, H.H.; Santana-Michel, F.J.; Guzmán-Hernández, L. Flora de Manantlán. SIDA Bot. Misc. 1995, 13, 1–312. [Google Scholar]
- Tanksley, S.D.; McCouch, S.R. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef]
- Beebe, S.; Rengifo, J.; Gaitan, E.; Duque, M.C.; Tohme, J. Diversity and origin of Andean landraces of common bean. Crop Sci. 2001, 41, 854–862. [Google Scholar] [CrossRef]
- Bitocchi, E.; Bellucci, E.; Giardini, A.; Rau, D.; Rodriguez, M.; Biagetti, E.; Santilocchi, R.; Spagnoletti Zeuli, P.; Gioia, T.; Logozzo, G.; et al. Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 2012. [Google Scholar] [CrossRef]
- Gepts, P.; Debouck, D.G. Origin, Domestication, and Evolution of the Common Bean (Phaseolus vulgaris L.). In Common Beans: Research for Crop Improvement; Schoonhoven, A.V., Ed.; Commonwealth Agricultural Bureaux International: Wallingford, UK, 1991; pp. 7–53. [Google Scholar]
- Acevedo, M.; Steadman, J.R.; Rosas, J.C.; Venegas, J. Coevolution of the bean rust pathogen Uromyces appendiculatus with its wild, weedy and domesticated hosts (Phaseolus spp.) at a center of diversity. Annu. Rep. Bean Improv. Coop. (USA) 2008, 51, 22–23. [Google Scholar]
- Acevedo, M.; Steadman, J.R.; Rosas, J.C.; Venegas, J. Characterization of virulence diversity of the bean rust pathogen Uromyces appendiculatus in wild bean populations as a tool for effective resistance gene deployment. Annu. Rep. Bean Improv. Coop. (USA) 2005, 48, 132–133. [Google Scholar]
- Keneni, G.; Bekele, E.; Getu, E.; Imtiaz, M.; Damte, T.; Mulatu, B.; Dagne, K. Breeding food legumes for resistance to storage insect pests: potential and limitations. Sustainability 2011, 3, 1399–1415. [Google Scholar] [CrossRef]
- Osborn, T.C.; Alexander, D.C.; Sun, S.M.; Cardona, C.; Bliss, F.A. Insecticidal activity and lectin homology of arcelin seed protein. Science 1988, 240, 207–210. [Google Scholar]
- Osborn, T.C.; Hartweck, L.M.; Harmsen, R.H.; Vogelzang, R.D.; Kmiecik, K.A.; Bliss, F.A. Registration of Phaseolus vulgaris genetic stocks with altered seed protein compositions. Crop Sci. 2003, 43, 1570–1571. [Google Scholar] [CrossRef]
- Kornegay, J.; Cardona, C.; Posso, C.E. Inheritance of resistance to Mexican bean weevil in common bean determined by bioassay and biochemical tests. Crop Sci. 1993, 33, 589–594. [Google Scholar] [CrossRef]
- Beaver, J.S.; Zapata, M.; Alameda, M.; Porch, T.G. Registration of PR0401-259 and PR0650-31 dry bean germplasm lines. J. Plant Regist. 2012, 6, 81–84. [Google Scholar] [CrossRef]
- Acevedo, M.; Steadman, J.R.; Rosas, J.C.; Venegas, J. New sources of resistance to bean rust and implications for host-pathogen coevolution. Annu. Rep. Bean Improv. Coop. (USA) 2006, 49, 77–78. [Google Scholar]
- Mkwaila, W.; Terpstra, K.A.; Ender, M.; Kelly, J.D. Identification of QTL for agronomic traits and resistance to white mold in wild and landrace germplasm of common bean. Plant Breed. 2011, 130, 665–672. [Google Scholar] [CrossRef]
- Kelly, J.; Long, B.; Blakely, N.; Terpstra, K. Dry bean yield trials, 2004. Available online: http://www.psm.msu.edu/VarietyTrials/Acrobat/04_DryBean_Report.pdf (accessed on 6 May 2013).
- Wright, E.M.; Kelly, J.D. Mapping QTL for seed yield and canning quality following processing of black bean (Phaseolus vulgaris L.). Euphytica 2011, 179, 471–484. [Google Scholar] [CrossRef]
- Ferwerda, F.H.; Bassett, M.J. Barriers to interspecific hybridization in crosses between Phaseolus coccineus L. (G35172) and Phaseolus vulgaris L. Annu. Rep. Bean Improv. Coop.(USA) 2000, 43, 21–22. [Google Scholar]
- Singh, S.P.; Gutiérrez, J.A. Geographical distribution of the DL1 and DL2 genes causing hybrid dwarfism in Phaseolus vulgaris L., their association with seed size, and their significance to breeding. Euphytica 1984, 33, 337–345. [Google Scholar] [CrossRef]
- Acevedo, M.; Steadman, J.R.; Rosas, J.C. Uromyces appendiculatus in Honduras: Pathogen diversity and host resistance screening. Plant Dis. 2013, 97, 652–661. [Google Scholar] [CrossRef]
- Singh, S.P.; Terán, H.; Beaver, J.S. Scarlet runner bean germplasm accessions G 35006 and G 35172 possess resistance to multiple diseases of common bean. Annu. Rep. Bean Improv. Coop.(USA) 2009, 52, 20–21. [Google Scholar]
- Osorno, J.M.; Muñoz, C.G.; Beaver, J.S.; Ferwerda, F.H.; Bassett, M.J.; Miklas, P.N.; Olczyk, T.; Bussey, B. Two genes from Phaseolus coccineus confer resistance to Bean Golden Yellow Mosaic Virus in common bean. J. Am. Soc. Hort. Sci. 2007, 132, 530–533. [Google Scholar]
- Schwartz, H.F.; Otto, K.; Terán, H.; Lema, M. Inheritance of white mold resistance in Phaseolus vulgaris × P. coccineus crosses. Plant Dis. 2006, 90, 1167–1170. [Google Scholar] [CrossRef]
- McCoy, S.; Higgins, B.; Steadman, J.R. Use of multi site screening to identify and verify partial resistance to white mold in common bean in 2011. Annu. Rep. Bean Improv. Coop. (USA) 2012, 55, 153–154. [Google Scholar]
- Wilkinson, R.E. Incorporation of Phaseolus coccineus germplasm may facilitate production of high yielding P. vulgaris lines. Annu. Rep. Bean Improv. Coop.(USA) 1983, 26, 28–29. [Google Scholar]
- Miklas, P.N.; Zapata, M.; Beaver, J.S.; Grafton, K.F. Registration of four dry bean germplasms resistant to common bacterial blight: ICB-3, ICB-6, ICB-8 and ICB-1. Crop Sci. 1999, 39, 594. [Google Scholar] [CrossRef]
- Schmit, V.; Baudoin, J.P. Screening for resistance to Ascochyta blight in populations of Phaseolus coccineus L. and P. polyanthus Greenman. Field Crops Res. 1992, 30, 155–165. [Google Scholar] [CrossRef]
- Mahuku, G.S.; Jara, C.E.; Cajiao, C.; Beebe, S. Sources of resistance to Colletotrichum lindemuthianum in the secondary gene pool of Phaseolus vulgaris and in crosses of primary and secondary gene pools. Plant Dis. 2002, 86, 1383–1387. [Google Scholar] [CrossRef]
- Freytag, G.F.; Bassett, M.J.; Zapata, M. Registration of XR-235-1-1 bean germplasm. Crop Sci. 1982, 22, 1268–1269. [Google Scholar] [CrossRef]
- Zapata, M.; Freytag, G.; Wilkinson, R. Release of five common bean germplasm lines resistant to common bacterial blight W-BB-11, W-BB-20-1, W-BB-35, W-BB-52, and W-BB. J. Agric. Univ. Puerto Rico 2004, 88, 91–95. [Google Scholar]
- Beaver, J.S.; Porch, T.G.; Zapata, M. Registration of ‘Verano’ white bean. J. Plant Regist. 2008, 2, 187–189. [Google Scholar] [CrossRef]
- Ferwerda, F.H. The Investigation of Genetic Barriers to Interspecific Crosses between Phaseolus acutifolius A. Gray, Phaseolus coccineus L. and Phaseolus vulgaris L. and the Inheritance of Resistance to Bean Golden Mosaic Virus from P. coccineus L. Ph.D. Thesis, University of Florida, Gainsville, FL, USA, August 2001. [Google Scholar]
- Koinange, E.M.K.; Gepts, P. Hybrid weakness in wild Phaseolus vulgaris L. J. Hered. 1982, 83, 135–139. [Google Scholar]
- Butare, L.; Rao, I.; Lepoivre, P.; Polania, J.; Cajiao, C.; Cuasquer, J.; Beebe, S. New genetic sources of resistance in the genus Phaseolus to individual and combined aluminium toxicity and progressive soil drying stresses. Euphytica 2011, 181, 385–404. [Google Scholar] [CrossRef]
- Nabhan, G.P. Tepary beans: The effects of domestication on adaptation to arid environments. Arid Lands Nwsl. 1979, 10, 11–16. [Google Scholar]
- Porch, T.G.; Ramirez, V.H.; Santana, D.; Harmsen, E.W. Evaluation of common bean for drought tolerance in Juana Diaz, Puerto Rico. J. Agron. Crop. Sci. 2009, 195, 328–334. [Google Scholar] [CrossRef]
- Federici, C.T.; Ehdaie, B.; Waines, J.D. Domesticated and wild tepary bean: Field performance with and without drought-stress. Agron. J. 1990, 82, 896–900. [Google Scholar] [CrossRef]
- Markhart, A.H. Comparative water relation of Phaseolus vulgaris L. and Phaseolus acutifolius Gray. J. Plant Physiol. 1985, 77, 113–117. [Google Scholar] [CrossRef]
- Bhardwaj, H.L.; Rangappa, M.; Hamama, A.A. Planting date and genotype effects on tepary bean productivity. HortScience 2002, 2, 317–318. [Google Scholar]
- Thomas, C.V.; Manshardt, R.M.; Waines, J.G. Teparies as a source of useful traits for improving common beans. Desert Plants 1983, 5, 43–48. [Google Scholar]
- Ferwerda, F.H.; Bassett, M.J.; Beaver, J.S. Viability of seed of reciprocal interspecific crosses between Phaseolus vulgaris L. and Phaseolus acutifolius A. Grey. Annu. Rep. Bean Improv. Coop. (USA) 2003, 46, 29–30. [Google Scholar]
- Pratt, R.C. Gene transfer between tepary and common bean. Desert Plants 1983, 5, 57–63. [Google Scholar]
- Haghighi, K.R.; Ascher, P.D. Fertile, intermediate hybrids between Phaseolus vulgaris and P. acutifolius from congruity backcrossing. Sex. Plant Reprod. 1988, 1, 51–58. [Google Scholar]
- Anderson, N.O.; Ascher, P.D.; Haghighi, K. Congruity backcrossing as a means of creating genetic variability in self pollinated crops: seed morphology of Phaseolus vulgaris L. and P. acutifolius A. Gray hybrids. Euphytica 1996, 87, 211–224. [Google Scholar] [CrossRef]
- Mejía-Jiménez, A.; Muñoz, C.; Jacobson, H.J.; Roca, W.M.; Singh, S.P. Interspecific hybridization between common and tepary beans: Increased hybrid embryo growth, fertility and efficiency of hybridization through recurrent and congruity backcrossing. Theor. Appl. Genet. 1994, 88, 324–331. [Google Scholar]
- Singh, S.P.; Debouck, D.G.; Roca, W.W. Interspecific hybridization between Phaseolus vulgaris L. and P. parvifolius Freytag. Annu. Rep. Bean Improv. Coop. (USA) 1998, 4, 7–8. [Google Scholar]
- Scott, M.E.; Michaels, T.E. Xanthomonas resistance of Phaseolus interspecific cross selections confirmed by field performance. HortScience 1992, 27, 348–350. [Google Scholar]
- Singh, S.P.; Muñoz, C.G. Resistance to common bacterial blight among Phaseolus species and common bean improvement. Crop Sci. 1999, 39, 80–89. [Google Scholar] [CrossRef]
- McElroy, J.B. Breeding Dry Beans, Phaseolus vulgaris L., for Common Bacterial Blight Resistance Derived from Phaseolus acutifolius A. Gray. Ph.D. Thesis, Cornell University, Ithaca, NY, USA, 1985. [Google Scholar]
- Mutlu, N.; Urrea, C.A.; Miklas, P.N.; Pastor-Corrales, M.A.; Steadman, J.R.; Lindgren, D.T.J.; Reiser, J.; Vidaver, A.K.; Coyne, D.P. Registration of common bacterial blight, rust and bean common mosaic resistant Great Northern common bean germplasm line ABC-Weihing. J. Plant Regist. 2008, 2, 53–55. [Google Scholar] [CrossRef]
- Mutlu, N.; Miklas, P.N.; Steadman, J.R.; Vidaver, A.M.; Lindgren, D.T.; Reiser, J.; Coyne, D.P.; Pator-Corrales, M.A. Registration of common bacterial blight resistant pinto bean germplasm line ABCP-8. Crop Sci. 2005, 45, 806–807. [Google Scholar] [CrossRef]
- Fourie, D.; Herselman, L. Application of molecular markers in breeding for bean common blight resistance in South Africa. Afr. Crop Sci. J. 2011, 19, 369–376. [Google Scholar]
- Costa, J.G.C.; Rava, C.A. Linhagens de feijoeiro comum com fenótipos agronômicos favoráveis e resistência ao crestamento bacteriano comum e antracnose. Ciênc. Agrotec. Lavras 2003, 27, 1176–1182. [Google Scholar] [CrossRef]
- Osorno, J.M.; Grafton, K.; Vanderwal, A.; Gegner, S. A new small red bean with improved resistance to common bacterial blight: Registration of ‘Rio Rojo’. J. Plant Regist. 2013, 8. in press. [Google Scholar]
- Kusolwa, P.M.; Myers, J.R. Seed storage proteins ARL2 and its variants from the APA locus of wild tepary bean G40199 confers resistance to Acanthoscellides obtectus when expressed in common beans. Afr. Crop Sci. J. 2011, 19, 255–265. [Google Scholar]
- Miklas, P.N.; Schwartz, H.F.; Salgado, M.O.; Beaver, J.S. Reaction of select tepary bean to Ashy Stem Blight and Fusarium Wilt. HortScience 1998, 33, 136–139. [Google Scholar]
- Miklas, P.N.; Santiago, J. Reaction of select tepary bean to Bean Golden Mosaic Virus. HortScience 1996, 31, 430–432. [Google Scholar]
- Pastor-Corrales, M.A.; Steadman, J.R.; Urrea, C.A.; Blair, M.W.; Venegas, J.P. The domesticated tepary bean accession G 40022 has broader resistance to the highly variable bean rust pathogen than the known rust resistance genes in common bean. Annu. Rep. Bean Improv. Coop. (USA) 2011, 54, 124–125. [Google Scholar]
- Mogotsi, K.K. Phaseolus acutifolius A. Gray. In Plant Resources of Tropical Africa 1. Cereals and Pulses; Brink, M., Belay, G., Eds.; PROTA Foundation: Wageningen, The Netherlands, 2006; pp. 133–137. [Google Scholar]
- Singh, S.P.; Debouck, D.G.; Roca, W.W. Successful interspecific hybridization between Phaseolus vulgaris L. and P. costaricensis Freytag & Debouck. Annu. Rep. Bean Improv. Coop. (USA) 1997, 40, 40–41. [Google Scholar]
- Singh, S.P.; Terán, H.; Schwartz, H.F.; Otto, K.; Debouck, D.G.; Roca, W.; Lema, M. White mold-resistant, interspecific common bean breeding line VRW 32 derived from Phaseolus costaricensis. J. Plant Regist. 2013, 7, 95–99. [Google Scholar] [CrossRef]
- Montero-Rojas, M.; Ortiz, M.; Beaver, J.S.; Siritunga, D. Genetic, morphological and cyanogen content evaluation of a new collection of Caribbean Lima bean (Phaseolus lunatus L.) landraces. Genet. Resour. Crop Evol. 2013. [Google Scholar] [CrossRef]
- Hyten, D.L.; Song, Q.; Fickus, E.W.; Quigley, C.V.; Lim, J.-S.; Choi, I.-Y.; Hwang, E.-Y.; Pastor-Corrales, M.P.; Cregan, P.B. High-throughput SNP discovery and assay development in common bean. BMC Genomics 2010, 11, 475. [Google Scholar] [CrossRef]
- Phytozome. Available online: http://www.phytozome.net/commonbean (accessed on 6 May 2013).
- Le, S.Q.; Durbin, R. SNP detection and genotyping from low-coverage sequencing data on multiple diploid samples. Genome Res. 2010, 21, 952–960. [Google Scholar]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping by sequencing (GBS) approach for high diversity species. PLoS One 2011, 6, e19379. [Google Scholar]
- Kidd, J.M.; Graves, T.; Newman, T.; Fulton, R.; Hayden, H.S.; Malig, M.; Kallicki, J.; Kaul, R.; Wilson, R.K.; Eichler, E.E. A human genome structural variation sequencing resource reveals insights into mutation mechanisms. Cell 2011, 143, 837–847. [Google Scholar]
- Cokus, S.J.; Feng, S.; Hang, X.; Chen, Z.; Merriman, B.; Haudenshcild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef]
- Rech, E.L.; Vianna, G.R.; Aragão, F.J.L. High-efficiency transformation by biolistics of soybean, common bean and cotton transgenic plants. Nat. Protoc. 2008, 3, 410–418. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Porch, T.G.; Beaver, J.S.; Debouck, D.G.; Jackson, S.A.; Kelly, J.D.; Dempewolf, H. Use of Wild Relatives and Closely Related Species to Adapt Common Bean to Climate Change. Agronomy 2013, 3, 433-461. https://doi.org/10.3390/agronomy3020433
Porch TG, Beaver JS, Debouck DG, Jackson SA, Kelly JD, Dempewolf H. Use of Wild Relatives and Closely Related Species to Adapt Common Bean to Climate Change. Agronomy. 2013; 3(2):433-461. https://doi.org/10.3390/agronomy3020433
Chicago/Turabian StylePorch, Timothy G., James S. Beaver, Daniel G. Debouck, Scott A. Jackson, James D. Kelly, and Hannes Dempewolf. 2013. "Use of Wild Relatives and Closely Related Species to Adapt Common Bean to Climate Change" Agronomy 3, no. 2: 433-461. https://doi.org/10.3390/agronomy3020433
APA StylePorch, T. G., Beaver, J. S., Debouck, D. G., Jackson, S. A., Kelly, J. D., & Dempewolf, H. (2013). Use of Wild Relatives and Closely Related Species to Adapt Common Bean to Climate Change. Agronomy, 3(2), 433-461. https://doi.org/10.3390/agronomy3020433




