Pod Shattering: A Homologous Series of Variation Underlying Domestication and an Avenue for Crop Improvement
Abstract
:1. Introduction
2. Milestone in Plant Domestication: Pod Shattering as Part of the Domestication Syndrome
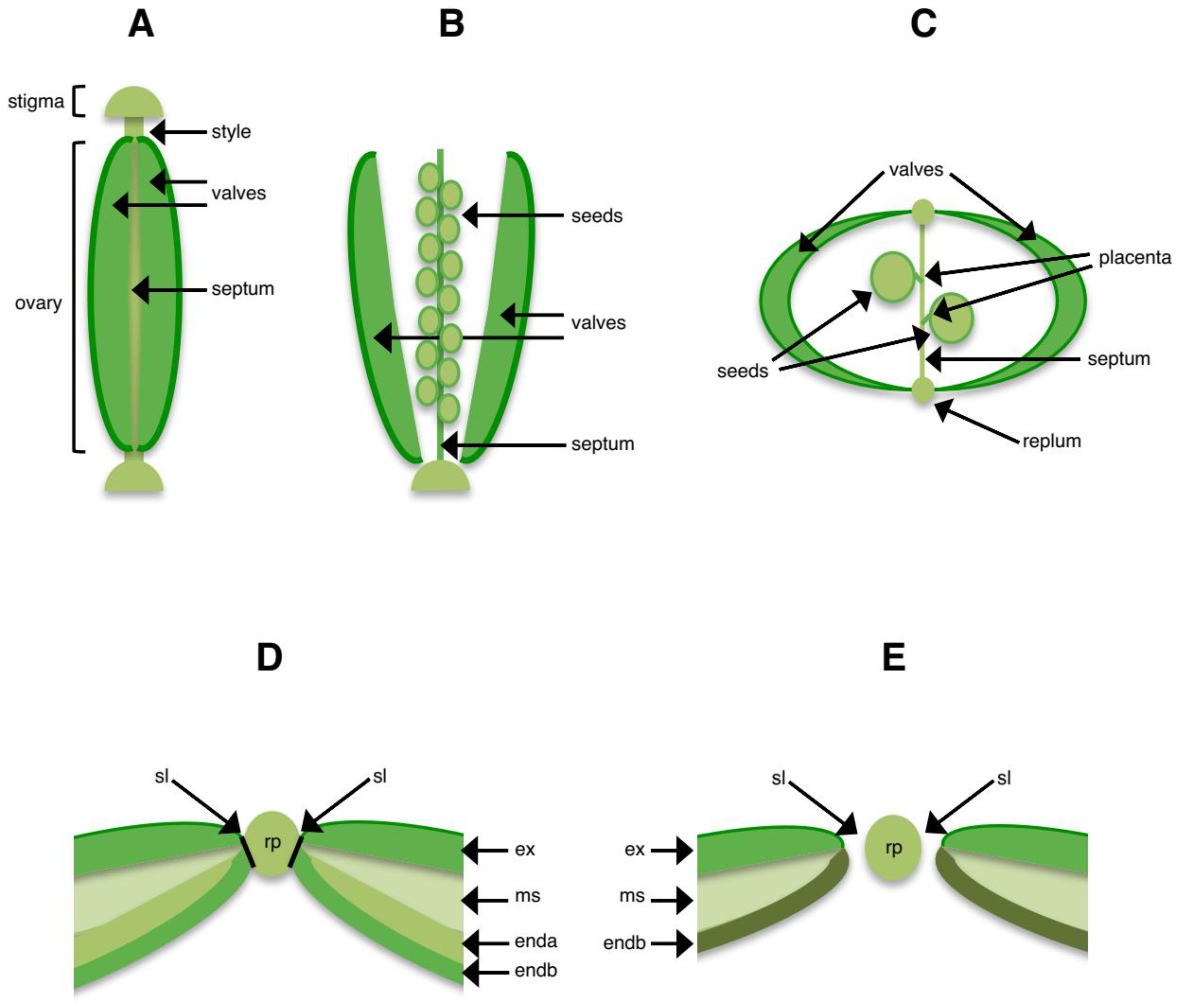
3. Structures Involved in Shattering
4. Molecular Basis of Shattering
4.1. Shattering in Cereal Crops
4.2. Shattering in Arabidopsis and Other Crucifers
4.3. Shattering in Legumes
5. Crop Losses and the Impact of Environmental Factors on Pod Shattering
6. Efforts to Control Pod Shattering
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ballester, P.; Ferrándiz, C. Shattering fruits: Variations on a dehiscent theme. Curr. Opin. Plant Biol. 2017, 35, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Howe, H.F.; Smallwood, J. Ecology of seed dispersal. Annu. Rev. Ecol. Syst. 1982, 13, 201–228. [Google Scholar] [CrossRef]
- Abbo, S.; Gopher, A. Near Eastern Plant Domestication: A History of Thought. Trends Plant Sci. 2017, 22, 491–511. [Google Scholar] [CrossRef] [PubMed]
- Abbo, S.; Van-Oss, R.P.; Gopher, A.; Saranga, Y.; Ofner, I.; Peleg, Z. Plant domestication versus crop evolution: A conceptual framework for cereals and grain legumes. Trends Plant Sci. 2014, 19, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Abbo, S.; Saranga, Y.; Peleg, Z.; Kerem, Z.; Lev-Yadun, S.; Gopher, A. Reconsidering domestication of legumes versus cereals in the ancient Near East. Q. Rev. Biol. 2009, 84, 29–50. [Google Scholar] [CrossRef] [PubMed]
- Zohary, D. Pulse domestication and cereal domestication: How different are they? Econ. Bot. 1989, 43, 31–34. [Google Scholar] [CrossRef]
- Hajjar, R.; Hodgkin, T. The use of wild relatives in crop improvement: A survey of developments over the last 20 years. Euphytica 2007, 156, 1–13. [Google Scholar] [CrossRef]
- Brumlop, S.; Reichenbecher, W.; Tappeser, B.; Finckh, M.R. What is the SMARTest way to breed plants and increase agrobiodiversity? Euphytica 2013, 194, 53–66. [Google Scholar] [CrossRef]
- Warschefsky, E.; Varma Penmetsa, R.; Cook, D.R.; Von Wettberg, E.J.B. Back to the wilds: Tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. Am. J. Bot. 2014, 101, 1791–1800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darwin, C. The Variation of Animals and Plants under Domestication; John Murray: London, UK, 1868; Volume 8. [Google Scholar]
- Vavilov, N.I. The Origin, Variation, Immunity and Breeding of Cultivated Plants; Chester, K.S., Ed.; Chronica Botanica: Waltham, MA, USA, 1951; Volume 72, p. 482. [Google Scholar]
- Vavilov, N.I.; Vavylov, M.I.; Dorofeev, V.F. Origin and Geography of Cultivated Plants; Cambridge University Press: Cambridge, UK, 1992. [Google Scholar]
- Koinange, E.M.K.; Singh, S.P.; Gepts, P. Genetic control of the domestication syndrome in common bean. Crop Sci. 1996, 36, 1037–1045. [Google Scholar] [CrossRef]
- Konishi, S.; Izawa, T.; Lin, S.Y.; Ebana, K.; Fukuta, Y.; Sasaki, T.; Yano, M. An SNP caused loss of seed shattering during rice domestication. Science 2006, 312, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, D.S.; Liu, H.S.; Yin, C.S.; Li, X.; Liang, W.; Yuan, Z.; Xu, B.; Chu, H.W.; Wang, J.; et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 2006, 18, 2999–3014. [Google Scholar] [CrossRef] [PubMed]
- Ladizinsky, G. The genetics of several morphological traits in the lentil. J. Hered. 1979, 70, 135–137. [Google Scholar] [CrossRef]
- Simons, K.J.; Fellers, J.P.; Zhang, Z.C.; Faris, J.D. Molecular Characterization of the Major Wheat Domestication Gene Q. Genetics 2006, 172, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Weeden, N.F.; Brauner, S.; Przyborowski, J.A. Genetic Analysis of Pod Dehiscence in Pea (Pisum Sativum L.). Cell. Mol. Biol. Lett. 2002, 7, 657–663. [Google Scholar] [PubMed]
- Lin, Z.; Li, X.; Shannon, L.; Yeh, C.; Wang, M.; Bai, G.; Peng, Z.; Li, J.; Trick, H.; Clemente, T.; et al. Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 2012, 44, 720–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paterson, A.H.; Lin, Y.R.; Li, Z.; Schertz, K.F.; Doebley, J.F.; Pinson, S.R.M.; Liu, S.C.; Stansel, J.W.; Irvine, J.E. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 1995, 269, 1714–1718. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.P. Legumes of the World; Royal Botanic Gardens Kew: London, UK, 2005. [Google Scholar]
- Azani, N.; Babineau, M.; Bailey, C.D.; Banks, H.; Barbosa, A.R.; Pinto, R.B.; Boatwright, J.S.; Borges, L.M.; Brown, G.K.; Bruneau, A.; et al. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny—The Legume Phylogeny Working Group (LPWG). Taxon 2017, 66, 44–77. [Google Scholar] [CrossRef]
- Foyer, C.H.; Lam, H.M.; Nguyen, H.T.; Siddique, K.H.M.; Varshney, R.K.; Colmer, T.D.; Cowling, W.; Bramley, H.; Mori, T.A.; Hodgson, J.M.; et al. Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2016, 2, 16112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Considine, M.J.; Siddique, K.H.M.; Foyer, C.H. Nature’s pulse power: Legumes, food security and climate change. J. Exp. Bot. 2017, 68, 1815–1818. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.A.; Buchmann, N.; Sprent, J.; Buckley, T.N.; Turnbull, T.L. Crops, Nitrogen, Water: Are Legumes Friend, Foe, or Misunderstood Ally? Trends Plant Sci. 2018, 23, 539–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuller, D.Q.; Harvey, E.L. The archaeobotany of Indian pulses: Identification, processing and evidence for cultivation. Environ. Archaeol. 2006, 11, 219–246. [Google Scholar] [CrossRef]
- Krieg, C.P.; Kassa, M.T.; von Wettberg, E.J.B. Germplasm Characterization and Trait Discovery. In The Pigeonpea Genome; Springer: Basel, Switzerland, 2017; pp. 65–79. [Google Scholar]
- Prosperi, J.M.; Jenczewski, E.; Muller, M.H.; Fourtier, S.; Sampoux, J.P.; Ronfort, J. Alfalfa domestication history, genetic diversity and genetic resources. Legume Perspectives 2014, 4, 13–14. [Google Scholar]
- Gladstones, J.S. (Ed.) Lupins as crop plants. Field Crop abstr. 1970, 23, 123–148. [Google Scholar]
- Caracuta, V.; Barzilai, O.; Khalaily, H.; Milevski, I.; Paz, Y.; Vardi, J.; Regev, L.; Boaretto, E. The onset of faba bean farming in the Southern Levant. Sci. Rep. 2015, 5, 14370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caracuta, V.; Vardi, J.; Paz, Y.; Boaretto, E. Farming legumes in the pre-pottery Neolithic: New discoveries from the site of Ahihud (Israel). PLoS ONE 2017, 12, e0177859. [Google Scholar] [CrossRef] [PubMed]
- Fuller, D.Q.; Denham, T.; Arroyo-Kalin, M.; Lucas, L.; Stevens, C.J.; Qin, L.; Allaby, R.G.; Purugganan, M.D. Convergent evolution and parallelism in plant domestication revealed by an expanding archaeological record. Proc. Natl. Acad. Sci. USA 2014, 111, 6147–6152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purugganan, M.D.; Fuller, D.Q. The Nature of Selection During Plant Domestication. Nature 2009, 457, 843–848. Available online: https://www.nature.com/articles/nature07895 (accessed on 2 June 2018).
- Meyer, R.S.; Purugganan, M.D. Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 2013, 14, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The molecular genetics of crop domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Bitocchi, E.; Bellucci, E.; Giardini, A.; Rau, D.; Rodriguez, M.; Biagetti, E.; Santilocchi, R.; Zeuli, P.S.; Gioia, T.; Logozzo, G.; et al. Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 2013, 197, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammer, K. Das Domestikationssyndrom. Die Kult. 1984, 32, 11–34. [Google Scholar] [CrossRef]
- Vaughan, D.A.; Balázs, E.; Harrison, J.S.H. From Crop Domestication to Super—Domestication. Ann. Bot. 2007, 100, 893–901. [Google Scholar] [CrossRef] [PubMed]
- Fuller, D.Q. Contrasting patterns in crop domestication and domestication rates: Recent archaeobotanical insights from the Old World. Ann. Bot. 2007, 100, 903–924. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Martin, B.; Milla, R.; Martin-Robles, N.; Arc, E.; Kranner, I.; Becerril, J.M.; Garcia-Plazaola, J.I. Side-effects of domestication: Cultivated legume seeds contain similar tocopherols and fatty acids but less carotenoids than their wild counterparts. BMC Plant Biol. 2014, 14, 1599. [Google Scholar] [CrossRef]
- Kerem, Z.; Lev-Yadun, S.; Gopher, A.; Weinberg, P.; Abbo, S. Chickpea domestication in the Neolithic Levant through the nutritional perspective. J. Archaeol. Sci. 2007, 34, 1289–1293. [Google Scholar] [CrossRef]
- Zohary, D. Unconscious selection and the evolution of domesticated plants. Econ. Bot. 2004, 58, 5–10. [Google Scholar] [CrossRef]
- Abd El-Moneim, A.M. Selection for Non-Shattering Common Vetch, Vicia sativa L. Plant Breed. 1993, 110, 168–171. [Google Scholar] [CrossRef]
- Gioia, T.; Logozzo, G.; Kami, J.; Spagnoletti Zeuli, P.; Gepts, P. Identification and characterization of a homologue to the Arabidopsis INDEHISCENT gene in common bean. J. Hered. 2012, 104, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Kaga, A.; Isemura, T.; Tomooka, N.; Vaughan, D.A. The genetics of domestication of the azuki bean (Vigna angularis). Genetics 2008, 178, 1013–1036. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.; Muehlbauer, F.J. Gene mapping in Lentil with Recombinant Inbred Lines. J. Hered. 1994, 85, 306–310. [Google Scholar] [CrossRef]
- Boersma, J.G.; Nelson, M.N.; Sivasithamparam, K.; Yang, H. Development of sequence-specific PCR markers linked to the Tardus gene that reduces pod shattering in narrow-leafed lupin (Lupinus angustifolius L.). Mol. Breed 2009, 23, 259–267. [Google Scholar] [CrossRef]
- Funatsuki, H.; Hajika, M.; Hagihara, S.; Yamada, T.; Tanaka, Y.; Tsuji, H.; Ishimoto, M.; Fujino, K. Confirmation of the location and the effects of a major QTL controlling pod dehiscence, qPDH1, in soybean. Breed. Sci. 2008, 58, 63–69. [Google Scholar] [CrossRef] [Green Version]
- Kongjaimun, A.; Kaga, A.; Tomooka, N.; Somta, P.; Vaughan, D.A.; Srinives, P. The genetics of domestication of yardlong bean, Vigna unguiculata (L.) Walp. ssp. unguiculata cv.-gr. sesquipedalis. Ann. Bot. 2012, 109, 1185–1200. [Google Scholar] [CrossRef] [PubMed]
- Weeden, N.F. Genetic changes accompanying the domestication of Pisum sativum: Is there a common genetic basis to the domestication syndrome for legumes? Ann. Bot. 2007, 100, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.W.; Leslie, M.E.; Liljegren, S.J. Plant separation: 50 ways to leave your mother. Curr. Opin. Plant Biol. 2006, 9, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Wang, Y.Z. Seed shattering: From models to crops. Front. Plant Sci. 2015, 6, 476. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Yang, H.; Zhang, L.; Wang, X.; Liu, G.; Wang, H.; Hua, W. A large replum-valve joint area is associated with increased resistance to pod shattering in rapeseed. J. Plant Res. 2015, 128, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Lenser, T.; Theißen, G. Molecular mechanisms involved in convergent crop domestication. Trends Plant Sci. 2013, 18, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Dardick, C.; Callahan, A.M. Evolution of the fruit endocarp: Molecular mechanisms underlying adaptations in seed protection and dispersal strategies. Front. Plant Sci. 2014, 5, 284. [Google Scholar] [CrossRef] [PubMed]
- Østergaard, L.; Borkhardt, B.; Ulvskov, P. Dehiscence. In Annual Plant Reviews; Roberts, J.A., Gonzalez-Carranza, Z., Eds.; John Wiley & Sons: Chichester, UK, 2007; Volume 25, pp. 137–163. [Google Scholar]
- Ferrándiz, C.; Pelaz, S.; Yanofsky, M.F. Control of carpel and fruit development in Arabidopsis. Annu. Rev. Biochem. 1999, 68, 321–354. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.A.; Elliott, K.A.; Gonzalez-Carranza, Z.H. Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 2002, 53, 131–158. [Google Scholar] [CrossRef] [PubMed]
- Meakin, P.J.; Roberts, J.A. Dehiscence of fruit in oilseed rape (Brassica napus L.) II. The role of cell wall degrading enzymes and ethylene. J. Exp. Bot. 1990, 41, 1003–1011. [Google Scholar] [CrossRef]
- Meakin, P.J.; Roberts, J.A. Dehiscence of fruit in oilseed rape (Brassica napus L.) I. Anatomy of pod dehiscence. J. Exp. Bot. 1990, 41, 955–1002. [Google Scholar] [CrossRef]
- Morgan, C.L.; Bruce, D.M.; Child, R.; Ladbrooke, Z.L.; Arthur, A.E. Genetic variation for pod shatter resistance among lines of oilseed rape developed from synthetic B. napus. Field. Crop. Res. 1998, 58, 153–165. [Google Scholar] [CrossRef]
- Sorefan, K.; Girin, T.; Liljegren, S.; Ljung, K.; Robles, P.; Galvin-Ampudia, C.; Offringa, R.; Friml, J.; Yanofsky, M.F.; Ostergaard, L. A regulated auxin minimum is required for seed dispersal. Nature 2009, 459, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Chauvaux, N.; Child, R.; John, K.; Ulvskov, P.; Borkhardt, B.; Prinsen, E.; Van Onckelen, H.A. The role of auxin in cell separation in the dehiscence zone of oilseed rape pods. J. Exp. Bot. 1997, 48, 1423–1429. [Google Scholar] [CrossRef] [Green Version]
- Marsch-Martínez, N.; Ramos-Cruz, D.; Irepan Reyes-Olalde, J.; Lozano-Sotomayor, P.; Zúñiga-Mayo, V.M.; De Folter, S. The role of cytokinin during Arabidopsis gynoecia and fruit morphogenesis and patterning. Plant J. 2012, 72, 222–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christiansen, L.C.; Dal Degan, F.; Ulvskov, P.; Borkhardt, B. Examination of the dehiscence zone in soybean pods and isolation of a dehiscence-related endopolygalacturonase gene. Plant Cell Environ. 2002, 25, 479–490. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Shen, Y.Y.; Wu, X.M.; Wang, J.B. The basis of pod dehiscence: Anatomical traits of the dehiscence zone and expression of eight pod shatter-related genes in four species of Brassicaceae. Biol. Plant. 2016, 60, 343–354. [Google Scholar] [CrossRef]
- Tiwari, S.; Bhatia, V. Characters of pod anatomy associated with pod shatering in soybean. Ann. Bot. 1995, 76, 483–485. [Google Scholar] [CrossRef]
- Dong, Y.; Yang, X.; Liu, J.; Wang, B.H.; Liu, B.L.; Wang, Y.Z. Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat. Commun. 2014, 5, 3352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glémin, S.; Bataillon, T. A comparative view of the evolution of grasses under domestication. New Phytol. 2009, 183, 273–290. [Google Scholar] [CrossRef] [PubMed]
- Sang, T. Genes and Mutations Underlying Domestication Transitions in Grasses. Plant Physiol. 2009, 149, 63–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Olsen, K.M. Chapter Three—To Have and to Hold: Selection for Seed and Fruit Retention During Crop Domestication. Curr. Top. Dev. Biol. 2016, 119, 63–109. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Gill, B.S. Multiple genetic pathways for seed shattering in the grasses. Funct. Integr. Genom. 2006, 6, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Estornell, L.H.; Agusti, J.; Merelo, P.; Talón, M.; Tadeo, F.R. Elucidating mechanisms underlying organ abscission. Plant Sci. 2013, 199, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lu, D.; Li, C.; Luo, J.; Zhu, B.; Zhu, J.; Shangguan, Y.; Wang, Z.; Sang, T.; Zhou, B.; et al. Genetic Control of Seed Shattering in Rice by the APETALA2 Transcription Factor SHATTERING ABORTION1. Plant Cell 2012, 24, 1034–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.; Cho, L.H.; Kim, S.L.; Choi, H.; Koh, H.J.; An, G. The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J. 2014, 79, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Faris, J.D.; Fellers, J.P.; Brooks, S.A.; Gill, B.S. A bacterial artificial chromosome contig spanning the major domestication locus Q in wheat and identification of a candidate gene. Genetics 2003, 164, 311–321. [Google Scholar] [PubMed]
- Kato, K.; Miura, H.; Sawada, S. QTL mapping of genes controlling ear emergence time and plant height on chromosome 5A of wheat. Theor. Appl. Genet. 1999, 98, 472–477. [Google Scholar] [CrossRef]
- Muramatsu, M. Dosage effect of the spelta gene q of hexaploid wheat. Genetics 1963, 48, 469–482. [Google Scholar] [PubMed]
- Tang, H.; Cuevas, H.E.; Das, S.; Sezen, U.U.; Zhou, C.; Guo, H.; Goff, V.H.; Ge, Z.; Clemente, T.E.; Paterson, A.H. Seed shattering in a wild sorghum is conferred by a locus unrelated to domestication. Proc. Natl. Acad. Sci. USA 2013, 110, 15824–15829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnaud, N.; Girin, T.; Sorefan, K.; Fuentes, S.; Wood, T.A.; Lawrenson, T.; Sablowski, R.; Østergaard, L. Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 2010, 24, 2127–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liljegren, S.; Ditta, G.; Eshed, Y.; Savidge, B. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 2000, 404, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Liljegren, S.J.; Roeder, A.H.K.; Kempin, S.A.; Gremski, K.; Østergaard, L.; Guimil, S.; Reyes, D.K.; Yanofsky, M.F. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 2004, 116, 843–853. [Google Scholar] [CrossRef]
- Rajani, S.; Sundaresan, V. The Arabidopsis myc/bHLH gene alcatraz enables cell separation in fruit dehiscence. Curr. Biol. 2001, 11, 1914–1922. [Google Scholar] [CrossRef]
- Roeder, A.H.K.; Ferrándiz, C.; Yanofsky, M.F. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 2003, 13, 1630–1635. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Ferrándiz, C.; Yanofsky, M.F.; Martienssen, R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 1998, 125, 1509–1517. [Google Scholar] [PubMed]
- Dinneny, J.R.; Weigel, D.; Yanofsky, M.F. A genetic framework for fruit patterning in Arabidopsis thaliana. Development 2005, 132, 4687–4696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savidge, B. Temporal Relationship between the Transcription of Two Arabidopsis MADS Box Genes and the Floral Organ Identity Genes. Plant Cell 1995, 7, 721–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ripoll, J.J.; Roeder, A.H.K.; Ditta, G.S.; Yanofsky, M.F. A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 2011, 138, 5167–5176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrášek, J.; Mravec, J.; Bouchard, R.; Blakeslee, J.; Abas, M.; Seifertova, D.; Wisnieska, J.; Tadele, Z.; Kubes, M.; Covanova, M.; et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 2006, 312, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Girin, T.; Paicu, T.; Stephenson, P.; Fuentes, S.; Körner, E.; O’Brien, M.; Sorefan, K.; Wood, T.A.; Balanzá, V.; Ferrándiz, C.; et al. INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell 2011, 23, 3641–3653. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Demura, T.; Ye, Z.H. SND1, a NAC Domain Transcription Factor, Is a Key Regulator of Secondary Wall Synthesis in Fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsuda, N.; Ohme-Takagi, M. NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 2008, 56, 768–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, M.; Kay, P.; Wilson, S.; Swain, S.M. ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 Are Polygalacturonases Required for Cell Separation during Reproductive Development in Arabidopsis. Plant Cell 2009, 21, 216–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofhuis, H.; Moulton, D.; Lessinnes, T.; Routier-Kierzkowska, A.L.; Bomphrey, R.J.; Mosca, G.; Reinhardt, H.; Sarchet, P.; Gan, X.; Tsiantis, M.; et al. Morphomechanical innovation drives explosive seed dispersal. Cell 2016, 166, 222–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, M.A.; Mian, M.A.R.; Carter, T.E., Jr.; Ashley, D.A.; Boerma, H.R. Pod dehiscence of soybean: Identification of quantitative trait loci. J. Hered. 1997, 88, 152–154. [Google Scholar] [CrossRef]
- Funatsuki, H.; Ishimoto, M.; Tsuji, H.; Kawaguchi, K.; Hajika, M.; Fujino, K. Simple sequence repeat markers linked to a major QTL controlling pod shattering in soybean. Plant Breed 2006, 125, 195–197. [Google Scholar] [CrossRef]
- Funatsuki, H.; Suzuki, M.; Hirose, A.; Inaba, H.; Yamada, T.; Hajika, M.; Komatsu, K.; Katayama, T.; Sayama, T.; Ishimoto, M.; et al. Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc. Natl. Acad. Sci. USA 2014, 111, 17797–17802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, S.T.; Kwak, M.; Kim, H.K.; Choung, M.G.; Han, W.Y.; Baek, I.Y.; Kim, M.Y.; Van, K.; Lee, S.H. Population-specific QTLs and their different epistatic interactions for pod dehiscence in soybean [Glycine max (L.) Merr.]. Euphytica 2009, 166, 15. [Google Scholar] [CrossRef]
- Liu, B.; Fujita, T.; Yan, Z.; Sakamoto, S.; Xu, D.; Abe, J. QTL Mapping of Domestication-related Traits in Soybean (Glycine max). Ann. Bot. 2007, 100, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Fujino, K.; Funatsuki, H.A. Major Soybean QTL, qPDH1, Controls Pod Dehiscence without Marked Morphological Change. Plant Prod. Sci. 2009, 12, 217–223. [Google Scholar] [CrossRef]
- Suzuki, M.; Fujino, K.; Nakamoto, Y.; Ishimoto, M.; Funatsuki, H. Fine mapping and development of DNA markers for the qPDH1 locus associated with pod dehiscence in soybean. Mol. Breed. 2010, 25, 407–418. [Google Scholar] [CrossRef]
- Yamada, T.; Funatsuki, H.; Hagihara, S.; Fujita, S.; Tanaka, Y.; Tsuji, H.; Ishimoto, M.; Fujino, K.; Hajika, M.A. major QTL, qPDH1, is commonly involved in shattering resistance of soybean cultivars. Breed. Sci. 2009, 59, 435–440. [Google Scholar] [CrossRef]
- Blixt, S. Mutation genetics in Pisum. In Agri Hortique Genetica; Food and Agriculture Organization of the United Nations: Rome, Italy, 1972; p. 30. [Google Scholar]
- Aliboh, V.O.; Kehinde, O.B.; Fawole, I. Inheritance of leaf mark, pod dehiscence and dry pod colour in crosses between wild and cultivated cowpeas. Afr. Crop Sci. J. 1996, 4, 283–288. [Google Scholar]
- Andargie, M.; Pasquet, R.S.; Gowda, B.S.; Muluvi, G.M.; Timko, M.P. Construction of a SSR-based genetic map and identification of QTL for domestication traits using recombinant inbred lines from a cross between wild and cultivated cowpea (V. unguiculata (L.) Walp.). Mol. Breed. 2011, 28, 413–420. [Google Scholar] [CrossRef]
- Mohammed, M.S.; Russom, Z.; Abdul, S.D. Inheritance of hairiness and pod shattering, heritability and correlation studies in crosses between cultivated cowpea (Vigna unguiculata (L.) Walp.) and its wild (var. pubescens) relative. Euphytica 2009, 171, 397–407. [Google Scholar] [CrossRef]
- Suanum, W.; Somta, P.; Kongjaimun, A.; Yimram, T.; Kaga, A.; Tomooka, N.; Takahashi, Y.; Srinives, P. Co-localization of QTLs for pod fiber content and pod shattering in F2 and backcross populations between yardlong bean and wild cowpea. Mol. Breed 2016, 36, 80. [Google Scholar] [CrossRef]
- Fratini, R.; Durán, Y.; Garcia, P.; De La Vega, M.P. Identification of quantitative trait loci (QTL) for plant structure, growth habit and yield in lentil. Spanish J. Agric. Res. 2007, 5, 348–356. [Google Scholar] [CrossRef]
- Isemura, T.; Kaga, A.; Konishi, S.; Ando, T.; Tomooka, N.; Han, O.K.; Vaughan, D.A. Genome dissection of traits related to domestication in azuki bean (Vigna angularis) and comparison with other warm-season legumes. Ann. Bot. 2007, 100, 1053–1071. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Dong, D.; Luo, D.; Zhou, Q.; Chai, X.; Zhang, J.; Xie, W.; Liu, W.; Dong, Y.; Wang, Y.; et al. Transcriptome Analyses Reveal Candidate Pod Shattering-Associated Genes Involved in the Pod Ventral Sutures of Common Vetch (Vicia sativa L.). Front. Plant Sci. 2017, 8, 649. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.N.; Phan, H.T.T.; Ellwood, S.R.; Moolhuijzen, P.M.; Hane, J.; Williams, A.; O’Lone, C.E.; Fosu-Nyarko, J.; Scobie, M.; Cakir, M.; et al. The first gene-based map of Lupinus angustifolius L.-location of domestication genes and conserved synteny with Medicago truncatula. Theor. Appl. Genet. 2006, 113, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Hradilová, I.; Trněný, O.; Válková, M.; Cechová, M.; Janská, A.; Prokešová, L.; Aamir, K.; Krezdorn, N.; Rotter, B.; Winter, P.; et al. A combined comparative transcriptomic, metabolomic, and anatomical analyses of two key domestication traits: Pod dehiscence and seed dormancy in pea (Pisum sp.). Front. Plant Sci. 2017, 8, 542. [Google Scholar] [CrossRef] [PubMed]
- Hagerty, C.H.; Cuesta-Marcos, A.; Cregan, P.; Song, Q.; McClean, P.; Myers, J.R. Mapping snap bean pod and color traits, in a dry bean× snap bean recombinant inbred population. J. Am. Soc. Hortic. Sci. 2016, 141, 131–138. [Google Scholar]
- Nanni, L.; Bitocchi, E.; Bellucci, E.; Rossi, M.; Rau, D.; Attene, G.; Gepts, P.; Papa, R. Nucleotide diversity of a genomic sequence similar to SHATTERPROOF (PvSHP1) in domesticated and wild common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2011, 123, 1341–1357. [Google Scholar] [CrossRef] [PubMed]
- Ladizinsky, G. Seed dispersal in relation to the domestication of Middle East legumes. Econ. Bot. 1979, 33, 284–289. [Google Scholar] [CrossRef]
- Kazan, K.; Muehlbauer, F.J.; Weeden, N.E.; Ladizinsky, G. Inheritance and linkage relationships of morphological and isozyme loci in chickpea (Cicer arietinum L.). Theor. Appl. Genet. 1993, 86, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Gaur, P.M.; Slinkard, A.E. Genetic control and linkage relations of additional isozyme markers in chick-pea. Theor. Appl. Genet. 1990, 80, 648–656. [Google Scholar] [CrossRef] [PubMed]
- Von Wettberg, E.J.B.; Chang, P.L.; Başdemir, F.; Carrasquila-Garcia, N.; Korbu, L.B.; Moenga, S.M.; Bedada, G.; Greenlon, A.; Moriuchi, K.S.; Singh, V.; et al. Ecology and genomics of an important crop wild relative as a prelude to agricultural innovation. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Gan, Y.; Blackshaw, R.E.; May, W.E.; Vera, C.; Johnson, E.N. Yield Stability and Seed Shattering Characteristics of Brassica juncea Canola in the Northern Great Plains. Crop Sci. 2016, 56, 1296–1305. [Google Scholar] [CrossRef]
- Cavalieri, A.; Lewis, D.W.; Gulden, R.H. Pod drop and pod shatter are not closely related in canola. Crop Sci. 2014, 54, 1184–1188. [Google Scholar] [CrossRef]
- Summers, J.E.; Bruce, D.M.; Vancanneyt, G.; Redig, P.; Werner, C.P.; Morgan, C.; Child, R.D. Pod shatter resistance in the resynthesized Brassica napus line DK142. J. Agric. Sci. 2003, 140, 43–52. [Google Scholar] [CrossRef]
- Baloch, U.K. Wheat: Post-harvest operations; Food and Agriculture Organization of the United Nations: Rome, Italy, 1999. [Google Scholar]
- Grover, D.K.; Singh, J.M. Post-harvest Losses in Wheat Crop in Punjab: Past and Present. Agric. Econ. Res. Rev. 2013, 26, 293–297. [Google Scholar]
- Tukamuhabwa, P.; Dashiell, K.; Rubaihayo, P.; Nanasirye, M. Determination of field yield loss and effect of environment on pod shattering in soybean. Afr. Crop Sci. J. 2002, 10, 203–209. [Google Scholar]
- Zhang, L.; Boahen, S. Evaluation of critical shattering time of early-maturity soybeans under early soybean production system. Agric. Biol. J. 2010, 1, 440–447. [Google Scholar] [CrossRef]
- Gan, Y.; Malhi, S.S.; Brandt, S.A.; McDonald, C.L. Assessment of seed shattering resistance and yield loss in five oilseed crops. Can. J. Plant Sci. 2008, 88, 267–270. [Google Scholar] [CrossRef] [Green Version]
- Price, J.S.; Hobson, R.N.; Neale, M.A.; Bruce, D.M. Seed losses in commercial harvesting of oilseed rape. J. Agric. Eng. Res. 1996, 65, 183–191. [Google Scholar] [CrossRef]
- Vera, C.L.; Downey, R.K.; Woods, S.M.; Raney, J.P.; McGregor, D.I.; Elliott, R.H.; Johnson, E.N. Yield and quality of canola seed as affected by stage of maturity at swathing. Can. J. Plant Sci. 2007, 87, 13–26. [Google Scholar] [CrossRef] [Green Version]
- Van Gastel, A.; Bishaw, Z.; Niane, A.; Gregg, B.; Gan, Y. Chickpea seed production. In Chickpea Breeding and Management; Yadav, S.S., Chen, W., Eds.; CABI (Centre for Agriculture and Bioscience International) International: Oxford, UK, 2007; pp. 417–444. [Google Scholar]
- Philbrook, B.D.; Oplinger, E.S. Soybean Field Losses as Influenced by Harvest Delays. Agron. J. 1989, 81, 251–258. [Google Scholar] [CrossRef]
- Erskine, W. Selection for pod retention and pod indehiscence in lentils. Euphytica 1985, 34, 105–112. [Google Scholar] [CrossRef]
- Tiwari, S.; Bhatnager, P. Minimizing pod shattering in soybean. In India Farming; ICAR: Indore, India, 1989; Volume 39, pp. 23–24. [Google Scholar]
- Tsuchiya, T. Physiological and Genetic Analysis of Pod Shattering in Soybeans. Jpn. Agric. Res. Q. 1987, 21, 166–175. [Google Scholar]
- Child, R.D.; Summers, J.E.; Babij, J.; Farrent, J.W.; Bruce, D.M. Increased resistance to pod shatter is associated with changes in the vascular structure in pods of a resynthesized Brassica napus line. J. Exp. Bot. 2003, 54, 1919–1930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margheim, J.F.; Baltensperger, D.D.; Wilson, R.G.; Lyon, D.J.; Hein, G.L.; Harveson, R.M.; Burgener, P.A.; Krall, J.M.; Cecil, J.T.; Rickertsen, J.R.; et al. Chickpea Production in the High Plains; University of Nebraska–Lincoln: Lincoln, NE, USA, 2004. [Google Scholar]
- Gulden, R.H.; Cavalieri, A.; Syrovy, L.D.; Shirtliffe, S.J. Pod drop in Brassica napus is linked to weight-adjusted pod-retention resistance. Field. Crop. Res. 2017, 205, 34–44. [Google Scholar] [CrossRef]
- Østergaard, L.; Kempin, S.A.; Bies, D.; Klee, H.J.; Yanofsky, M.F. Pod shatter-resistant Brassica fruit produced by ectopic expression of the FRUITFULL gene. Plant Biotechnol. J. 2006, 4, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Sintim, H.; Zheljazkov, V.; Obour, A.K. Managing harvest time to control pod shattering in oilseed camelina. Agon. J. 2016, 108, 656–661. [Google Scholar] [CrossRef]
- Li, X.R.; Deb, J.; Kumar, S.V.; Østergaard, L. Temperature Modulates Tissue-Specification Program to Control Fruit Dehiscence in Brassicaceae. Mol. Plant 2018, 11, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Kuai, J.; Sun, Y.; Liu, T.; Zhang, P.; Zhou, M.; Wu, J.; Zhou, G. Physiological mechanisms behind differences in pod shattering resistance in rapeseed (Brassica napus L.) varieties. PLoS ONE 2016, 11, e0157341. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Gan, Y.; Poppy, L. Evaluation of on-farm crop management decisions on canola productivity. Can. J. Plant Sci. 2014, 94, 131–139. [Google Scholar] [CrossRef]
- Alberti, P.K. Development of Best Management Practices for Production of Ethiopian Mustard (Brassica carinata) in South Dakota. Master’s Thesis, South Dakota State University, Brookings, SD, USA, 18 December 2017. [Google Scholar]
- Kumar, D.; Kalita, P. Reducing postharvest losses during storage of grain crops to strengthen food security in developing countries. Foods 2017, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Lusser, M.; Parisi, C.; Plan, D.; Rodríguez-Cerezo, E. Deployment of new biotechnologies in plant breeding. Nat. Biotechnol. 2012, 30, 231–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaart, J.G.; van de Wiel, C.C.M.; Lotz, L.A.P.; Smulders, M.J.M. Opportunities for Products of New Plant Breeding Techniques. Trends Plant Sci. 2016, 21, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Klee, H.J.; Horsch, R.B.; Hinchee, M.A.; Hein, M.B.; Hoffmann, N.L. The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Genes Dev. 1987, 1, 86–96. [Google Scholar] [CrossRef]
- Beetham, P.R.; Kipp, P.B.; Sawycky, X.L.; Arntzen, C.J.; May, G.D. A tool for functional plant genomics: Chimeric RNA/DNA oligonucleotides cause in vivo gene-specific mutations. Proc. Natl. Acad. Sci. USA 1999, 96, 8774–8778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Townsend, J.A.; Wright, D.A.; Winfrey, R.J.; Fu, F.; Maeder, M.L.; Joung, J.K.; Voytas, D.F. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 2009, 459, 442–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schouten, H.J.; Jacobsen, E. Cisgenesis and intragenesis, sisters in innovative plant breeding. Trends Plant Sci. 2008, 13, 260–261. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 1231143. [Google Scholar] [CrossRef] [PubMed]
- Van de Wiel, C.C.M.; Schaart, J.G.; Lotz, L.A.P.; Smulders, M.J.M. New traits in crops produced by genome editing techniques based on deletions. Plant Biotechnol. Rep. 2017, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Altpeter, F.; Springer, N.M.; Bartley, L.E.; Blechl, A.E.; Brutnell, T.P.; Citovsky, V.; Conrad, L.J.; Gelvin, S.B.; Jackson, D.P.; Kausch, A.P.; et al. Advancing crop transformation in the era of genome editing. Plant Cell 2016, 28, 1510–1520. [Google Scholar] [CrossRef] [PubMed]
- Fuller, D.Q.; Allaby, R. Seed dispersal and crop domestication: Shattering, germination and seasonality in evolution under cultivation. Annu. Plant Rev. 2009, 38, 238–295. [Google Scholar] [CrossRef]
- Jenkins, E.S.; Paul, W.; Coupe, S.A.; Bell, S.J.; Davies, E.C.; Roberts, J.A. Characterization of an mRNA encoding a polygalacturonase expressed during pod development in oilseed rape (Brassica napus L.). J. Exp. Bot. 1996, 47, 111–115. [Google Scholar] [CrossRef]
- Petersen, M.; Sander, L.; Child, R.; van Onckelen, H.; Ulvskov, P.; Borkhardt, B. Isolation and characterisation of a pod dehiscence zone-specific polygalacturonase from Brassica napus. Plant Mol. Biol. 1996, 31, 517–527. [Google Scholar] [CrossRef] [PubMed]


| Species Name | Identified QTL | Method | Reference |
|---|---|---|---|
| Vicia sativa L. (common vetch) | unknown | cross breeding | [44] |
| Vigna angularis (Willd.) Ohwi & H. Ohashi (azuki bean) | Pdt | linkage mapping, QTL analysis | [46,110] |
| Lupinus angustifolius L. (narrow-leafed lupin) | Lentus, Tardus | linkage mapping, QTL analysis | [112] |
| Lens culinaris Medikus (lentil) | Pi | cross breeding, linkage mapping, QTL analysis | [16,47,109] |
| Vigna unguiculata (L.) Walp. (cowpea) | Dhp/Pdd, Pdt | cross breeding, linkage mapping, QTL analysis | [50,105,106,107,108] |
| Pisum sativum L. (pea) | Dpo1/2, Np/Gp | linkage mapping, QTL analysis | [18,51,104,113] |
| Phaseolus vulgaris L. (common bean) | St | linkage mapping, genetic association | [13,45,114,115] |
| Glycine max L. (soybean) | Pdh1, SHAT1-5 | linkage mapping, QTL analysis, genetic association | [49,69,96,97,98,99,100,101,102,103] |
| Species Examined | Reference | |
|---|---|---|
| Agricultural factors | ||
| Harvest method | Brassica napus L. | [128,129] |
| Harvest time | Brassica napus L. | [128,129] |
| Cicer arietinum L. | [130] | |
| Glycine max L. | [131] | |
| Lens culinaris Medikus | [132] | |
| Triticum aestivum L. | [124] | |
| Seeding time | Brassica napus L. | [129] |
| Environmental factors | ||
| Precipitation | Brassica napus L. | [129] |
| Glycine max L. | [133] | |
| Wind | Brassica napus L. | [129] |
| Temperature | Cicer arietinum L. | [130] |
| Glycine max L. | [134] | |
| Humidity | Brassica napus L. | [129,134] |
| Glycine max L. | [133] | |
| Structural factors | ||
| Plant architecture | Brassica napus L. | [122] |
| Vascular bundle size | Brassica napus L. | [135] |
| Pod structure | Brassica napus L. | [122] |
| Seed moisture content | Cicer arietinum L. | [136] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogutcen, E.; Pandey, A.; Khan, M.K.; Marques, E.; Penmetsa, R.V.; Kahraman, A.; Von Wettberg, E.J.B. Pod Shattering: A Homologous Series of Variation Underlying Domestication and an Avenue for Crop Improvement. Agronomy 2018, 8, 137. https://doi.org/10.3390/agronomy8080137
Ogutcen E, Pandey A, Khan MK, Marques E, Penmetsa RV, Kahraman A, Von Wettberg EJB. Pod Shattering: A Homologous Series of Variation Underlying Domestication and an Avenue for Crop Improvement. Agronomy. 2018; 8(8):137. https://doi.org/10.3390/agronomy8080137
Chicago/Turabian StyleOgutcen, Ezgi, Anamika Pandey, Mohd Kamran Khan, Edward Marques, R. Varma Penmetsa, Abdullah Kahraman, and Eric J. B. Von Wettberg. 2018. "Pod Shattering: A Homologous Series of Variation Underlying Domestication and an Avenue for Crop Improvement" Agronomy 8, no. 8: 137. https://doi.org/10.3390/agronomy8080137
APA StyleOgutcen, E., Pandey, A., Khan, M. K., Marques, E., Penmetsa, R. V., Kahraman, A., & Von Wettberg, E. J. B. (2018). Pod Shattering: A Homologous Series of Variation Underlying Domestication and an Avenue for Crop Improvement. Agronomy, 8(8), 137. https://doi.org/10.3390/agronomy8080137








