Preliminary Genetic Map of a New Recombinant Inbred Line Population for Narrow-leafed Lupin (Lupinus angustifolius L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Material
2.2. Phenotyping Type of Growth
2.3. DNA Extraction
2.4. SSR and ISSR Genotyping
2.5. DArTseq Genotyping
2.6. Genetic Map Construction
2.7. Homology Search within Lupin and Legume Gene Sequences
3. Results
3.1. ISSR and SSR Genotyping
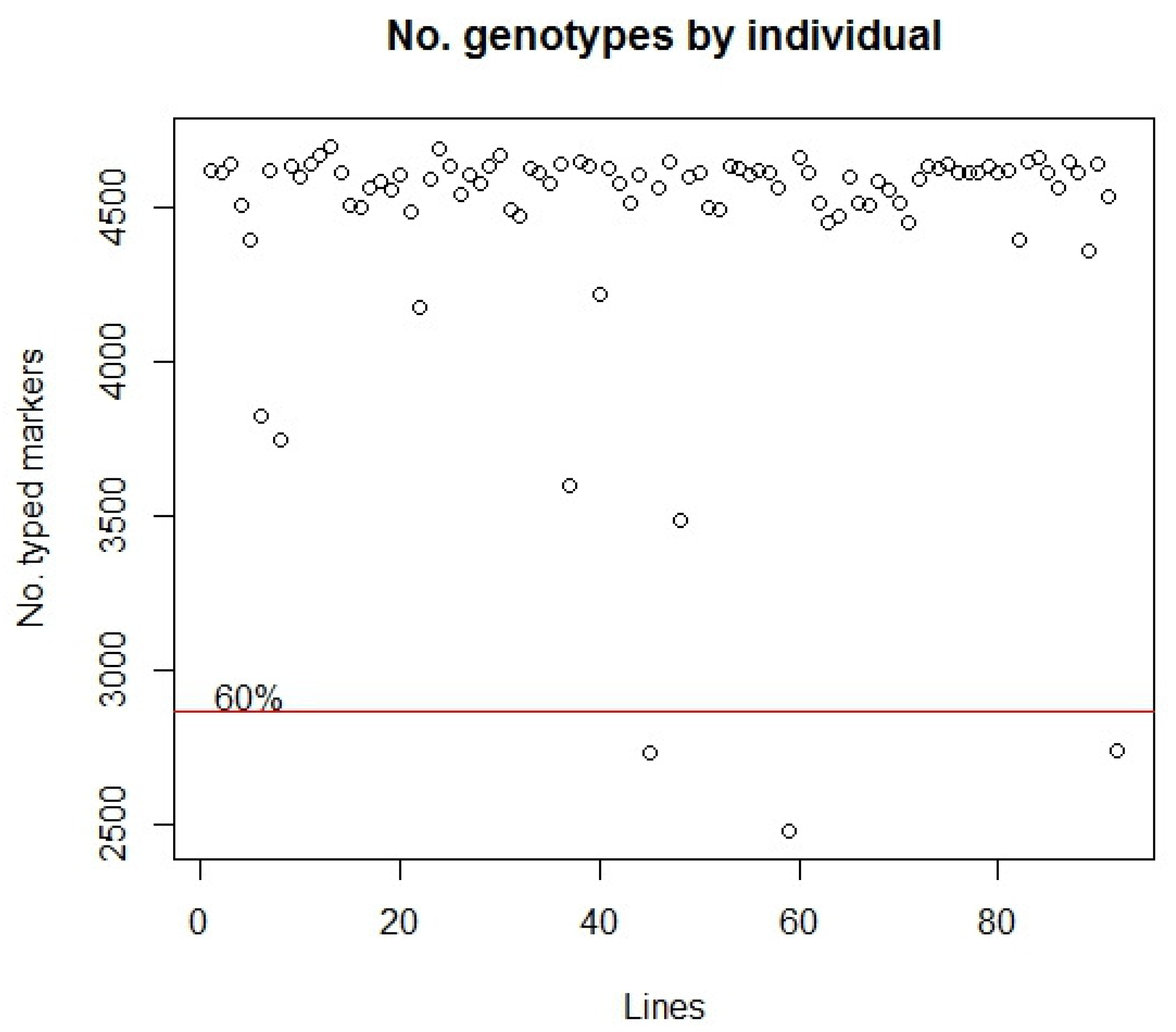
3.2. DArTseq Genotyping
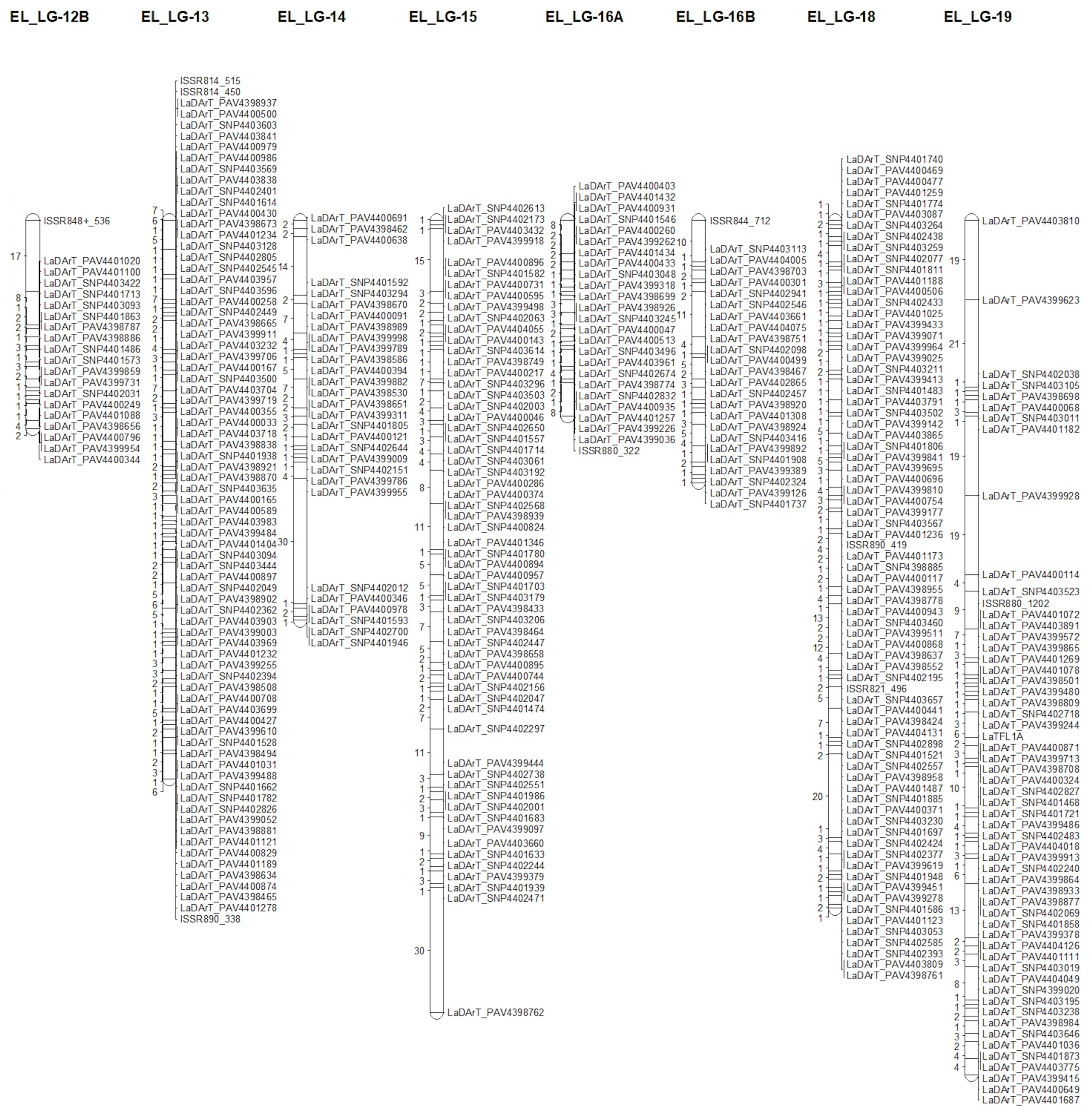
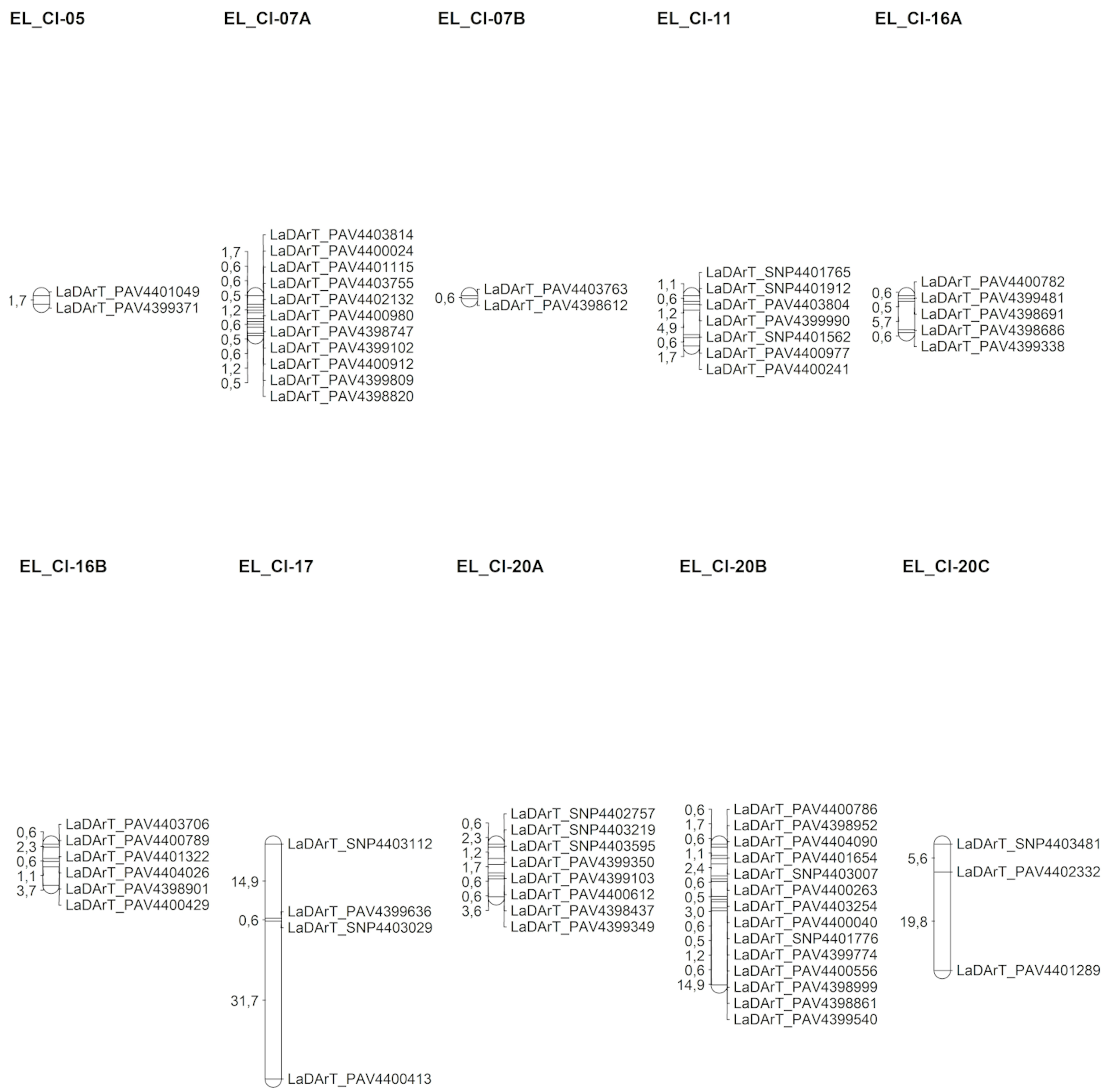
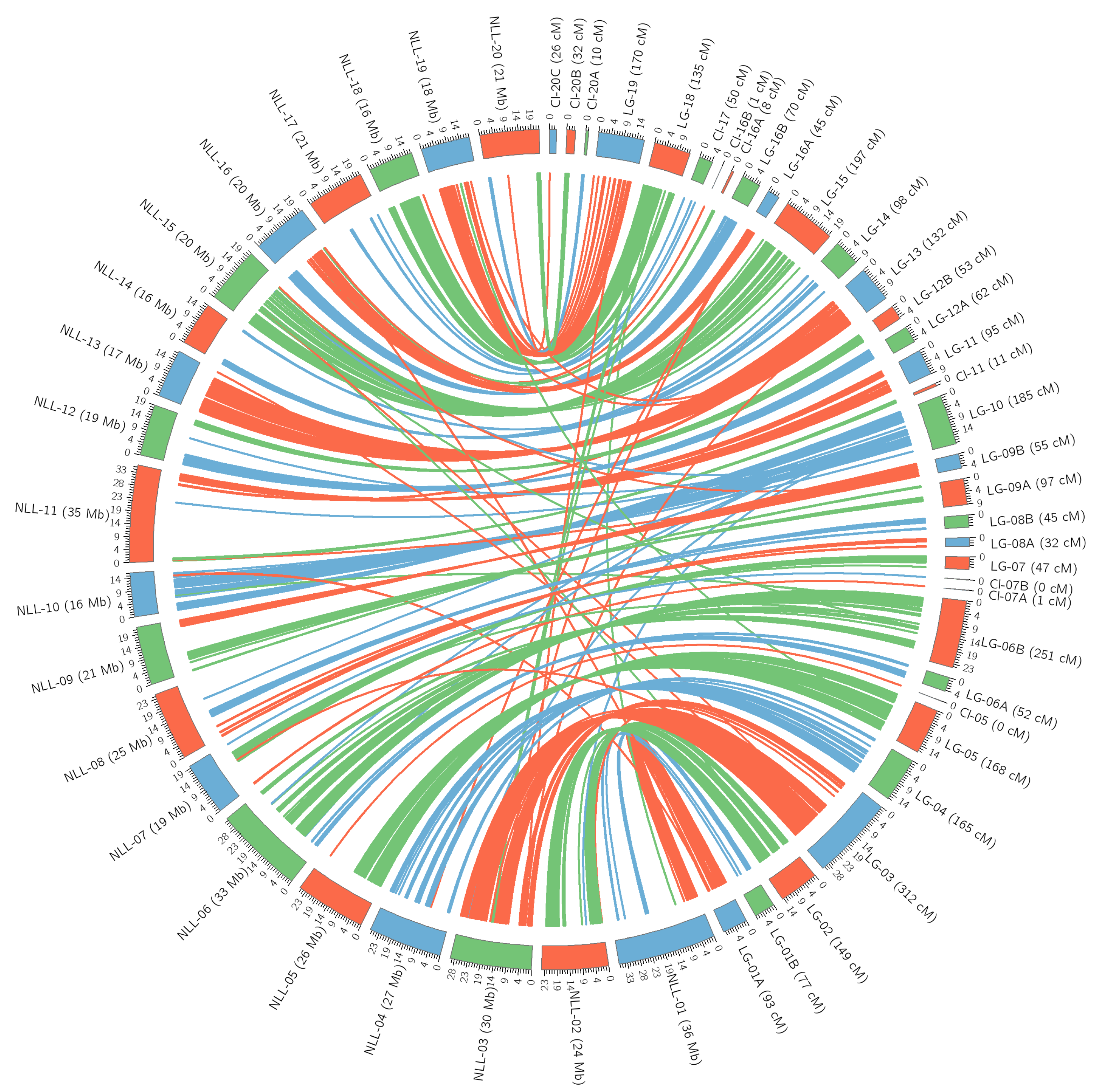
3.3. Genetic Map Construction
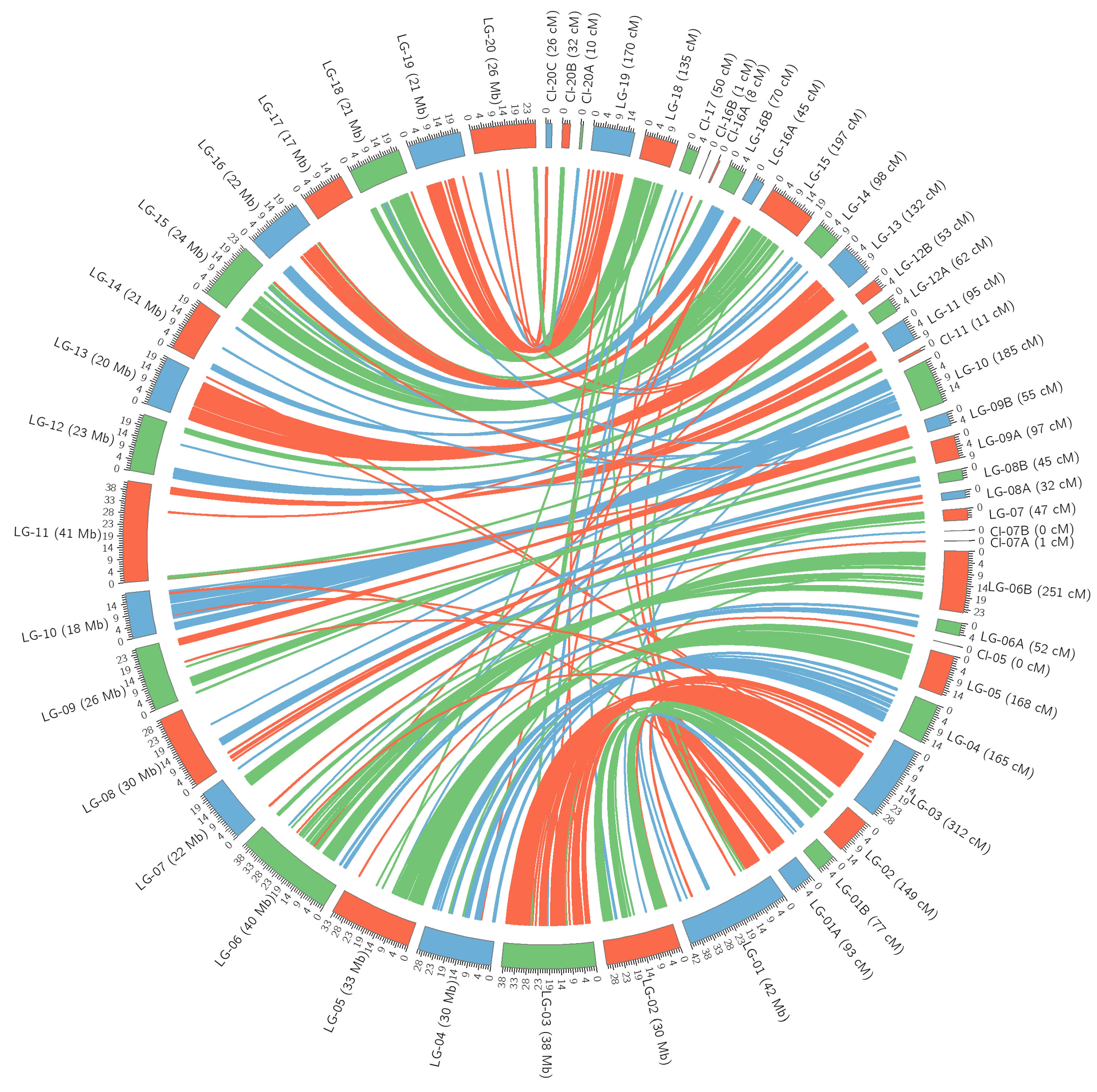
3.4. Analysis of Unplaced Scaffolds from Reference Genome
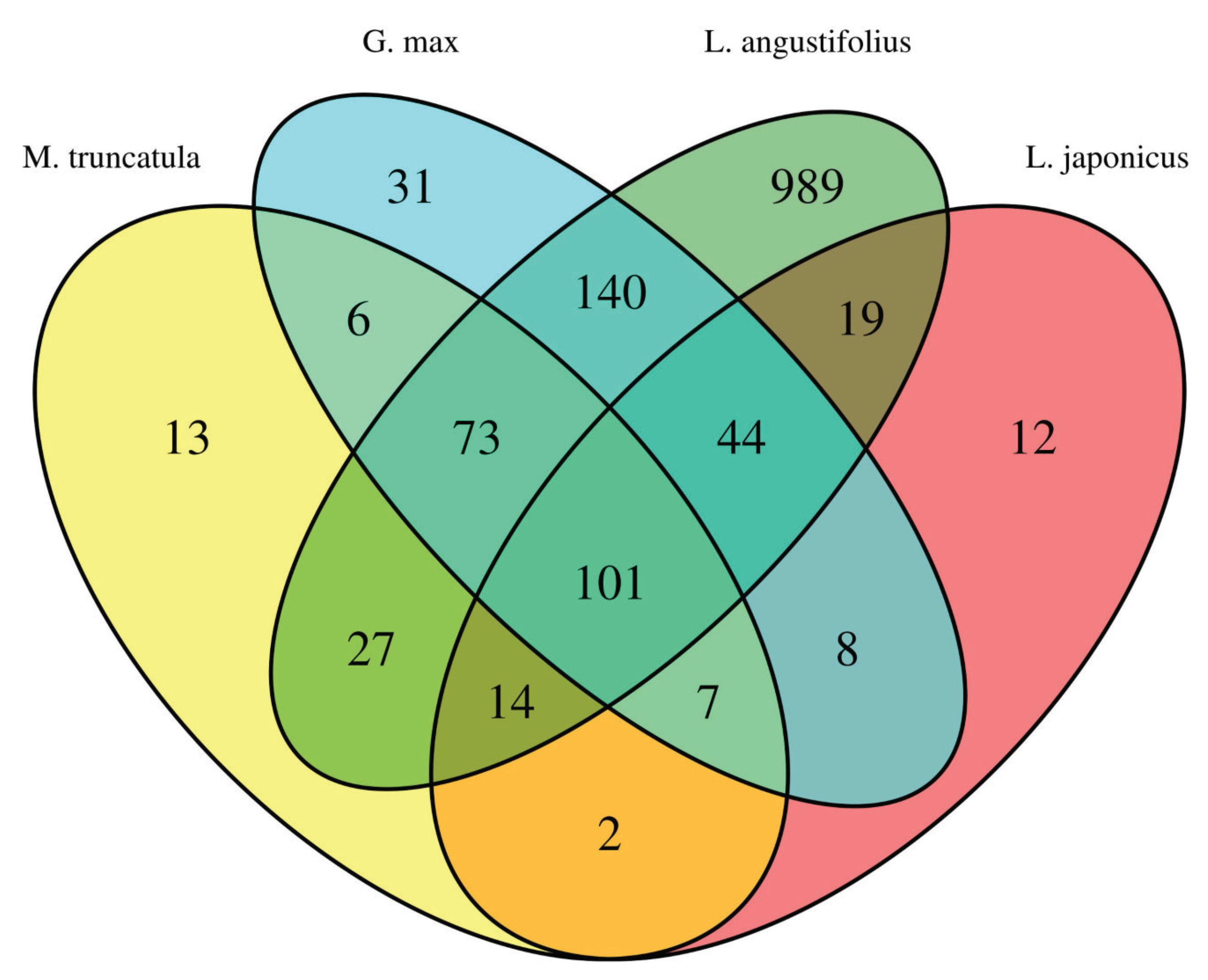
3.5. Homology between DArTseq Markers and Model Legume Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lucas, M.M.; Stoddard, F.L.; Annicchiarico, P.; Frias, J.; Martinez-Villaluenga, C.; Sussmann, D.; Duranti, M.; Seger, A.; Zander, P.M.; Pueyo, J.J. The future of lupin as a protein crop in Europe. Front. Plant Sci. 2015, 6, 705. [Google Scholar] [CrossRef]
- Legocki, A.; Bothe, H.; Pühler, A. Biological Fixation of Nitrogen for Ecology and Sustainable Agriculture; Springer Science & Business Media: Berlin, Germany, 2013; Volume 39. [Google Scholar]
- Lee, Y.P.; Mori, T.A.; Sipsas, S.; Barden, A.; Puddey, I.B.; Burke, V.; Hall, R.S.; Hodgson, J.M. Lupin-enriched bread increases satiety and reduces energy intake acutely. Am. J. Clin. Nutr. 2006, 84, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.S.; Thomas, S.J.; Johnson, S.K. Australian sweet lupin flour addition reduces the glycaemic index of a white bread breakfast without affecting palatability in healthy human volunteers. Asia Pac. J. Clin. Nutr. 2005, 14, 91. [Google Scholar] [PubMed]
- Hayashi, M.; Miyahara, A.; Sato, S.; Kato, T.; Yoshikawa, M.; Taketa, M.; Hayashi, M.; Pedrosa, A.; Onda, R.; Imaizumi-Anraku, H.; et al. Construction of a genetic linkage map of the model legume Lotus japonicus using an intraspecific F2 population. DNA Res. 2001, 8, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Nakamura, Y.; Kaneko, T.; Asamizu, E.; Kato, T.; Nakao, M.; Sasamoto, S.; Watanabe, A.; Ono, A.; Kawashima, K.; et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008, 15, 227–239. [Google Scholar] [CrossRef]
- Choi, H.K.; Kim, D.; Uhm, T.; Limpens, E.; Lim, H.; Mun, J.H.; Kalo, P.; Penmetsa, R.V.; Seres, A.; Kulikova, O.; et al. A sequence-based genetic map of Medicago truncatula and comparison of marker colinearity with M. sativa. Genetics 2004, 166, 1463–1502. [Google Scholar] [CrossRef]
- Branca, A.; Paape, T.D.; Zhou, P.; Briskine, R.; Farmer, A.D.; Mudge, J.; Bharti, A.K.; Woodward, J.E.; May, G.D.; Gentzbittel, L.; et al. Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proc. Natl. Acad. Sci. USA 2011, 108, E864–E870. [Google Scholar] [CrossRef]
- Lark, K.; Weisemann, J.; Matthews, B.; Palmer, R.; Chase, K.; Macalma, T. A genetic map of soybean (Glycine max L.) using an intraspecific cross of two cultivars:‘Minosy’and ‘Noir 1’. Theor. Appl. Genet. 1993, 86, 901–906. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178. [Google Scholar] [CrossRef]
- Hane, J.K.; Ming, Y.; Kamphuis, L.G.; Nelson, M.N.; Garg, G.; Atkins, C.A.; Bayer, P.E.; Bravo, A.; Bringans, S.; Cannon, S.; et al. A comprehensive draft genome sequence for lupin (Lupinus angustifolius), an emerging health food: Insights into plant–microbe interactions and legume evolution. Plant Biotechnol. J. 2017, 15, 318–330. [Google Scholar] [CrossRef]
- Boersma, J.G.; Pallotta, M.; Li, C.; Buirchell, B.J.; Sivasithamparam, K.; Yang, H. Construction of a genetic linkage map using MFLP and identification of molecular markers linked to domestication genes in narrow-leafed lupin (Lupinus angustifolius L.). Cell. Mol. Biol. Lett. 2005, 10, 331–344. [Google Scholar] [PubMed]
- Nelson, M.N.; Phan, H.T.; Ellwood, S.R.; Moolhuijzen, P.M.; Hane, J.; Williams, A.; Clare, E.; Fosu-Nyarko, J.; Scobie, M.; Cakir, M.; et al. The first gene-based map of Lupinus angustifolius L.-location of domestication genes and conserved synteny with Medicago truncatula. Theor. Appl. Genet. 2006, 113, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.N.; Moolhuijzen, P.M.; Boersma, J.G.; Chudy, M.; Lesniewska, K.; Bellgard, M.; Oliver, R.P.; Święcicki, W.; Wolko, B.; Cowling, W.A.; et al. Aligning a new reference genetic map of Lupinus angustifolius with the genome sequence of the model legume, Lotus japonicus. DNA Res. 2010, 17, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Kroc, M.; Koczyk, G.; Święcicki, W.; Kilian, A.; Nelson, M.N. New evidence of ancestral polyploidy in the Genistoid legume Lupinus angustifolius L.(narrow-leafed lupin). Theor. Appl. Genet. 2014, 127, 1237–1249. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Tao, Y.; Zheng, Z.; Zhang, Q.; Zhou, G.; Sweetingham, M.W.; Howieson, J.G.; Li, C. Draft genome sequence, and a sequence-defined genetic linkage map of the legume crop species Lupinus angustifolius L. PLoS ONE 2013, 8, e64799. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Jian, J.; Wang, P.; Li, C.; Tao, Y.; Li, X.; Renshaw, D.; Clements, J.; Sweetingham, M.; Yang, H. Construction of an ultra-high density consensus genetic map, and enhancement of the physical map from genome sequencing in Lupinus angustifolius. Theor. Appl. Genet. 2018, 131, 209–223. [Google Scholar] [CrossRef]
- Von Mark, V.C.; Kilian, A.; Dierig, D.A. Development of DArT marker platforms and genetic diversity assessment of the US collection of the new oilseed crop lesquerella and related species. PLoS ONE 2013, 8, e64062. [Google Scholar]
- Zou, J.; Raman, H.; Guo, S.; Hu, D.; Wei, Z.; Luo, Z.; Long, Y.; Shi, W.; Fu, Z.; Du, D.; et al. Constructing a dense genetic linkage map and mapping QTL for the traits of flower development in Brassica carinata. Theor. Appl. Genet. 2014, 127, 1593–1605. [Google Scholar] [CrossRef]
- Sánchez-Sevilla, J.F.; Horvath, A.; Botella, M.A.; Gaston, A.; Folta, K.; Kilian, A.; Denoyes, B.; Amaya, I. Diversity Arrays Technology (DArT) marker platforms for diversity analysis and linkage mapping in a complex crop, the octoploid cultivated strawberry (Fragaria × ananassa). PLoS ONE 2015, 10, e0144960. [Google Scholar] [CrossRef]
- Ren, R.; Ray, R.; Li, P.; Xu, J.; Zhang, M.; Liu, G.; Yao, X.; Kilian, A.; Yang, X. Construction of a high-density DArTseq SNP-based genetic map and identification of genomic regions with segregation distortion in a genetic population derived from a cross between feral and cultivated-type watermelon. Mol. Genet. Genom. 2015, 290, 1457–1470. [Google Scholar] [CrossRef]
- Iqbal, M.M.; Huynh, M.; Udall, J.A.; Kilian, A.; Adhikari, K.N.; Berger, J.D.; Erskine, W.; Nelson, M.N. The first genetic map for yellow lupin enables genetic dissection of adaptation traits in an orphan grain legume crop. BMC Genet. 2019, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Galek, R. Studies on the Variability of Some Morphological and Functional Characters of Lupinus with Particular Consideration Intra and Interspecific Hybrids. Monografie (Poland) 2010, 1–122. [Google Scholar]
- Doyle, J. DNA protocols for plants Molecular techniques in taxonomy. NATO ASI Ser. 1991, 57, 283–293. [Google Scholar]
- Clements, J.; Galek, R.; Kozak, B.; Michalczyk, D.J.; Piotrowicz-Cieślak, A.I.; Sawicka-Sienkiewicz, E.; Stawiński, S.; Zalewski, D. Diversity of selected Lupinus angustifolius L. genotypes at the phenotypic and DNA level with respect to microscopic seed coat structure and thickness. PLoS ONE 2014, 9, e102874. [Google Scholar] [CrossRef]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C.; et al. Diversity arrays technology: A generic genome profiling technology on open platforms. In Data Production and Analysis in Population Genomics; Springer: Berlin/Heidelberg, Germany, 2012; pp. 67–89. [Google Scholar]
- Cichy, K.A.; Fernandez, A.; Kilian, A.; Kelly, J.D.; Galeano, C.H.; Shaw, S.; Brick, M.; Hodkinson, D.; Troxtell, E. QTL analysis of canning quality and color retention in black beans (Phaseolus vulgaris L.). Mol. Breed. 2014, 33, 139–154. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: http://www.R-project.org (accessed on 16 October 2019).
- Arends, D.; Prins, P.; Jansen, R.C.; Broman, K.W. R/qtl: High-throughput multiple QTL mapping. Bioinformatics 2010, 26, 2990–2992. [Google Scholar] [CrossRef]
- Taylor, J.; Butler, D. Package ASMap: Efficient Genetic Linkage Map Construction andDiagnosis. J. Stat. Softw. 2017, 79, 1–29. [Google Scholar] [CrossRef]
- Kosambi, D.D. The estimation of map distances from recombination values. In DD Kosambi; Springer: Berlin/Heidelberg, Germany, 2016; pp. 125–130. [Google Scholar]
- Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Qian, W.; Ge, S.; Hong, D.Y. Genetic variation within and among populations of a wild rice Oryza granulata from China detected by RAPD and ISSR markers. Theor. Appl. Genet. 2001, 102, 440–449. [Google Scholar] [CrossRef]
- Venkat, S.K.; Bommisetty, P.; Patil, M.S.; Reddy, L.; Chennareddy, A. The genetic linkage maps of Anthurium species based on RAPD, ISSR and SRAP markers. Sci. Hortic. 2014, 178, 132–137. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, L.; Qi, J.; Chen, H.; Tao, A.; Xu, J.; Lin, L.; Fan, P. Genetic linkage map construction for white jute (Corchorus capsularis L.) using SRAP, ISSR and RAPD markers. Plant Breed. 2014, 133, 777–781. [Google Scholar] [CrossRef]
- Sahu, A.R.; Mishra, R.R.; Rath, S.C.; Panigrahi, J. Construction of interspecific genetic linkage map of pigeonpea using SCoT, RAPD, ISSR markers and simple inherited trait loci. Nuclear 2015, 58, 23–31. [Google Scholar] [CrossRef]
- Yang, H.; Li, C.; Lam, H.M.; Clements, J.; Yan, G.; Zhao, S. Sequencing consolidates molecular markers with plant breeding practice. Theor. Appl. Genet. 2015, 128, 779–795. [Google Scholar] [CrossRef] [PubMed]
- Sieber, A.N.; Longin, C.F.H.; Würschum, T. Molecular characterization of winter durum wheat (Triticum durum) based on a genotyping-by-sequencing approach. Plant Genet. Resour. 2017, 15, 36–44. [Google Scholar] [CrossRef]
- Aitken, K.S.; McNeil, M.D.; Berkman, P.J.; Hermann, S.; Kilian, A.; Bundock, P.C.; Li, J. Comparative mapping in the Poaceae family reveals translocations in the complex polyploid genome of sugarcane. BMC Plant Biol. 2014, 14, 190. [Google Scholar] [CrossRef]
- Baloch, F.S.; Alsaleh, A.; Shahid, M.Q.; Çiftçi, V.; de Miera, L.E.S.; Aasim, M.; Nadeem, M.A.; Aktaş, H.; Özkan, H.; Hatipoğlu, R. A whole genome DArTseq and SNP analysis for genetic diversity assessment in durum wheat from central fertile crescent. PLoS ONE 2017, 12, e0167821. [Google Scholar] [CrossRef]
- Gawroński, P.; Pawełkowicz, M.; Tofil, K.; Uszyński, G.; Sharifova, S.; Ahluwalia, S.; Tyrka, M.; Wędzony, M.; Kilian, A.; Bolibok-Brągoszewska, H. DArT markers effectively target gene space in the rye genome. Front. Plant Sci. 2016, 7, 1600. [Google Scholar] [CrossRef]
- Ho, W.K.; Chai, H.H.; Kendabie, P.; Ahmad, N.S.; Jani, J.; Massawe, F.; Kilian, A.; Mayes, S. Integrating genetic maps in bambara groundnut [Vigna subterranea (L) Verdc.] and their syntenic relationships among closely related legumes. BMC Genom. 2017, 18, 192. [Google Scholar] [CrossRef]
- Millan, T.; Winter, P.; JŘngling, R.; Gil, J.; Rubio, J.; Cho, S.; Cobos, M.; Iruela, M.; Rajesh, P.; Tekeoglu, M.; et al. A consensus genetic map of chickpea (Cicer arietinum L.) based on 10 mapping populations. Euphytica 2010, 175, 175–189. [Google Scholar] [CrossRef]
- Yadav, P.; Saxena, K.; Hingane, A.; Kumar, C.S.; Kandalkar, V.; Varshney, R.K.; Saxena, R.K. An “Axiom Cajanus SNP Array” based high density genetic map and QTL mapping for high-selfing flower and seed quality traits in pigeonpea. BMC Genom. 2019, 20, 235. [Google Scholar] [CrossRef] [PubMed]
- Patil, G.; Mian, R.; Vuong, T.; Pantalone, V.; Song, Q.; Chen, P.; Shannon, G.J.; Carter, T.C.; Nguyen, H.T. Molecular mapping and genomics of soybean seed protein: A review and perspective for the future. Theor. Appl. Genet. 2017, 130, 1975–1991. [Google Scholar] [CrossRef] [PubMed]
- Dorrance, A.E. Management of Phytophthora sojae of soybean: A review and future perspectives. Can. J. Plant Pathol. 2018, 40, 210–219. [Google Scholar] [CrossRef]
- Książkiewicz, M.; Nazzicari, N.; Yang, H.; Nelson, M.N.; Renshaw, D.; Rychel, S.; Ferrari, B.; Carelli, M.; Tomaszewska, M.; Stawiński, S.; et al. A high-density consensus linkage map of white lupin highlights synteny with narrow-leafed lupin and provides markers tagging key agronomic traits. Sci. Rep. 2017, 7, 15335. [Google Scholar] [CrossRef]


| Primer Number | Number of Amplification Products | Number of Polymorphic Products | Number of Polymorphic Products with Segregation 1:1 | Size of Amplification Product (bp) |
|---|---|---|---|---|
| 807 | 5 | 5 | 3 | 243, 463, 477 |
| 808 | 8 | 2 | 2 | 493, 1060 |
| 810 | 5 | 1 | 1 | 354 |
| 811 | 6 | 2 | 1 | 667 |
| 813 | 5 | 3 | 1 | 1780 |
| 814 | 10 | 10 | 2 | 450, 515 |
| 815 | 7 | 7 | 2 | 178, 1540 |
| 817 | 7 | 4 | 2 | 357, 1100 |
| 818 | 5 | 5 | 5 | 552, 607, 648, 684, 703 |
| 821 | 3 | 3 | 2 | 496, 638 |
| 823 | 5 | 4 | 3 | 164, 550, 1361 |
| 826 | 6 | 5 | 3 | 277, 471, 882 |
| 828 | 6 | 4 | 2 | 501, 955 |
| 836 | 7 | 6 | 3 | 425, 494, 521 |
| 840 | 8 | 4 | 2 | 216, 818 |
| 842 | 9 | 9 | 3 | 684, 834, 858 |
| 844 | 5 | 4 | 2 | 712, 812 |
| 848 | 5 | 3 | 2 | 536, 748 |
| 849 | 3 | 1 | 1 | 393 |
| 859 | 6 | 4 | 1 | 585 |
| 874 | 3 | 3 | 2 | 659, 1030 |
| 880 | 4 | 4 | 3 | 322, 961, 1200 |
| 884 | 10 | 5 | 2 | 425, 494 |
| 886 | 13 | 1 | 1 | 469 |
| 888 | 8 | 4 | 1 | 433 |
| 889 | 7 | 1 | 1 | 346 |
| 890 | 9 | 4 | 2 | 338, 419 |
| 892 | 3 | 3 | 1 | 1370 |
| 900 | 8 | 8 | 1 | 2230 |
| Marker | Number of Lines in ‘Emir’ Type | Number of Lines in ‘LAE-1’ Type | Number of Lines without Genotyping Results | p-Value for Test |
|---|---|---|---|---|
| SSR_11_260 | 52 | 40 | 0 | 0.211 |
| SSR_11_280 | 52 | 40 | 0 | 0.211 |
| LaSSR_003_289 | 45 | 47 | 0 | 0.835 |
| LaSSR_003_293 | 45 | 47 | 0 | 0.835 |
| LaSSR_015_235 | 35 | 57 | 0 | 0.022 |
| LaSSR_008_254 | 33 | 56 | 3 | 0.015 |
| LaSSR_008_264 | 35 | 54 | 3 | 0.044 |
| LaSSR_011_306 | 36 | 50 | 6 | 0.131 |
| LaSSR_011_312 | 36 | 50 | 6 | 0.131 |
| LaSSR_002_179 | 42 | 45 | 5 | 0.748 |
| LaSSR_002_211 | 37 | 50 | 5 | 0.163 |
| LaSSR_005_291 | 42 | 41 | 9 | 0.913 |
| LaSSR_005_295 | 42 | 41 | 9 | 0.913 |
| LaSSR_019_212 | 49 | 40 | 3 | 0.34 |
| LaSSR_019_216 | 49 | 40 | 3 | 0.34 |
| LaSSR_022_254 | 37 | 53 | 2 | 0.092 |
| LaSSR_022_259 | 39 | 51 | 2 | 0.206 |
| LaSSR_024_209 | 38 | 52 | 2 | 0.14 |
| LaSSR_024_213 | 38 | 52 | 2 | 0.14 |
| Linkage Group | Number of Unique Loci | Number of Markers | % Polymorphic Markers | Length (cM) | Average of Distances (cM) | Maximum Distance (cM) |
|---|---|---|---|---|---|---|
| EL_LG_01A | 34 | 124 | 27.42% | 84.13 | 2.55 | 13.92 |
| EL_LG_01B | 24 | 81 | 29.63% | 85.55 | 3.72 | 29.82 |
| EL_LG_02 | 74 | 305 | 24.26% | 142.94 | 1.96 | 19.38 |
| EL_LG_03 | 145 | 505 | 28.71% | 359.74 | 2.52 | 20.63 |
| EL_LG_04 | 49 | 180 | 27.22% | 174.58 | 3.64 | 28.03 |
| EL_LG_05 | 84 | 273 | 30.77% | 194.68 | 2.35 | 14.92 |
| EL_LG_06A | 17 | 101 | 16.83% | 55.67 | 3.48 | 24.8 |
| EL_LG_06B | 81 | 314 | 25.8% | 223.78 | 2.8 | 23.33 |
| EL_LG_07 | 21 | 76 | 27.63% | 52.34 | 2.62 | 17.05 |
| EL_LG_08A | 12 | 57 | 21.05% | 31.3 | 2.85 | 15.96 |
| EL_LG_08B | 19 | 100 | 19% | 48.96 | 2.72 | 12.96 |
| EL_LG_09A | 24 | 88 | 27.27% | 100.66 | 4.38 | 28.03 |
| EL_LG_09B | 32 | 150 | 21.33% | 55.05 | 1.78 | 6.36 |
| EL_LG_10 | 66 | 244 | 27.05% | 183.46 | 2.82 | 20.63 |
| EL_LG_11 | 35 | 119 | 29.41% | 88.84 | 2.61 | 9.44 |
| EL_LG_12A | 26 | 69 | 37.68% | 63.16 | 2.53 | 12.03 |
| EL_LG_12B | 20 | 149 | 13.42% | 49.51 | 2.61 | 17.05 |
| EL_LG_13 | 77 | 399 | 19.3% | 133.99 | 1.76 | 7.1 |
| EL_LG_14 | 29 | 109 | 26.61% | 96.33 | 3.44 | 29.82 |
| EL_LG_15 | 60 | 171 | 35.09% | 189.61 | 3.21 | 29.82 |
| EL_LG_16A | 25 | 117 | 21.37% | 47.38 | 1.97 | 8.64 |
| EL_LG_16B | 25 | 99 | 25.25% | 62.96 | 2.62 | 10.28 |
| EL_LG_18 | 75 | 305 | 24.59% | 165.27 | 2.23 | 20.63 |
| EL_LG_19 | 57 | 200 | 28.5% | 205.01 | 3.66 | 20.63 |
| EL_Cl_05 | 2 | 5 | 40% | 1.74 | 1.74 | 1.74 |
| EL_Cl_07A | 11 | 71 | 15.49% | 8.02 | 0.8 | 1.74 |
| EL_Cl_07B | 2 | 11 | 18.18% | 0.57 | 0.57 | 0.57 |
| EL_Cl_11 | 7 | 52 | 13.46% | 10.13 | 1.69 | 4.95 |
| EL_Cl_16A | 5 | 11 | 45.45% | 7.35 | 1.84 | 5.65 |
| EL_Cl_16B | 6 | 11 | 54.55% | 8.26 | 1.65 | 3.62 |
| EL_Cl_17 | 4 | 10 | 40% | 47.23 | 15.74 | 31.74 |
| EL_Cl_20A | 8 | 43 | 18.6% | 10.57 | 1.51 | 3.62 |
| EL_Cl_20B | 14 | 49 | 28.57% | 28.28 | 2.18 | 14.92 |
| EL_Cl_20C | 4 | 4 | 100% | 25.37 | 12.69 | 10.28 |
| Total | 1174 | 4602 | 3042.42 |
| Linkage Groups | Corelation Coeficient r [11] | Corelation Coeficient r [17] |
|---|---|---|
| Cl_05 | ||
| Cl_07A | 0.97 | 0.97 |
| Cl_07B | 0.79 | 0.79 |
| Cl_11 | 0.96 | 0.96 |
| Cl_16A | 0.99 | 0.98 |
| Cl_16B | 0.39 | 0.39 |
| Cl_17 | 1.0 | 0.99 |
| Cl_20A | 0.96 | 0.96 |
| Cl_20B | 0.97 | 0.97 |
| Cl_20C | 1.0 | |
| LG_01A | 0.72 | 0.75 |
| LG_01B | 0.99 | 0.99 |
| LG_02 | 0.93 | 0.93 |
| LG_03 | 0.95 | 0.92 |
| LG_04 | 0.97 | 0.97 |
| LG_05 | 0.95 | 0.94 |
| LG_06A | 0.97 | 0.97 |
| LG_06B | 0.98 | 0.98 |
| LG_07 | 0.86 | 0.63 |
| LG_08A | 0.99 | 0.98 |
| LG_08B | 0.95 | 0.95 |
| LG_09A | 0.96 | 0.88 |
| LG_09B | 0.15 | 0.02 |
| LG_10 | 0.81 | 0.82 |
| LG_11 | 0.99 | 0.99 |
| LG_12A | 0.94 | 0.95 |
| LG_12B | 0.96 | 0.96 |
| LG_13 | 0.84 | 0.87 |
| LG_14 | 0.86 | 0.86 |
| LG_15 | 0.94 | 0.92 |
| LG_16A | 0.42 | 0.15 |
| LG_16B | 0.95 | 0.95 |
| LG_18 | 0.85 | 0.74 |
| LG_19 | 0.91 | 0.91 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozak, B.; Galek, R.; Zalewski, D.; Sawicka-Sienkiewicz, E. Preliminary Genetic Map of a New Recombinant Inbred Line Population for Narrow-leafed Lupin (Lupinus angustifolius L.). Agronomy 2019, 9, 653. https://doi.org/10.3390/agronomy9100653
Kozak B, Galek R, Zalewski D, Sawicka-Sienkiewicz E. Preliminary Genetic Map of a New Recombinant Inbred Line Population for Narrow-leafed Lupin (Lupinus angustifolius L.). Agronomy. 2019; 9(10):653. https://doi.org/10.3390/agronomy9100653
Chicago/Turabian StyleKozak, Bartosz, Renata Galek, Dariusz Zalewski, and Ewa Sawicka-Sienkiewicz. 2019. "Preliminary Genetic Map of a New Recombinant Inbred Line Population for Narrow-leafed Lupin (Lupinus angustifolius L.)" Agronomy 9, no. 10: 653. https://doi.org/10.3390/agronomy9100653
APA StyleKozak, B., Galek, R., Zalewski, D., & Sawicka-Sienkiewicz, E. (2019). Preliminary Genetic Map of a New Recombinant Inbred Line Population for Narrow-leafed Lupin (Lupinus angustifolius L.). Agronomy, 9(10), 653. https://doi.org/10.3390/agronomy9100653





