CRISPR/Cas9-Induced Mutagenesis of Semi-Rolled Leaf1,2 Confers Curled Leaf Phenotype and Drought Tolerance by Influencing Protein Expression Patterns and ROS Scavenging in Rice (Oryza sativa L.)
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Material and Growth Conditions
2.2. CRISPR/Cas9 Plasmid Construction and Generation of Mutant Plants
2.3. Genotyping of T0 Generation and Off-target Effect Detection
2.4. Morphophysiological and Microscopic Analysis
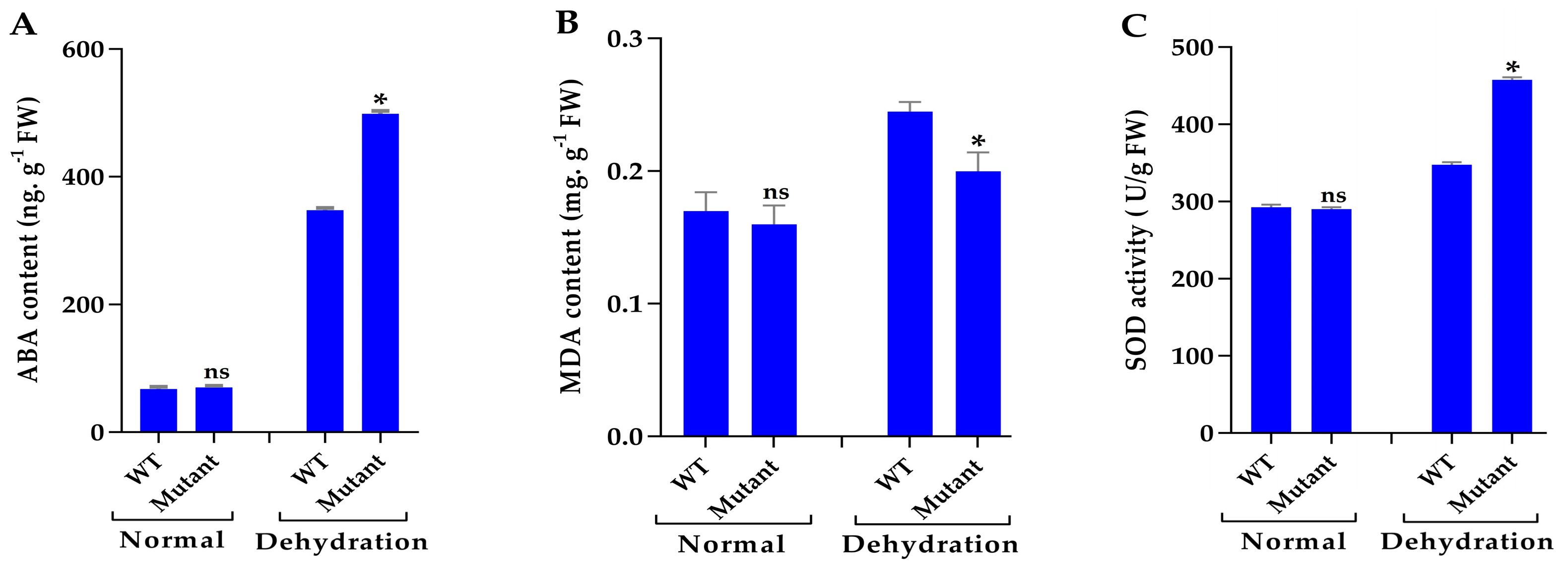
2.5. Measurement of ABA Contents, MDA Contents, and Antioxidant Enzyme Assays Under Drought Stress
2.6. Protein Extraction, Digestion, and Labeling
2.7. Protein–Protein Interaction (PPI) and Data Analysis
2.8. Expression Analysis of Target Genes and Validation of Proteomic Data
2.9. Development of Hybrids and Assessment of Main Agronomic Characters
3. Results
3.1. Assembly of Targets in pYLCRISPR/Cas9Pubi-H
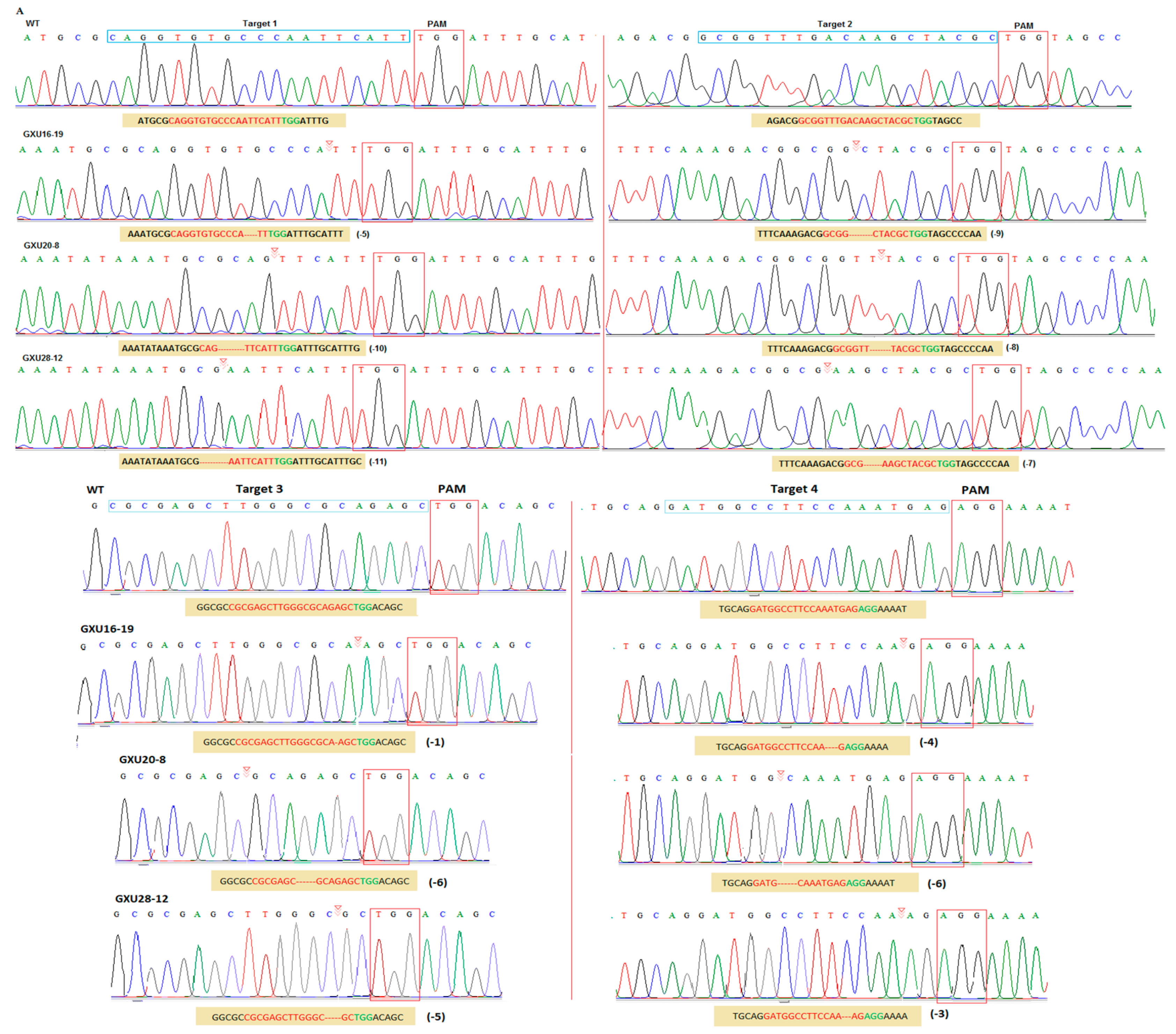
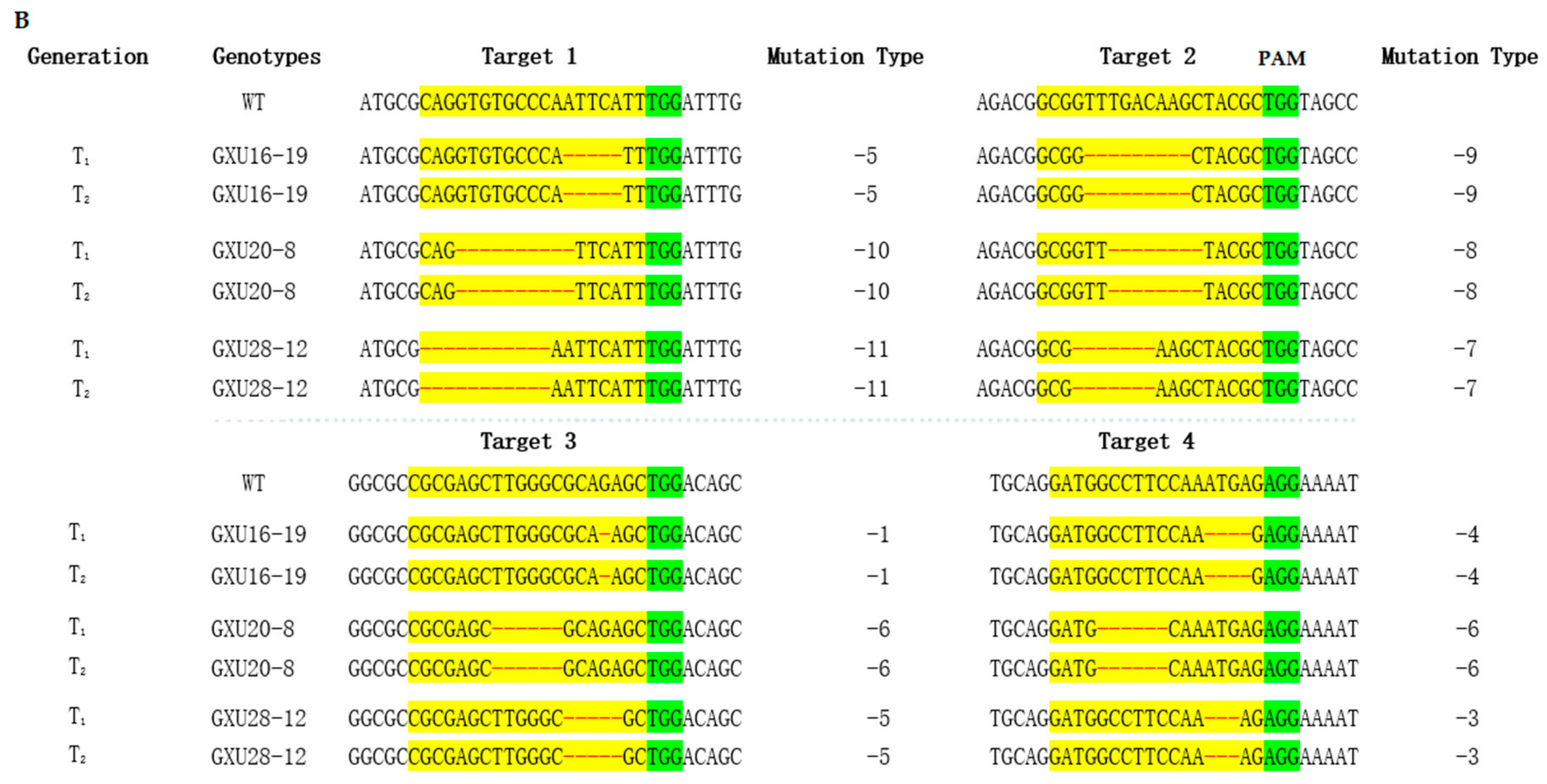
3.2. Frequency of Mutations and Off-target Analysis
3.3. Screening of T-DNA-Free Plants
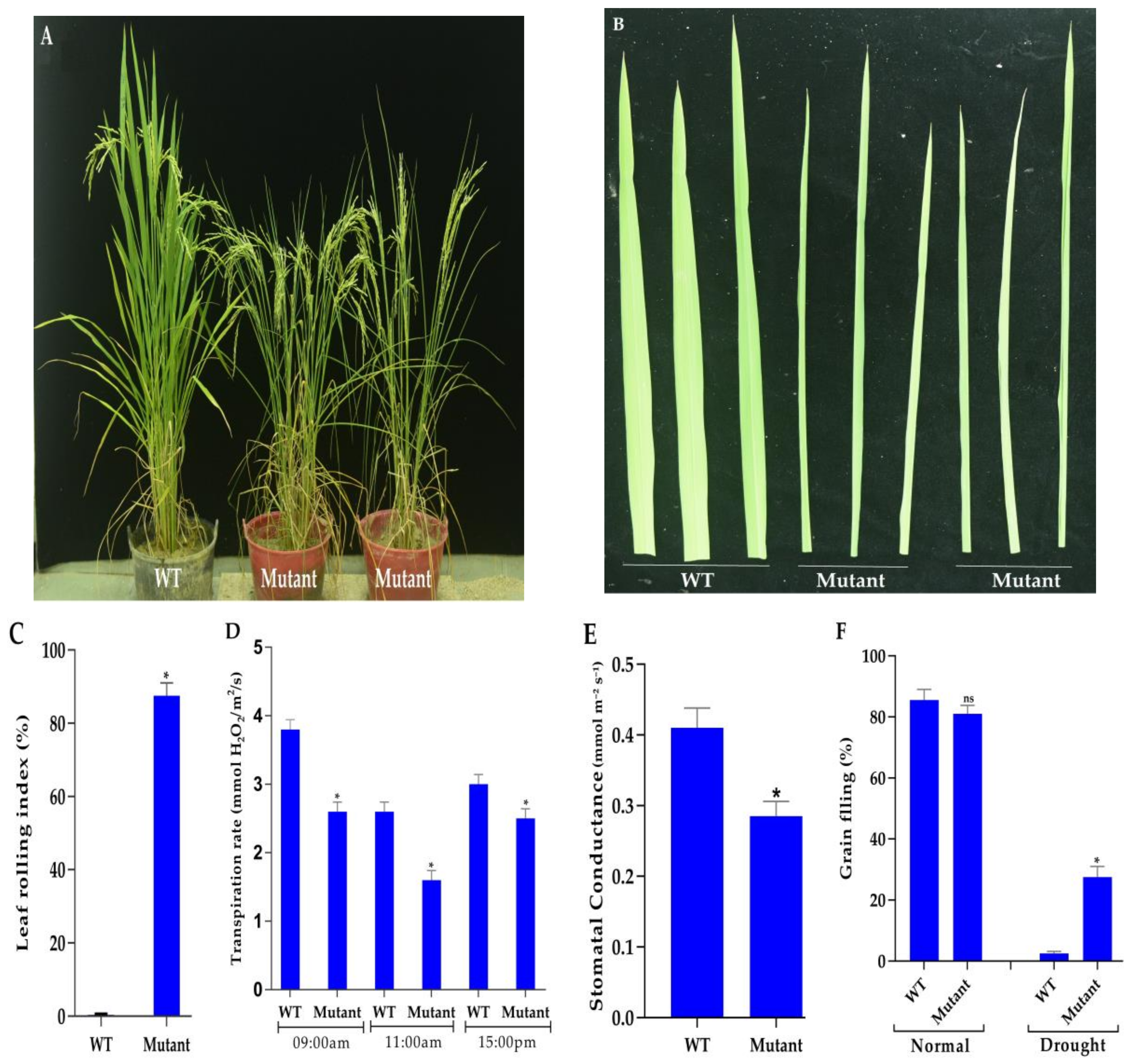
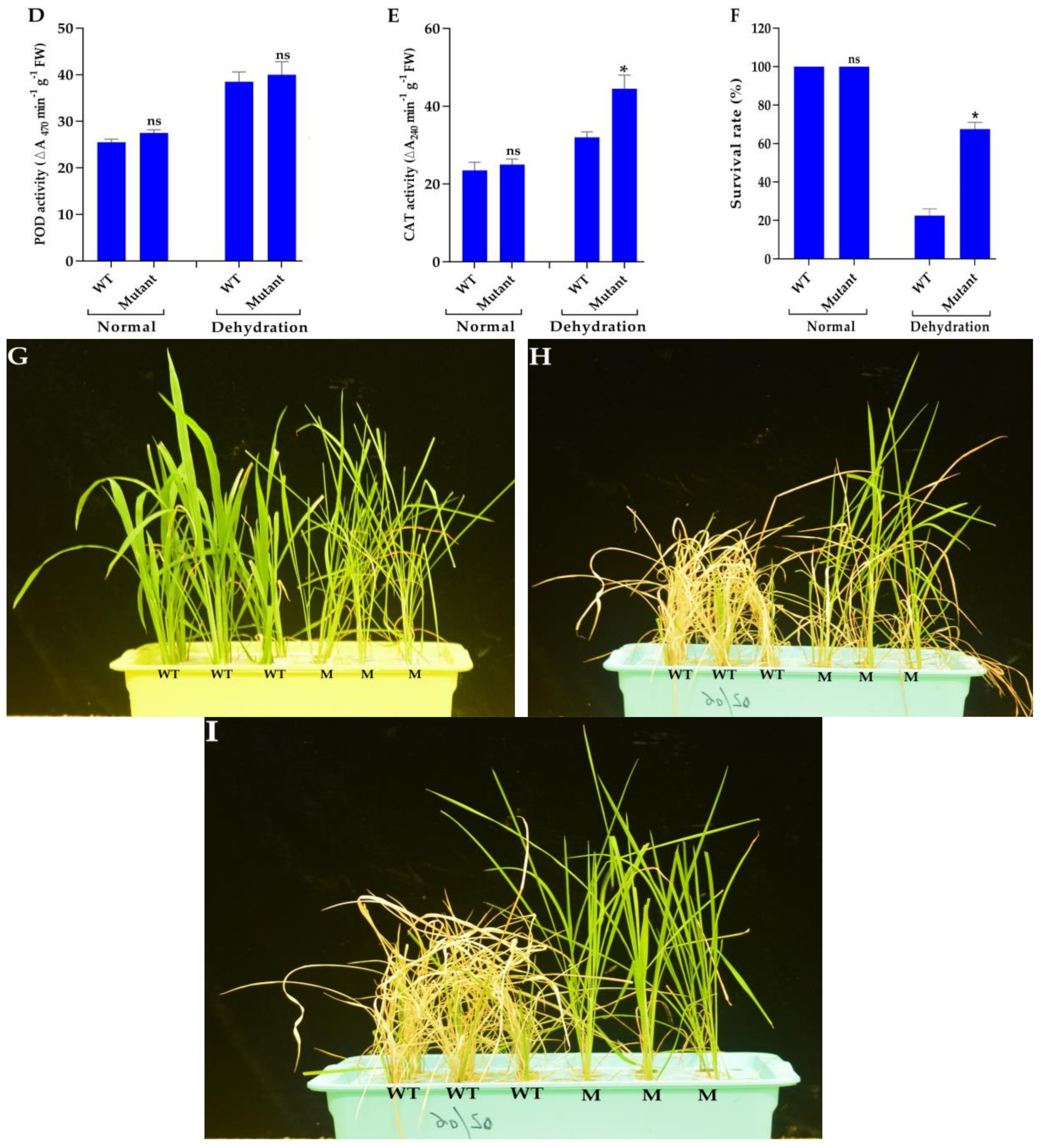
3.4. Phenotyping Under Normal and Drought Conditions
3.5. Measurement of Chlorophyll Contents
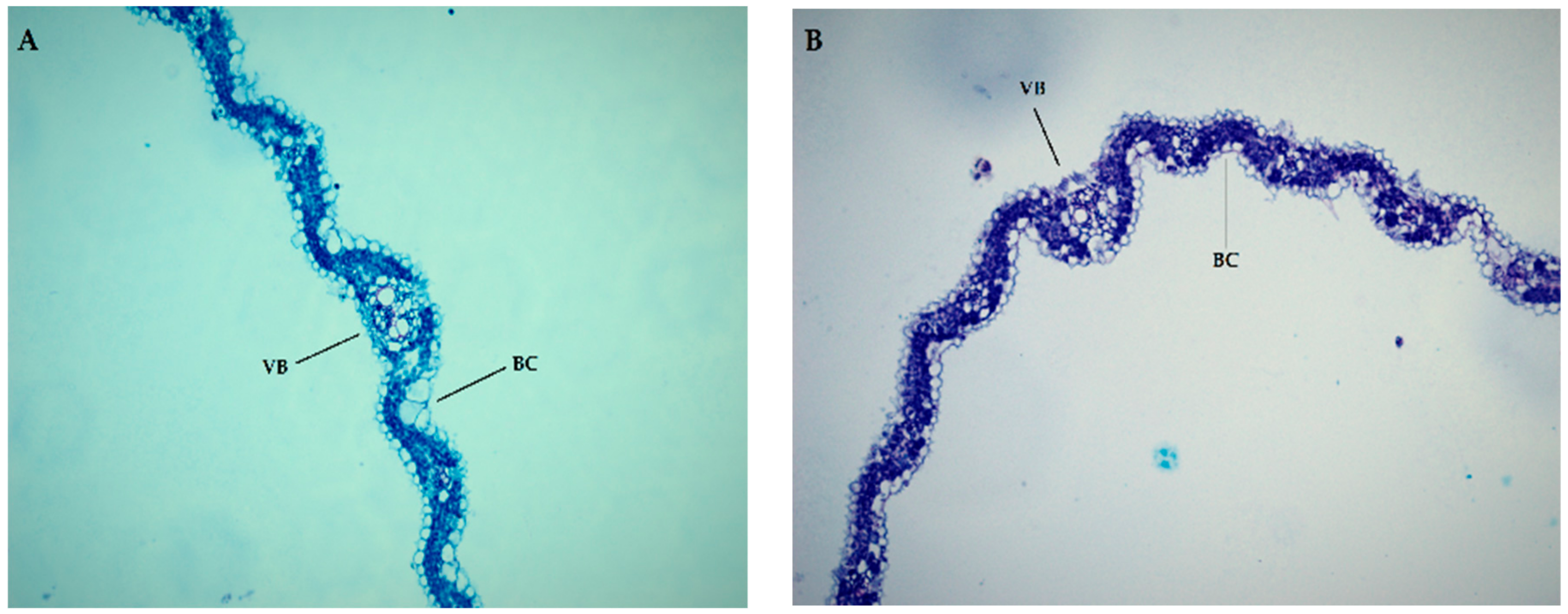
3.6. Microscopic Analysis
3.7. Mutant Plants Enhance ABA Content and Improve Antioxidant Enzyme Activities Under Drought Stress
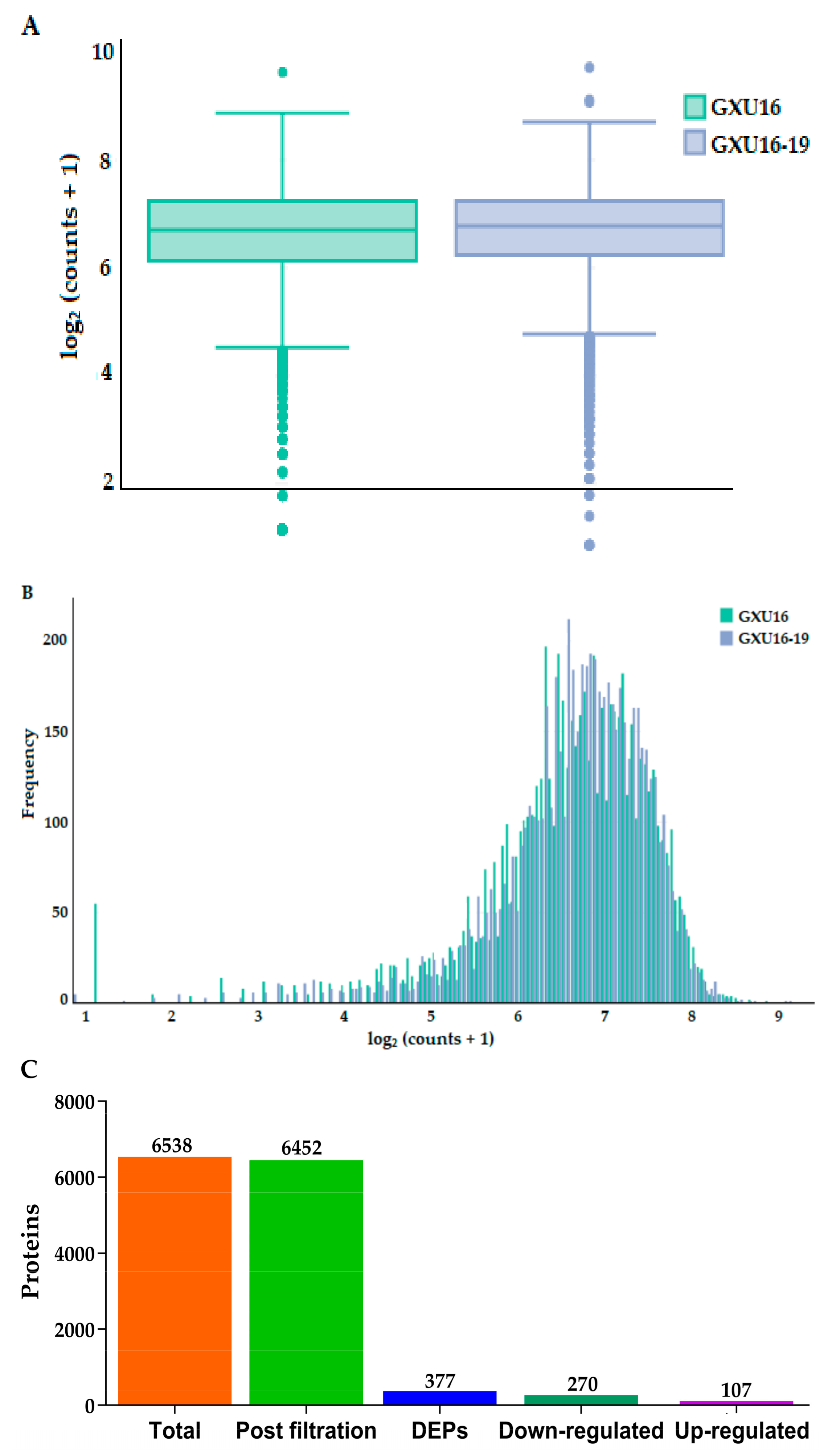
3.8. Protein Identification and Quantitation of Rolled Leaf Mutant and Wild Type
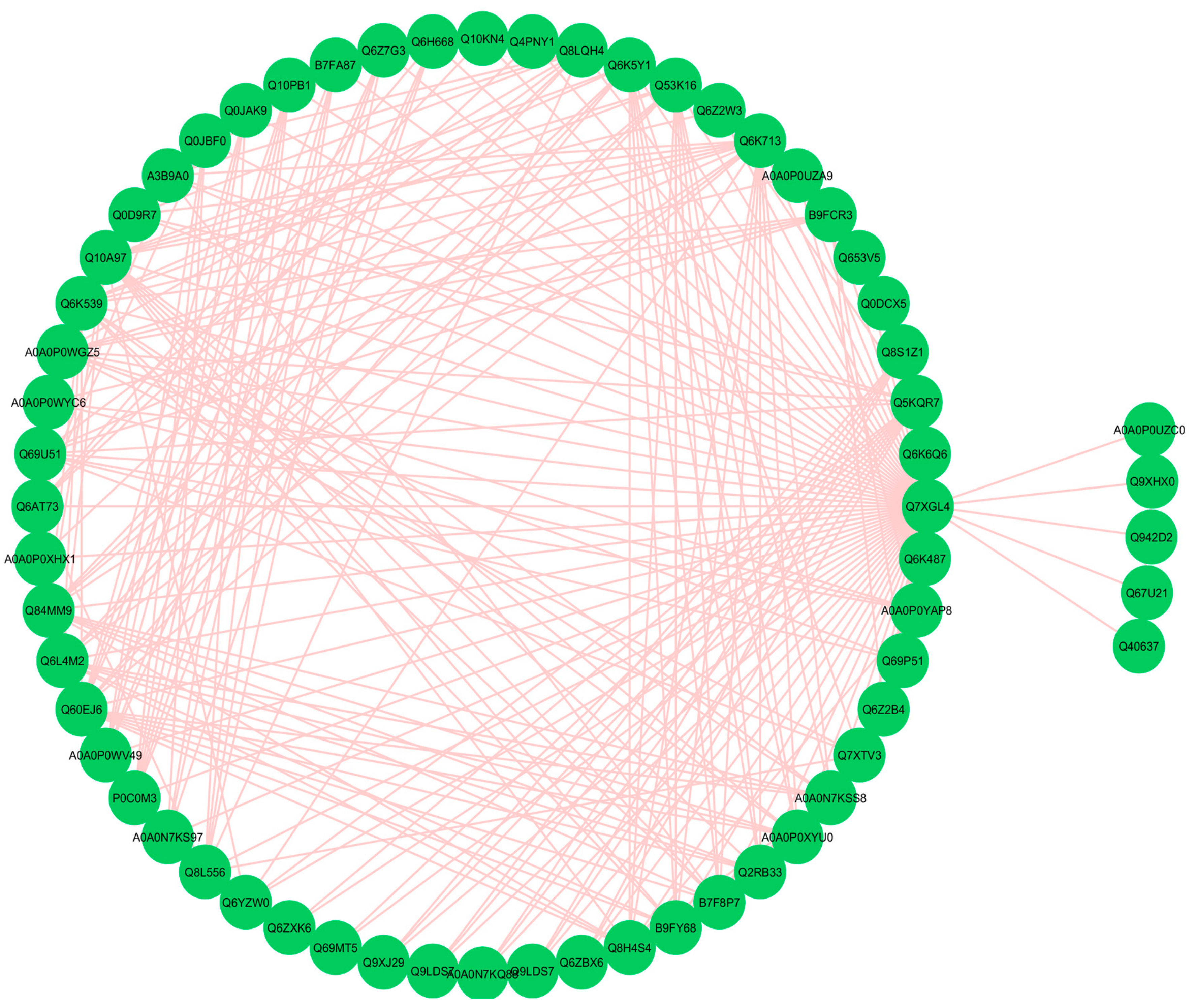
3.9. Functional Networks of the Differentially Expressed Proteins
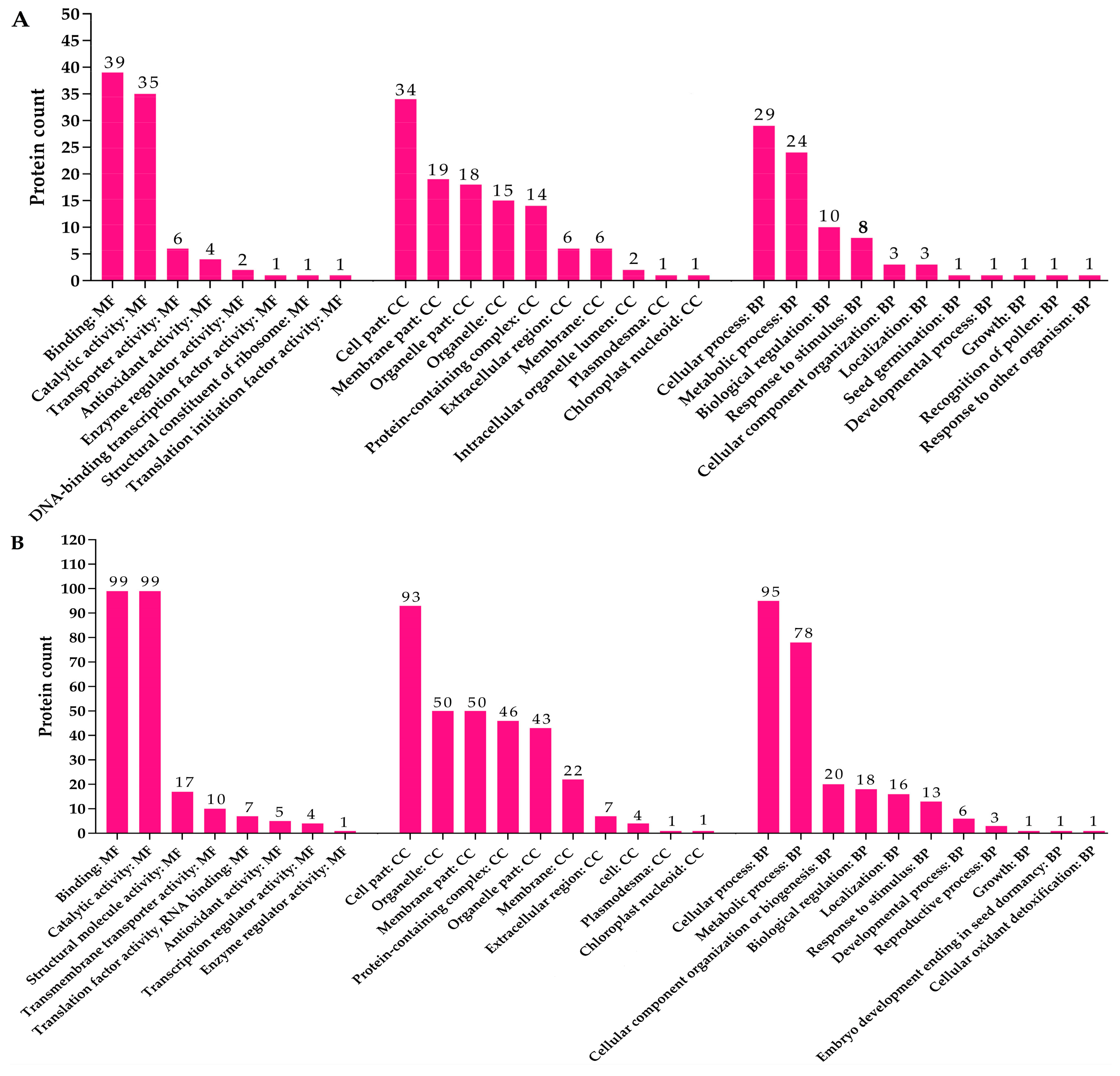
3.10. Gene Ontology (GO) Annotation of Up- and Downregulated Proteins
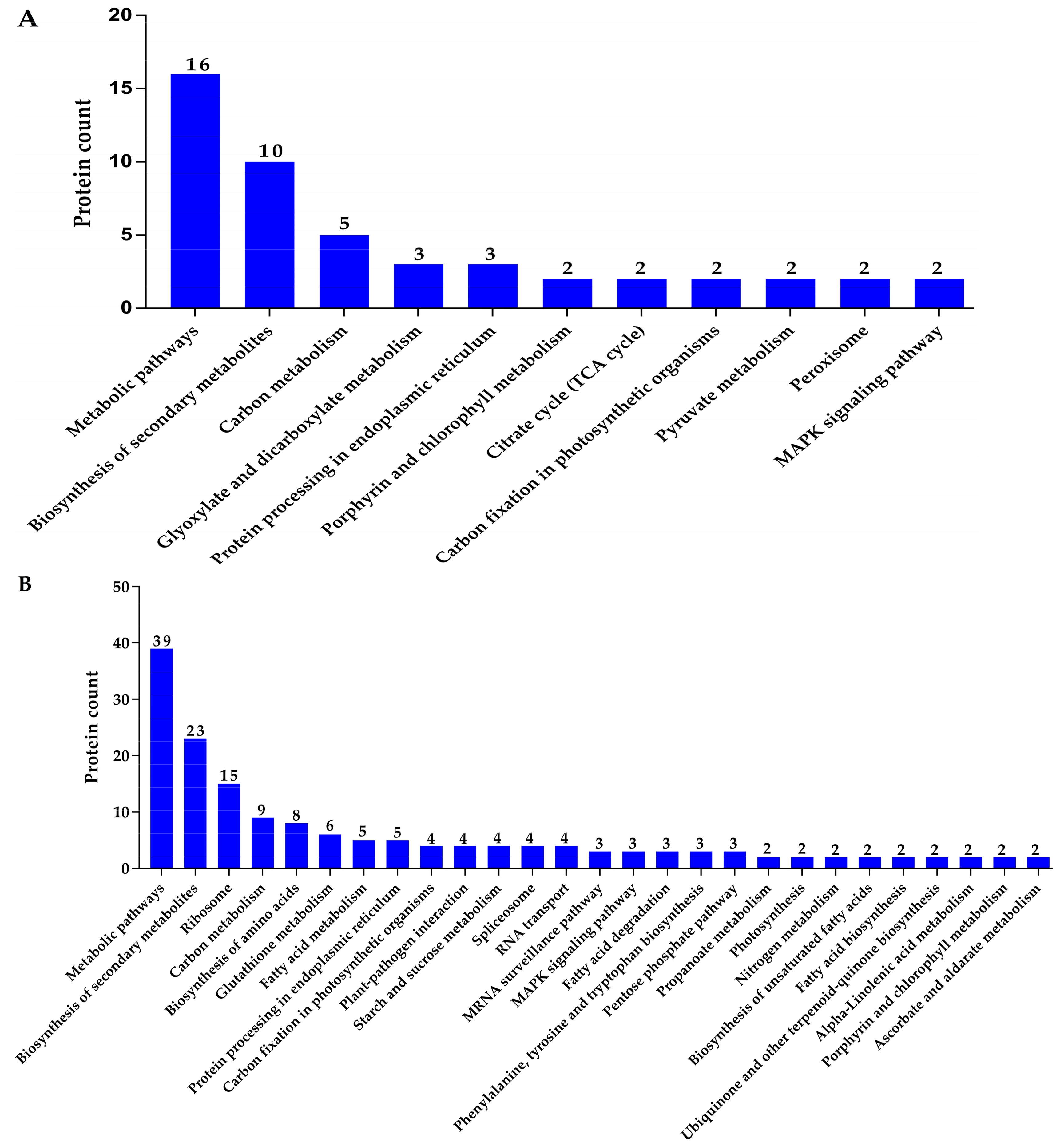
3.11. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis of Up- and Downregulated Proteins
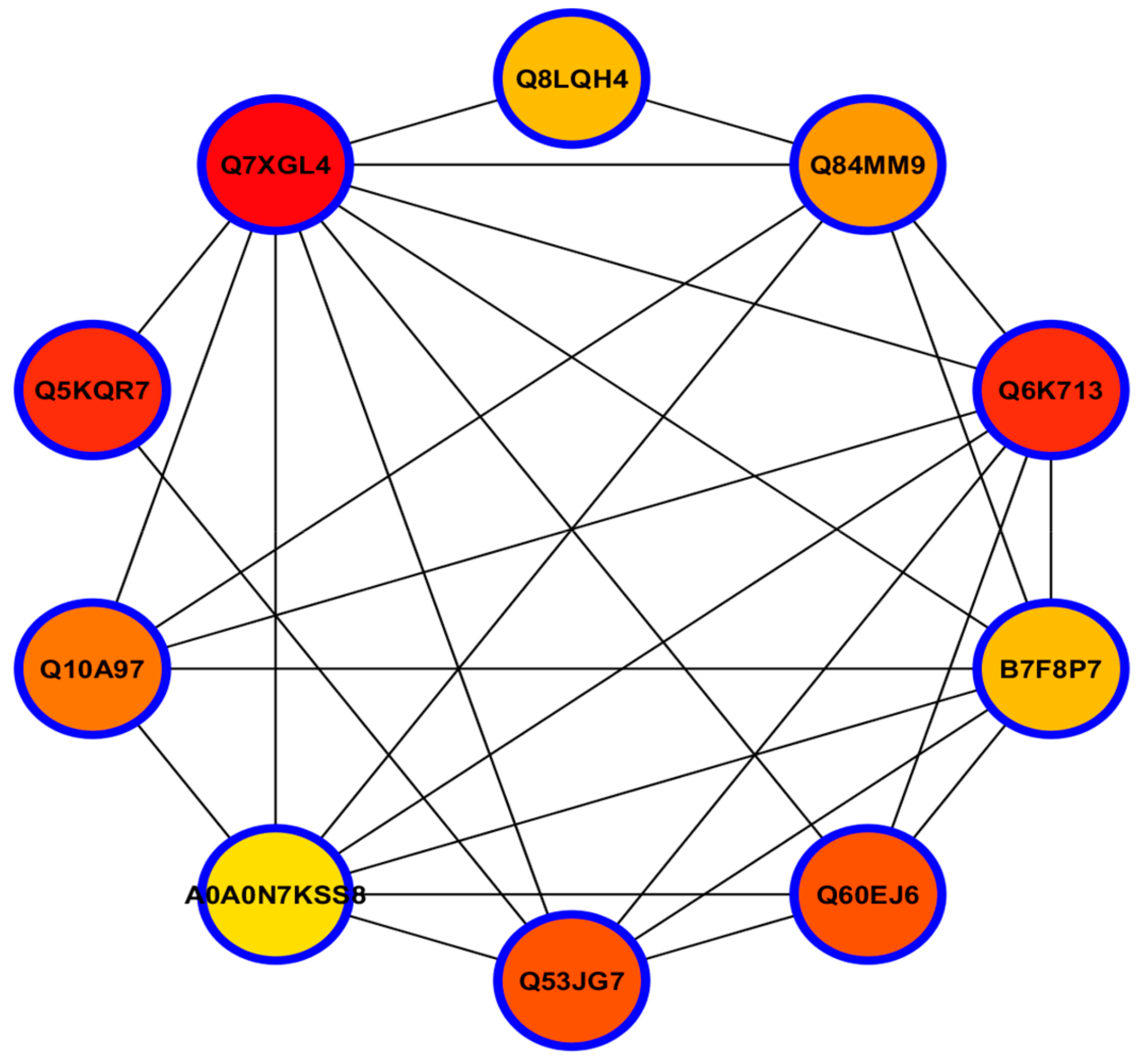
3.12. Hub-Protein Analysis
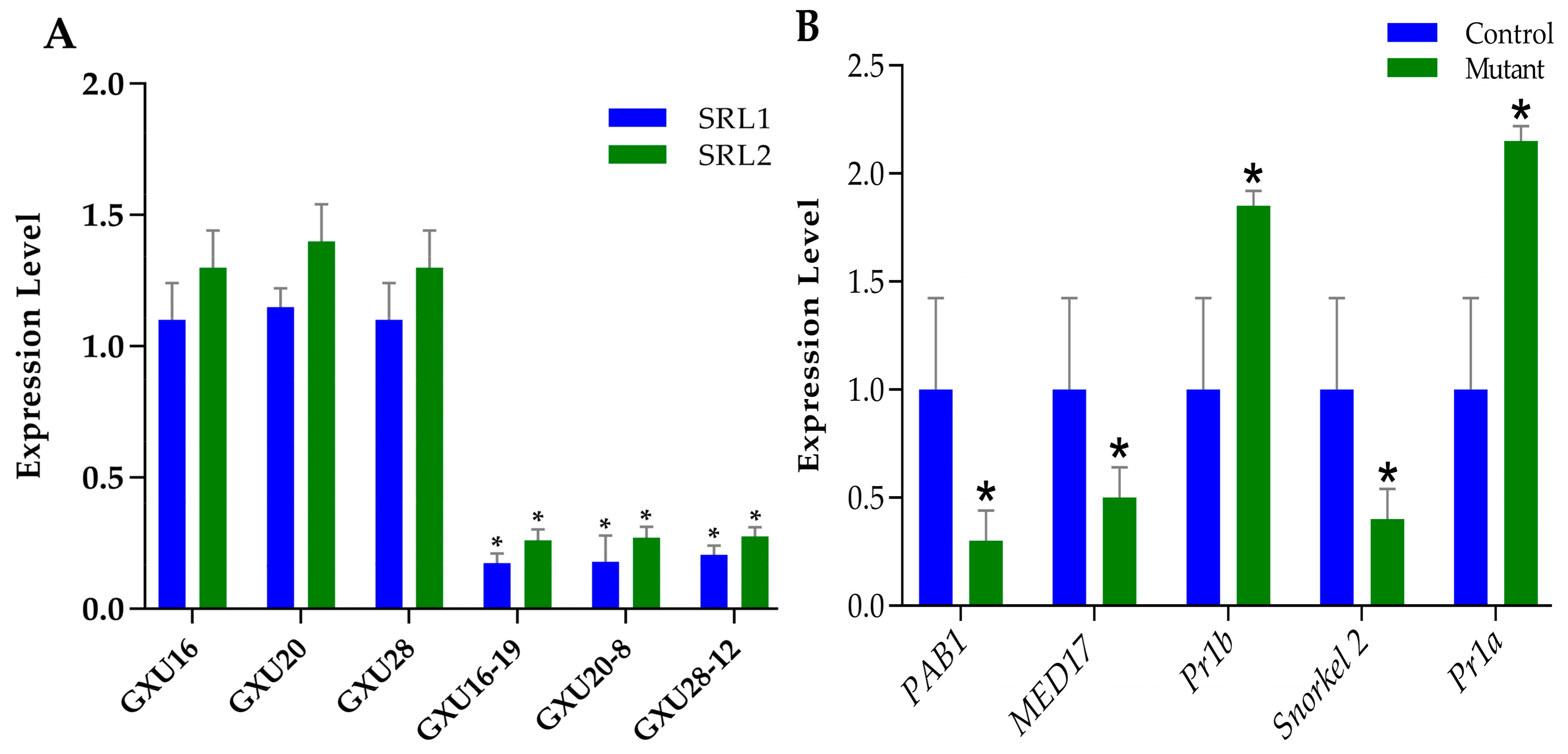
3.13. Expression Analysis of Target Genes Quantitative Real-Time-PCR Validation of DEPs
3.14. Performance of Semi-Rolled Leaf Hybrids Produced by Wild Type and Mutant Rolled Leaf Restorers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Accession Numbers
References
- Walter, A.; Silk, W.K.; Schurr, U. Environmental effects on spatial and temporal patterns of leaf and root growth. Annu. Rev. Plant Biol. 2009, 60, 279–304. [Google Scholar] [CrossRef]
- Zhu, D.; Lin, X.; Cao, W. Comparison of leaf photosynthetic characteristics among rice hybrids with different leaf rolling index. Zuo Wu Xue Bao 2001, 27, 329–333. [Google Scholar]
- Lang, Y.; Zhang, Z.; Gu, X.; Yang, J.; Zhu, Q. Physiological and ecological effects of crimpy leaf character in rice (Oryza sativa L.) I. Leaf orientation, canopy structure and light distribution. Zuo Wu Xue Bao 2004, 30, 806–810. [Google Scholar]
- Kadioglu, A.; Terzi, R. A dehydration avoidance mechanism: Leaf rolling. Bot. Rev. 2007, 73, 290–302. [Google Scholar] [CrossRef]
- Guo, T.; Wang, D.; Fang, J.; Zhao, J.; Yuan, S.; Xiao, L.; Li, X. Mutations in the Rice OsCHR4 Gene, Encoding a CHD3 Family Chromatin Remodeler, Induce Narrow and Rolled Leaves with Increased Cuticular Wax. Int. J. Mol. Sci. 2019, 20, 2567. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.J.; Zhang, G.H.; Qian, Q.; Xue, H.W. Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 2012, 159, 1488–1500. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Denning, G.; Mew, T. Hybrid Rice Breeding for Super High Yield; Denning, G.L., Mew, T.W., Eds.; Chinese Academy of Agricultural Sciences: Beijing, China, 1998; pp. 10–12. [Google Scholar]
- Chen, Z.; Zuo, S.; Zhang, Y.; Li, L.; Pan, X.; Ma, Y. Current progress in genetics research and breeding application of rolled leaf in rice. J. Yangzhou Univ. Agric. Life Sci. 2010, 31, 22–27. [Google Scholar]
- Xu, Y.; Wang, Y.; Long, Q.; Huang, J.; Wang, Y.; Zhou, K.; Zheng, M.; Sun, J.; Chen, H.; Chen, S. Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta 2014, 239, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shi, Z.Y.; Li, L.; Shen, G.Z.; Wang, X.Q.; An, L.S.; Zhang, J.L. Overexpression of ACL1 (abaxially curled leaf 1) increased bulliform cells and induced abaxial curling of leaf blades in rice. Mol. Plant 2010, 3, 807–817. [Google Scholar] [CrossRef]
- Chen, Q.; Xie, Q.; Gao, J.; Wang, W.; Sun, B.; Liu, B.; Zhu, H.; Peng, H.; Zhao, H.; Liu, C. Characterization of Rolled and Erect Leaf 1 in regulating leave morphology in rice. J. Exp. Bot. 2015, 66, 6047–6058. [Google Scholar] [CrossRef]
- Yang, S.Q.; Li, W.Q.; Miao, H.; Gan, P.F.; Qiao, L.; Chang, Y.L.; Shi, C.H.; Chen, K.M. REL2, a gene encoding an unknown function protein which contains DUF630 and DUF632 domains controls leaf rolling in rice. Rice 2016, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xiong, G.; Li, R.; Cui, J.; Tang, D.; Zhang, B.; Pauly, M.; Cheng, Z.; Zhou, Y. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 2009, 60, 1055–1069. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhu, L.; Zeng, D.; Gao, Z.; Guo, L.; Fang, Y.; Zhang, G.; Dong, G.; Yan, M.; Liu, J. Identification and characterization of NARROW ANDROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. 2010, 73, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Luan, W.; Liu, Y.; Zhang, F.; Song, Y.; Wang, Z.; Peng, Y.; Sun, Z. OsCD1 encodes a putative member of the cellulose synthase-like D sub-family and is essential for rice plant architecture and growth. Plant Biotech. J. 2011, 9, 513–524. [Google Scholar] [CrossRef]
- Fang, L.; Zhao, F.; Cong, Y.; Sang, X.; Du, Q.; Wang, D.; Li, Y.; Ling, Y.; Yang, Z.; He, G. Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotech. J. 2012, 10, 524–532. [Google Scholar] [CrossRef]
- Yang, C.; Li, D.; Liu, X.; Ji, C.; Hao, L.; Zhao, X.; Li, X.; Chen, C.; Cheng, Z.; Zhu, L. OsMYB103L, an R2R3-MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa L.). BMC Plant Biol. 2014, 14, 158. [Google Scholar] [CrossRef]
- Li, W.Q.; Zhang, M.J.; Gan, P.F.; Qiao, L.; Yang, S.Q.; Miao, H.; Wang, G.F.; Zhang, M.M.; Liu, W.T.; Li, H.F. CLD 1/SRL 1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J. 2017, 92, 904–923. [Google Scholar] [CrossRef]
- Liu, X.; Li, M.; Liu, K.; Tang, D.; Sun, M.; Li, Y.; Shen, Y.; Du, G.; Cheng, Z. Semi-Rolled Leaf2 modulates rice leaf rolling by regulating abaxial side cell differentiation. J. Exp. Bot. 2016, 67, 2139–2150. [Google Scholar] [CrossRef]
- Han, Y.; Luo, D.; Usman, B.; Nawaz, G.; Zhao, N.; Liu, F.; Li, R. Development of high yielding glutinous cytoplasmic male sterile rice (Oryza sativa L.) lines through CRISPR/Cas9 based mutagenesis of Wx and TGW6 and proteomic analysis of anther. Agronomy 2018, 8, 290. [Google Scholar] [CrossRef]
- Nawaz, G.; Han, Y.; Usman, B.; Liu, F.; Qin, B.; Li, R. Knockout of OsPRP1, a gene encoding proline-rich protein, confers enhanced cold sensitivity in rice (Oryza sativa L.) at the seedling stage. 3 Biotech 2019, 9, 254. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Bikard, D.; Jiang, W.; Samai, P.; Hochschild, A.; Zhang, F.; Marraffini, L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013, 41, 7429–7437. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.; Noirel, J.; Ow, S.Y.; Salim, M.; Pereira-Medrano, A.G.; Couto, N.; Pandhal, J.; Smith, D.; Pham, T.K.; Karunakaran, E. An insight into iTRAQ: Where do we stand now? Anal. Bioanal. Chem. 2012, 404, 1011–1027. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Islam, F.; Li, L.; Long, M.; Yang, C.; Jin, X.; Ali, B.; Mao, B.; Zhou, W. Complementary RNA-sequencing based transcriptomics and iTRAQ proteomics reveal the mechanism of the alleviation of quinclorac stress by salicylic acid in Oryza sativa ssp. japonica. Int. J. Mol. Sci. 2017, 18, 1975. [Google Scholar] [CrossRef]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef]
- Ma, X.; Chen, L.; Zhu, Q.; Chen, Y.; Liu, Y.G. Rapid decoding of sequence-specific nuclease-induced heterozygous and biallelic mutations by direct sequencing of PCR products. Mol. Plant 2015, 8, 1285–1287. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar] [CrossRef]
- Zhu, G.; Ye, N.; Zhang, J. Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol. 2009, 50, 644–651. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts: II. Role of electron transfer. Arch. Biochem. Biophys. 1968, 125, 850–857. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide dismutases: I. Occurrence in higher plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef]
- Aebi, H. Catalase in vitro. Methods Enzymol. 1984, 105, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Q.; Xu, X.Y.; Gong, Q.Q.; Xie, C.; Fan, W.; Yang, J.L.; Lin, Q.S.; Zheng, S.J. Root proteome of rice studied by iTRAQ provides integrated insight into aluminum stress tolerance mechanisms in plants. J. Proteom. 2014, 98, 189–205. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, J.C.; Cruz, R.T. Response of leaf water potential, stomatal resistance, and leaf rolling to water stress. Plant Physiol. 1980, 65, 428–432. [Google Scholar] [CrossRef] [PubMed]
- De Micco, V.; Aronne, G. Morpho-anatomical traits for plant adaptation to drought. Plant Res. Drought Stress 2012, 37–61. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, J. Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 2002, 53, 2401–2410. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.; Srivastava, A.K.; Pan, Y.; Bai, J.; Fang, J.; Shi, H.; Zhu, J.K. Knockdown of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol. 2018, 176, 2082–2094. [Google Scholar] [CrossRef]
- Han, Y.; Teng, K.; Nawaz, G.; Feng, X.; Usman, B.; Wang, X.; Luo, L.; Zhao, N.; Liu, Y.; Li, R. Generation of semi-dwarf rice (Oryza sativa L.) lines by CRISPR/Cas9-directed mutagenesis of OsGA20ox2 and proteomic analysis of unveiled changes caused by mutations. 3 Biotech 2019, 9, 387. [Google Scholar] [CrossRef]
- Ullah, N.; Yüce, M.; Gökçe, Z.N.Ö.; Budak, H. Comparative metabolite profiling of drought stress in roots and leaves of seven Triticeae species. BMC Gen. 2017, 18, 969. [Google Scholar] [CrossRef]
- Guo, R.; Shi, L.; Jiao, Y.; Li, M.; Zhong, X.; Gu, F.; Liu, Q.; Xia, X.; Li, H. Metabolic responses to drought stress in the tissues of drought-tolerant and drought-sensitive wheat genotype seedlings. AoB Plants 2018, 10, ply016. [Google Scholar] [CrossRef]
- Urano, K.; Maruyama, K.; Ogata, Y.; Morishita, Y.; Takeda, M.; Sakurai, N.; Suzuki, H.; Saito, K.; Shibata, D.; Kobayashi, M. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 2009, 57, 1065–1078. [Google Scholar] [CrossRef]
- Torras-Claveria, L.; Jáuregui, O.; Codina, C.; Tiburcio, A.F.; Bastida, J.; Viladomat, F. Analysis of phenolic compounds by high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry in senescent and water-stressed tobacco. Plant Sci. 2012, 182, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Mhamdi, A.; Foyer, C.H. The roles of reactive oxygen metabolism in drought: Not so cut and dried. Plant Phys. 2014, 164, 1636–1648. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.R.; Liu, B.; Feng, D.R.; Liu, H.Y.; He, Y.M.; Qi, K.B.; Wang, H.B.; Wang, J.F. MpAsr encodes an intrinsically unstructured protein and enhances osmotic tolerance in transgenic Arabidopsis. Plant Cell Rep. 2011, 30, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Vierling, E. The roles of heat shock proteins in plants. Annu. Rev. Plant Biol. 1991, 42, 579–620. [Google Scholar] [CrossRef]
- Torres, M.A.; Onouchi, H.; Hamada, S.; Machida, C.; Hammond-Kosack, K.E.; Jones, J.D. Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant. J. 1998, 14, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Shuai, B.; Reynaga-Pena, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef]
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jürgens, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F. Control of tillering in rice. Nature 2003, 422, 618. [Google Scholar] [CrossRef]
- Jisha, V.; Dampanaboina, L.; Vadassery, J.; Mithöfer, A.; Kappara, S.; Ramanan, R. Overexpression of an AP2/ERF type transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. PLoS ONE 2015, 10, e0127831. [Google Scholar] [CrossRef]
- Sharma, A.D.; Singh, P. Comparative studies on drought-induced changes in peptidyl prolyl cis–trans isomerase activity in drought-tolerant and susceptible cultivars of Sorghum bicolor. Curr. Sci. 2003, 84, 911–918. [Google Scholar]
- Fromont-Racine, M.; Senger, B.; Saveanu, C.; Fasiolo, F. Ribosome assembly in eukaryotes. Gene 2003, 313, 17–42. [Google Scholar] [CrossRef]
- Bai, J.; Yan, Q.; Jinghui, L.; Yuqing, W.; Rula, S.; Na, Z.; Ruizong, J. Proteomic response of oat leaves to long-term salinity stress. Environ. Sci. Pollut. Res. 2017, 24, 3387. [Google Scholar] [CrossRef] [PubMed]



| Genotypes | LT | PH | PN | PL (cm) | FLL (cm) | FLW (cm) | GNPP | SSR (%) | GW (g) | GL (mm) | GWD (mm) | GYPP (g) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GXU16 | Flat | 112.1 ± 3.4 | 7.5 ± 1.3 | 25.6 ± 1.4 | 51.0 ± 3.6 | 2.2 ± 0.1 * | 137 ± 12 | 90.2 ± 8.9 | 32.5 ± 1.3 | 10.0 ± 0.1 | 3.3 ± 0.2 | 30.0 ± 0.5 |
| GXU20 | Flat | 118.1 ± 2.6 | 7.8 ± 1.7 | 28.3 ± 1.5 | 48.5 ± 2.6 | 2.3 ± 0.3 | 139 ± 15 | 91.4 ± 5.3 | 34.5 ± 1.5 | 11.0 ± 0.3 | 3.4 ± 0.1 | 33.9 ± 0.1 |
| GXU28 | Flat | 125.2 ± 3.4 | 7.7 ± 2.0 | 27.4 ± 1.2 | 48.3 ± 1.9 | 2.4 ± 0.6 | 138 ± 10 | 90.5 ± 5.6 | 33.4 ± 1.4 | 11.0 ± 0.2 | 3.4 ± 0.3 | 32.8 ± 0.6 |
| GXU16-19 | Rolled | 80.1 ± 5.4 * | 10.8 ± 1.0 * | 17.4 ± 1.2 * | 37.5 ± 1.3 * | 1.6 ± 0.3 * | 115 ± 10 * | 72.3 ± 6.7 * | 28.5 ± 1.2 * | 9.1 ± 0.6 * | 3.0 ± 0.5 * | 25.4 ± 0.1 * |
| GXU20-8 | Rolled | 83.2 ± 4.3 * | 11.3 ± 1.0 * | 16.2 ± 1.2 * | 37.3 ± 1.8 * | 1.7 ± 0.5 * | 116 ± 11 * | 69.5 ± 5.3 * | 29.6 ± 1.5 * | 9.2 ± 0.8 * | 2.9 ± 0.4 * | 27.3 ± 0.1 * |
| GXU28-12 | Rolled | 87.3 ± 3.9 * | 10.4 ± 2.0 * | 17.5 ± 1.4 * | 36.9 ± 1.7 * | 1.7 ± 0.6 * | 109 ± 13 * | 67.6 ± 3.6 * | 28.7 ± 1.3 * | 9.2 ± 0.5 * | 2.9 ± 0.8 * | 21.9 ± 0.4 * |
| Genotypes | GXU16 | GXU20 | GXU28 | GXU16-19 | GXU20-8 | GXU28-12 |
|---|---|---|---|---|---|---|
| Chl a (mg·g−1) | 8.26 ± 0.11 | 8.13 ± 0.08 | 7.99 ± 0.07 | 4.25 ± 0.33 * | 4.11 ± 0.16 * | 4.14 ± 0.28 * |
| Chl b (mg·g−1) | 2.29 ± 0.03 | 2.75 ± 0.05 | 2.86 ± 0.12 | 1.52 ± 0.03 * | 1.49 ± 0.11 * | 1.39 ± 0.05 * |
| β-Car (mg·g−1) | 1.39 ± 0.06 | 1.47 ± 0.05 | 1.36 ± 0.04 | 0.98 ± 0.05 * | 1.02 ± 0.04 * | 1.06 ± 0.02 * |
| Chl a+Chl b (mg·g−1) | 10.54 ± 0.12 | 10.87± 0.09 | 10.85 ± 0.11 | 5.77 ± 0.07 * | 5.60 ± 0.08 * | 5.53 ± 0.03 * |
| Chl a/Chl b | 3.60 ± 0.12 | 2.95 ± 0.09 | 2.79 ± 0.06 | 2.79 ± 0.03 * | 2.75 ± 0.07 * | 2.97 ± 0.08 * |
| Protein ID | Gene Name/Locus | Annotation | Regulation |
|---|---|---|---|
| Proteins involved in abiotic stress | |||
| B8APG3 | OsI_11487 | Peroxidase | Up |
| B8ARU3 | OsI_18017 | Peroxidase | Up |
| A2YY59 | OsI_30285 | Superoxide dismutase | Up |
| A2ZBV1 | ASR5 | Abscisic stress-ripening protein 5 | Up |
| B8BFT1 | OsI_32804 | HSP domain-containing protein | Up |
| A2X294 | OsI_06318 | Hydrolase_4 domain-containing protein | Up |
| B8B3F7 | OsI_21871 | ParB domain-containing protein | Up |
| B8BBZ9 | OsI_28223 | NAD(P)-binding domain-containing protein | Up |
| Proteins involved in leaf rolling and development | |||
| Q8L3S3 | Os01g0572800 | Lateral organ boundaries (LOB) domain protein-like | Down |
| Q7XGL4 | CRLL2 | LOB domain protein 31 | Down |
| Q852M3 | LOC_Os03g57670 | LOB domain protein 4 | Down |
| Q8L4M5 | B1129G05.33 | LOB domain-containing protein | Down |
| A2X266 | OsI_06294 | Glycosyltransferase | Down |
| A2WLD5 | OsI_00644 | Glycosyltransferase | Down |
| B8BCU4 | OsI_31813 | Glycosyltransferase | Down |
| A2ZFX4 | OsI_36677 | Glycosyltransferase | Up |
| A2Y650 | OsI_20474 | Phosphoglycerate kinase | Down |
| A2Z498 | OsI_32475 | Peptidyl-prolyl cis-trans isomerase | Down |
| A2X9U8 | OsI_09019 | Peptidylprolyl isomerase | Down |
| A2XZE6 | OsI_18093 | Peptidyl-prolyl cis-trans isomerase | Down |
| A2YCR4 | OsI_22900 | Acyl-coenzyme A oxidase | Down |
| Genotypes | LT | PH | PN | PL (cm) | GNPP | SSR (%) | GW (g) | GL (mm) | GWD (mm) | GYPP (g) |
|---|---|---|---|---|---|---|---|---|---|---|
| GXU16 | Flat | 117.7 ± 4.2 | 7.6 ± 1.2 | 25.6 ± 1.4 | 134 ± 10 | 88.2 ± 8.7 | 32.5 ± 1.2 | 10.0 ± 0.1 | 3.1 ± 0.2 | 31.0 ± 0.3 |
| GXU20 | Flat | 119.3 ± 3.6 | 7.5 ± 1.6 | 28.3 ± 1.5 | 132 ± 12 | 89.4 ± 5.1 | 31.5 ± 1.6 | 11.0 ± 0.3 | 2.9 ± 0.1 | 33.9 ± 0.2 |
| GXU28 | Flat | 124.2 ± 3.7 | 7.7 ± 1.3 | 27.4 ± 1.2 | 133 ± 11 | 90.5 ± 5.2 | 31.4 ± 1.2 | 11.0 ± 0.2 | 3.0 ± 0.2 | 33.8 ± 0.5 |
| 36A/GXU16 | Flat | 114.6 ± 3.8 NS | 7.8 ± 1.6 NS | 26.1 ± 1.1 NS | 140 ± 18 NS | 83.3 ± 2.8 NS | 32.8 ± 1.0 NS | 10.0 ± 0.1 NS | 2.8 ± 0.1 NS | 29.8 ± 0.4 NS |
| 52A/GXU16 | Flat | 116.0 ± 3.6 NS | 8.0 ± 1.4 NS | 25.5 ± 1.3 NS | 145 ± 25 NS | 86.4 ± 2.6 NS | 33.2 ± 1.1 NS | 10.2 ± 0.2 NS | 2.8 ± 0.2 NS | 33.3 ± 0.4 NS |
| 36A/GXU20 | Flat | 115.2 ± 2.7 NS | 7.9 ± 2.1 NS | 26.3 ± 1.0 NS | 149 ± 21 NS | 83.8 ± 3.8 NS | 32.4 ± 1.2 NS | 10.0 ± 0.2 NS | 2.9 ± 0.2 NS | 31.9 ± 0.5 NS |
| 52A/GXU20 | Flat | 121.4 ± 5.0 NS | 8.8 ± 1.2 NS | 24.8 ± 1.6 NS | 134 ± 22 NS | 82.7 ± 4.4 NS | 32.5 ± 1.2 NS | 10.1 ± 0.3 NS | 2.8 ± 0.1 NS | 31.7 ± 0.4 NS |
| 36A/GXU28 | Flat | 115.2 ± 4.4 NS | 8.0 ± 0.9 NS | 25.5 ± 1.2 NS | 146 ± 21 NS | 85.4 ± 3.3 NS | 29.8 ± 1.3 NS | 10.0 ± 0.3 NS | 2.9 ± 0.2 NS | 29.7 ± 0.4 NS |
| 52A/GXU28 | Flat | 122.4 ± 5.9 NS | 8.2 ± 1.0 NS | 25.2 ± 1.6 NS | 138 ± 27 NS | 86.0 ± 3.2 NS | 29.4 ± 1.3 NS | 10.2 ± 0.3 NS | 2.9 ± 0.2 NS | 28.6 ± 0.6 NS |
| 36A/GXU16-19 | SR | 110.1 ± 5.5 NS | 8.8 ± 1.8 * | 28.3 ± 1.2 NS | 166 ± 12 * | 84.0 ± 3.8 NS | 31.6 ± 1.0 NS | 10.0 ± 0.1NS | 2.9 ± 0.1 NS | 38.7 ± 0.4 * |
| 52A/GXU16-19 | SR | 122.1 ± 3.6 NS | 10.2 ± 1.9 * | 27.6 ± 1.0 NS | 158 ± 12 * | 85.2 ± 4.2 NS | 32.4 ± 1.2 NS | 9.8 ± 0.2 NS | 2.8 ± 0.1 NS | 44.5 ± 0.4 * |
| 36A/GXU20-8 | SR | 112.5 ± 5.5 NS | 9.2 ± 1.8 * | 28.5 ± 1.4 NS | 168 ± 14 * | 85.4 ± 3.3 NS | 32.5 ± 1.0 NS | 10.1 ± 0.2 NS | 2.9 ± 0.1 NS | 42.9 ± 0.4 * |
| 52A/GXU20-8 | SR | 126.1 ± 5.0 NS | 10.5 ± 1.3 * | 27.0 ± 1.1 NS | 156 ± 18 * | 84.5 ± 4.9 NS | 32.6 ± 1.3 NS | 10.0 ± 0.1 NS | 2.8 ± 0.2 NS | 43.4 ± 0.4 * |
| 36A/GXU28-12 | SR | 117.8 ± 4.8 NS | 9.0 ± 1.1 * | 28.8 ± 1.0 NS | 170 ± 15 * | 86.7 ± 3.4 NS | 32.3 ± 1.1 NS | 9.9 ± 0.2 NS | 2.9 ± 0.2 NS | 40.6 ± 0.4 * |
| 52A/GXU28-12 | SR | 126.6 ± 3.4 NS | 10.0 ± 1.2 * | 27.2 ± 0.9 NS | 153 ± 14 * | 86.9 ± 6.0 NS | 31.5 ± 1.2 NS | 10.1 ± 0.2 NS | 2.8 ± 0.1 NS | 40.6 ± 0.4 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, S.; Qin, X.; Luo, L.; Han, Y.; Wang, X.; Usman, B.; Nawaz, G.; Zhao, N.; Liu, Y.; Li, R. CRISPR/Cas9-Induced Mutagenesis of Semi-Rolled Leaf1,2 Confers Curled Leaf Phenotype and Drought Tolerance by Influencing Protein Expression Patterns and ROS Scavenging in Rice (Oryza sativa L.). Agronomy 2019, 9, 728. https://doi.org/10.3390/agronomy9110728
Liao S, Qin X, Luo L, Han Y, Wang X, Usman B, Nawaz G, Zhao N, Liu Y, Li R. CRISPR/Cas9-Induced Mutagenesis of Semi-Rolled Leaf1,2 Confers Curled Leaf Phenotype and Drought Tolerance by Influencing Protein Expression Patterns and ROS Scavenging in Rice (Oryza sativa L.). Agronomy. 2019; 9(11):728. https://doi.org/10.3390/agronomy9110728
Chicago/Turabian StyleLiao, Shanyue, Xuemei Qin, Liang Luo, Yue Han, Xin Wang, Babar Usman, Gul Nawaz, Neng Zhao, Yaoguang Liu, and Rongbai Li. 2019. "CRISPR/Cas9-Induced Mutagenesis of Semi-Rolled Leaf1,2 Confers Curled Leaf Phenotype and Drought Tolerance by Influencing Protein Expression Patterns and ROS Scavenging in Rice (Oryza sativa L.)" Agronomy 9, no. 11: 728. https://doi.org/10.3390/agronomy9110728
APA StyleLiao, S., Qin, X., Luo, L., Han, Y., Wang, X., Usman, B., Nawaz, G., Zhao, N., Liu, Y., & Li, R. (2019). CRISPR/Cas9-Induced Mutagenesis of Semi-Rolled Leaf1,2 Confers Curled Leaf Phenotype and Drought Tolerance by Influencing Protein Expression Patterns and ROS Scavenging in Rice (Oryza sativa L.). Agronomy, 9(11), 728. https://doi.org/10.3390/agronomy9110728







