DNA Environment of Centromeres and Non-Homologous Chromosomes Interactions in Mouse
Abstract
1. Introduction
1.1. Pericentromeric Satellite DNA
1.2. Chromocenters and Ectopic Contacts of Pericentromeric Satellite DNA in Meiotic Prophase I
1.3. Pericentromeric Satellite DNA and Ectopic Recombination
2. Materials and Methods
2.1. Preparation of Synaptonemal Complexes (SCs)
2.2. Immunostaining Procedure
2.3. FISH Procedure
2.4. Microscopy
2.5. Analysis of SC Preparation
3. Results
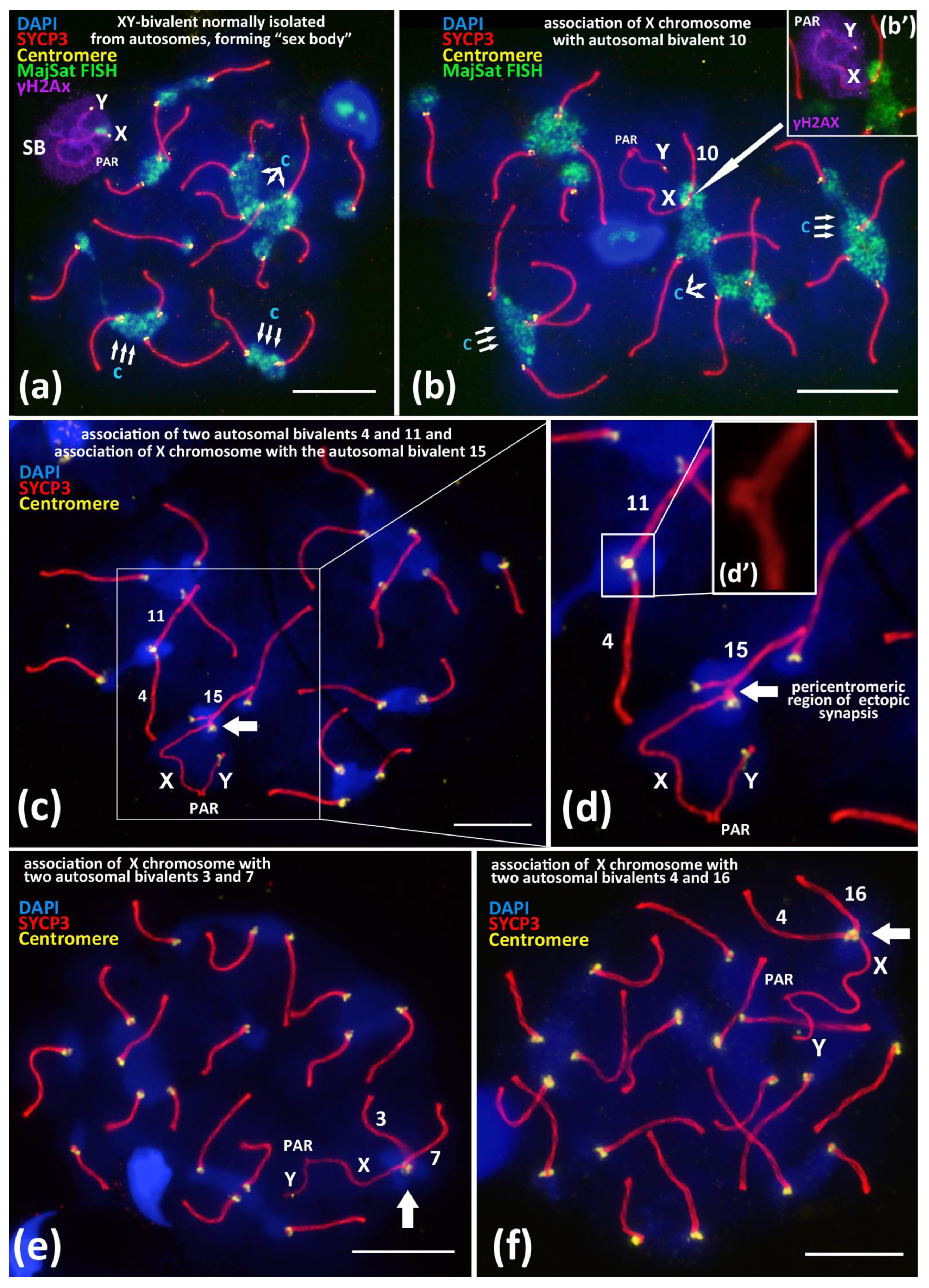
3.1. Associations of X Chromosome with Different Autosomal Bivalents
3.2. Meiotic Silencing of Sex Chromosomes and X-Autosomal Associations
3.3. Peculiarities of Morphology of Chromocenters in Meiosis
4. Discussion
4.1. Chromocenters and Associations between Non-Homologous Chromosomes in Meiosis
4.2. Sex Chromosomes Behaviour and Chromatin Inactivation during Meiotic Prophase I
4.3. Detailing the Mechanism of Associations between the X Chromosome and Different Autosomal Bivalents in Meiotic Prophase I
- Ectopic SC assembly. Non-homologous local pericentromeric side-by-side synapsis of the X chromosome and an autosomal univalent. This stage occurs during early meiosis and is characterized by the incomplete assembly of the autosomal bivalent, i.e., asynaptic fork at the pericentromeric region (Figure 2a);
- Correction of non-homologous synaptic X-autosomal association. This stage includes the complete assembly of the autosomal bivalent and the competitive replacement of the X chromosome from the short non-homologous SC-region (Figure 3b,c). Consequently, the X chromosome remains associated but end-to-end with only one autosomal chromosome (Figure 3b,c);
- Residual end-to-end association of the X chromosome with the centromeric region of the assembled autosomal bivalent. Such associations can persist until the diplotene stage (Figure 3d) and could also be disrupted before the end of prophase I (Supplementary Materials Figure S5a,b).
4.4. Formation of X-Autosomal Chromocenters and MSCI
4.5. Peculiarities of Chromocenters in Meiosis
- We have revealed that the pericentromeric chromatin of the X chromosome may be involved in ectopic interactions with the chromocenters of different autosomes (we detected 18 of 19 bivalents in such associations in our study) (Figure 1b–f and Figure 2; Figures S1–S5). This could be the basis for a mechanism for maintaining autosome-like DNA-repeat composition near the centromere of the X chromosome.
- We have shown that the pericentromeric chromatin of the X chromosome is spatially separated from the inactivated chromatin of the “sex body” (Figure 1b or Figure 3d’,e’; Figure S8). Thus, XY-chromatin inactivation (MSCI) does not interfere with the ectopic X-autosomal interactions of pericentromeric DNAs. This result indicates the high importance of chromocenters formation and functioning during meiosis.
- We have shown that the two-fold stretching of the meiotic nuclei chromatin on the glass surface does not destroy elongated chromocenters (Figure 4a–f; Figures S9 and S10). These “interbivalent chromatin fibers” contain highly repeated MajSat and MiSat DNA of pericentromeric regions of non-homologous chromosomes (Figure 4; Figures S7, S9 and S10). This finding is consistent with the data that centromere but not telomere clustering is the general and primordial meiotic mechanism in eukaryotes [26].
5. Conclusions
- We suggest the hypothetical model of associations between satellite DNA-enriched regions of autosomes and the centromeric region of the X chromosome. We have detailed the successive stages of correction of these non-homologous X-autosomal associations: early meiotic ectopic synaptonemal complex assembly followed by the competitive replacement of the X chromosome by autosome in later stages (Figure 2a–d).
- In the case of the X-autosomal association, the pericentromeric region of the X chromosome is integrated in the autosomal chromocenters enriched in MajSat DNA. The centromeric region of the X chromosome is free of γH2Ax-dependent chromatin inactivation (MSCI). Thus, the “sex body” and the proximal X-autosomal MajSat-rich chromocenter are spatially and functionally separated.
- Our results demonstrate the stability and remarkable sturdiness of the interbivalent chromatin fibers connecting the centromeric regions and that are enriched in satellite DNA in meiotic prophase I nuclei.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hartley, G.; O’Neill, R. Centromere Repeats: Hidden Gems of the Genome. Genes 2019, 10, 223. [Google Scholar] [CrossRef]
- Komissarov, A.S.; Gavrilova, E.V.; Demin, S.J.; Ishov, A.M.; Podgornaya, O.I. Tandemly repeated DNA families in the mouse genome. BMC Genom. 2011, 12, 531. [Google Scholar] [CrossRef]
- Wijchers, P.J.; Geeven, G.; Eyres, M.; Bergsma, A.J.; Janssen, M.; Verstegen, M.; De Laat, W. Characterization and dynamics of pericentromere-associated domains in mice. Genome Res. 2015, 25, 958–969. [Google Scholar] [CrossRef]
- Pardue, M.L.; Gall, J.G. Chromosomal localization of mouse satellite DNA. Science 1970, 168, 1356–1358. [Google Scholar] [CrossRef] [PubMed]
- Podgornaya, O.I.; Ostromyshenskii, D.I.; Enukashvily, N.I. Who needs this junk, or genomic dark matter. Biochemistry 2008, 83, 450–466. [Google Scholar] [CrossRef] [PubMed]
- Kipling, D.; Wilson, H.E.; Mitchell, A.R.; Taylor, B.A.; Cooke, H.J. Mouse centromere mapping using oligonucleotide probes that detect variants of the minor satellite. Chromosoma 1994, 103, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Pajpach, F.; Wu, T.; Shearwin-Whyatt, L.; Jones, K.; Grützner, F. Flavors of Non-Random Meiotic Segregation of Autosomes and Sex Chromosomes. Genes 2021, 12, 1338. [Google Scholar] [CrossRef]
- Wong, A.K.C.; Biddle, F.G.; Rattner, J.B. The chromosomal distribution of the major and minor satellite is not conserved in the genus Mus. Chromosoma 1990, 99, 190–195. [Google Scholar] [CrossRef]
- Garagna, S.; Redi, C.A.; Capanna, E.; Andayani, N.; Alfano, R.M.; Doi, P.; Viale, G. Genome distribution, chromosomal allocation and organization of the major and minor satellite DNA in 11 species and subspecies of the genus Mus. Cytogenet. Genome Res. 1993, 64, 247–255. [Google Scholar] [CrossRef]
- Guenatri, M.; Bailly, D.; Maison, C.; Almouzni, G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 2004, 166, 493–505. [Google Scholar] [CrossRef]
- Kalitisis, P.; Griffiths, B.; Choo, K.A. Mouse telocentric sequences reveal a high rate of homogenization and possible role in Robertsonian translocation. Proc. Natl. Acad. Sci. USA 2006, 103, 8786–8791. [Google Scholar] [CrossRef] [PubMed]
- Scherthan, H.; Weich, S.; Schwegler, H.; Heyting, C.; Härle, M.; Cremer, T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J. Cell Biol. 1996, 134, 1109–1125. [Google Scholar] [CrossRef]
- Zickler, D.; Kleckner, N. Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 1999, 33, 603–754. [Google Scholar] [CrossRef] [PubMed]
- Zickler, D.; Kleckner, N. A few of our favorite things: Pairing, the bouquet, crossover interference and evolution of meiosis. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2016; Volume 54, pp. 135–148. [Google Scholar] [CrossRef]
- Berrios, S. Nuclear architecture of mouse spermatocytes: Chromosome topology, heterochromatin, and nucleolus. Cytogenet. Genome Res. 2017, 151, 61–71. [Google Scholar] [CrossRef]
- Ur, S.N.; Corbett, K.D. Architecture and Dynamics of Meiotic Chromosomes. Annu. Rev. Genet. 2021, 55, 497–526. [Google Scholar] [CrossRef]
- Kazemi, P.; Taketo, T. Two telomeric ends of acrocentric chromosome play distinct roles in homologous chromosome synapsis in the fetal mouse oocyte. Chromosoma 2021, 130, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Koszul, R.; Kleckner, N. Dynamic chromosome movements during meiosis: A way to eliminate unwanted connections? Trends Cell Biol. 2009, 19, 716–724. [Google Scholar] [CrossRef]
- Stewart, M.N.; Dawson, D.S. Changing partners: Moving from non-homologous to homologous centromere pairing in meiosis. Trends Genet. 2008, 24, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Eyster, C.; Chuong, H.H.; Lee, C.Y.; Pezza, R.J.; Dawson, D. The pericentromeric heterochromatin of homologous chromosomes remains associated after centromere pairing dissolves in mouse spermatocyte meiosis. Chromosoma 2019, 128, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Klutstein, M.; Cooper, J.P. The Chromosomal Courtship Dance—homolog pairing in early meiosis. Curr. Opin. Cell Biol. 2014, 26, 123–131. [Google Scholar] [CrossRef]
- Scherthan, H. Telomere attachment and clustering during meiosis. Cell Mol. Life Sci. 2007, 64, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Scherthan, H. A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2001, 2, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Harper, L.; Golubovskaya, I.; Cande, W.Z. A bouquet of chromosomes. J. Cell Sci. 2004, 117, 4025–4032. [Google Scholar] [CrossRef] [PubMed]
- Zickler, D.; Kleckner, N. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 1998, 32, 619–697. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Agreiter, C.; Loidl, J. Spatial constraints on chromosomes are instrumental to meiotic pairing. J. Cell Sci. 2020, 133, 253724. [Google Scholar] [CrossRef]
- Berrios, S.; Manieu, C.; López-Fenner, J.; Ayarza, E.; Page, J.; González, M.; Fernández-Donoso, R. Robertsonian chromosomes and the nuclear architecture of mouse meiotic prophase spermatocytes. Biol. Res. 2014, 47, 1–13. [Google Scholar] [CrossRef]
- López-Fenner, J.; Berríos, S.; Manieu, C.; Page, J.; Fernández-Donoso, R. Bivalent Associations in Mus domesticus 2n = 40 Spermatocytes. Are They Random? Bull. Math. Biol. 2014, 76, 1941–1952. [Google Scholar] [CrossRef]
- Berríos, S.; Manterola, M.; Prieto, Z.; López-Fenner, J.; Page, J.; Fernández-Donoso, R. Model of chromosome associations in Mus domesticus spermatocytes. Biol. Res. 2010, 43, 275–285. [Google Scholar] [CrossRef]
- Peng, J.C.; Karpen, G.H. Epigenetic regulation of heterochromatic DNA stability. Curr. Opin. Genet. Dev. 2008, 18, 204–211. [Google Scholar] [CrossRef]
- Montgomery, E.A.; Huang, S.M.; Langley, C.H.; Judd, B.H. Chromosome rearrangement by ectopic recombination in Drosophila melanogaster: Genome structure and evolution. Genetics 1991, 129, 1085–1098. [Google Scholar] [CrossRef]
- Arnheim, N.; Krystal, M.; Schmickel, R.; Wilson, G.; Ryder, O.; Zimmer, E. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc. Natl. Acad. Sci. USA 1980, 77, 7323–7327. [Google Scholar] [CrossRef]
- Eickbush, T.H.; Eickbush, D.G. Finely orchestrated movements: Evolution of the ribosomal RNA genes. Genetics 2007, 175, 477–485. [Google Scholar] [CrossRef]
- Amaral, N.; Ryu, T.; Li, X.; Chiolo, I. Nuclear dynamics of heterochromatin repair. Trends Genet. 2017, 33, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Chiolo, I.; Minoda, A.; Colmenares, S.U.; Polyzos, A.; Costes, S.V.; Karpen, G.H. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 2011, 144, 732–744. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, A.A.; Jeggo, P.A. The heterochromatic barrier to DNA double strand break repair: How to get the entry visa. Int. J. Mol. Sci. 2012, 13, 11844–11860. [Google Scholar] [CrossRef]
- Waye, J.S.; Willard, H.F. Molecular analysis of a deletion polymorphism in alpha satellite of human chromosome 17: Evidence for homologous unequal crossing-over and subsequent fixation. Nucleic Acids Res. 1986, 14, 6915–6927. [Google Scholar] [CrossRef]
- Jakob, B.; Splinter, J.; Conrad, S.; Voss, K.O.; Zink, D.; Durante, M.; Taucher-Scholz, G. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011, 39, 6489–6499. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, C.; Soutoglou, E. Double strand break (DSB) repair in heterochromatin and heterochromatin proteins in DSB repair. DNA Repair 2014, 19, 163–168. [Google Scholar] [CrossRef]
- Tsouroula, K.; Furst, A.; Rogier, M.; Heyer, V.; Maglott-Roth, A.; Ferrand, A.; Reina-San-Martin, B.; Soutoglou, E. Temporal and spatial uncoupling of DNA double strand break repair pathways within mammalian heterochromatin. Mol. Cell 2016, 63, 293–305. [Google Scholar] [CrossRef]
- Navarro, J.; Vidal, F.; Guitart, M.; Egozcue, J. A method for the sequential study of synaptonemal complexes by light and electron microscopy. Hum. Genet. 1981, 59, 419–421. [Google Scholar] [CrossRef]
- Stepakov, A.; Galkina, S.; Bogomaz, D.; Gaginskaya, E.; Saifitdinova, A. Modified synthesis of 6-carboxyfluorescein (6-FAM): Application to probe labeling for conventional cytogenetics. Curr. J. Appl. Sci. Technol. 2015, 7, 423–428. [Google Scholar] [CrossRef]
- Bogdanov, Y.F.; Kolomiets, O.L. Synaptonemal Complex as an Indicator of the Dynamics of Meiosis and Chromosome Variation; KMK Press: Moscow, Russia, 2007; p. 359. [Google Scholar]
- Spangenberg, V.; Arakelyan, M.; Galoyan, E.; Pankin, M.; Petrosyan, R.; Stepanyan, I.; Grishaeva, T.; Danielyan, F.; Kolomiets, O. Extraordinary centromeres: Differences in the meiotic chromosomes of two rock lizards species Darevskia portschinskii and Darevskia raddei. PeerJ 2019, 7, e6360. [Google Scholar] [CrossRef]
- Turner, J.M. Meiotic sex chromosome inactivation. Development 2007, 134, 1823–1831. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.M. Meiotic silencing in mammals. Annu. Rev. Genet. 2015, 49, 395–412. [Google Scholar] [CrossRef]
- Burgoyne, P.S. Genetic homology and crossing over in the X and Y chromosomes of mammals. Hum. Genet. 1982, 61, 85–90. [Google Scholar] [CrossRef]
- Qiao, H.; Chen, J.K.; Reynolds, A.; Hoog, C.; Paddy, M.; Hunter, N. Interplay between synaptonemal complex, homologous recombination and centromeres during mammalian meiosis. PLoS Genet. 2012, 8, e1002790. [Google Scholar] [CrossRef] [PubMed]
- Solari, A.J. The behaviour of the XY pair in mammals. Int. Rev. Cytol. 1974, 38, 273–317. [Google Scholar] [CrossRef] [PubMed]
- Sciurano, R.B.; Solari, A.J. Ultrastructural and Immunofluorescent Methods for the Study of the XY Body as a Biomarker. Methods Mol. Biol. 2014, 1094, 137–149. [Google Scholar] [CrossRef]
- Waters, P.D.; Ruiz-Herrera, A. Meiotic executioner genes protect the Y from extinction. Trends Genet. 2020, 36, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Manterola, M.; Page, J.; Vasco, C.; Berríos, S.; Parra, M.T.; Viera, A.; Rufas, J.; Zuccotti, M.; Garagna, S.; Fernández-Donoso, R. A high incidence of meiotic silencing of unsynapsed chromatin is not associated with substantial pachytene loss in heterozygous male mice carrying multiple simple robertsonian translocations. PLoS Genet. 2009, 5, e1000625. [Google Scholar] [CrossRef]
- Rasmussen, S.W.; Holm, P.B. Chromosome pairing in autotetraploid Bombyx females. Mechanism for exclusive bivalent formation. Carlsberg Res. Commun. 1979, 44, 101–125. [Google Scholar] [CrossRef]
- Woglar, A.; Jantsch, V. Chromosome movement in meiosis I prophase of Caenorhabditis elegans. Chromosoma 2014, 123, 15–24. [Google Scholar] [CrossRef][Green Version]
- Pawlowski, W.P. Chromosome organization and dynamics in plants. Curr. Opin. Plant Biol. 2010, 13, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Oliver-Bonet, M.; Ko, E.; Martin, R.H. Male infertility in reciprocal translocation carriers: The sex body affair. Cytogenet. Genome Res. 2005, 111, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Homolka, D.; Ivanek, R.; Capkova, J.; Jansa, P.; Forejt, J. Chromosomal rearrangement interferes with meiotic X chromosome inactivation. Genome Res. 2007, 17, 1431–1437. [Google Scholar] [CrossRef]
- Kolomiets, O.L.; Abuduev, N.K.; Mazurova, T.F.; Bragina, E.E.; Kurilo, S.Y.D.L.; Bogdanov, Y.F. Damaging effect of antibiotics on the structure of synaptonemal complexes of meiotic mouse chromosomes. Russ. J. Genet. 2001, 37, 141–149. [Google Scholar] [CrossRef]
- Voet, T.; Liebe, B.; Labaere, C.; Marynen, P.; Scherthan, H. Telomere-independent homologue pairing and checkpoint escape of accessory ring chromosomes in male mouse meiosis. J. Cell Biol. 2003, 162, 795–808. [Google Scholar] [CrossRef]
- Saifitdinova, A.F.; Timofejeva, L.P.; Zhurov, V.G.; Gaginskaya, E.R. A highly repeated FCP centromeric sequence from chaffinch (Fringilla coelebs: Aves) genome is revealed within interchromosomal connectives during mitosis. Tsitologiia 2000, 42, 581–586. [Google Scholar]
- Saifitdinova, A.F.; Derjusheva, S.E.; Malykh, A.G.; Zhurov, V.G.; Andreeva, T.F.; Gaginskaya, E.R. Centromeric tandem repeat from the chaffinch genome: Isolation and molecular characterization. Genome 2001, 44, 96–103. [Google Scholar] [CrossRef]
- Kuznetsova, I.S.; Enukashvily, N.I.; Noniashvili, E.M.; Shatrova, A.N.; Aksenov, N.D.; Zenin, V.V.; Podgornaya, O.I. Evidence for the existence of satellite DNA-containing connection between metaphase chromosomes. J. Cell. Biochem. 2007, 101, 1046–1061. [Google Scholar] [CrossRef]



| Strain | X-Autosomal Associations N (%) | X-Two Autosomes Associations * N | Number of Nuclei Studied |
|---|---|---|---|
| BALB/c | 57 (6.1%) | 2 | 935 |
| CBA | 48 (5.6%) | 0 | 855 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spangenberg, V.; Losev, M.; Volkhin, I.; Smirnova, S.; Nikitin, P.; Kolomiets, O. DNA Environment of Centromeres and Non-Homologous Chromosomes Interactions in Mouse. Cells 2021, 10, 3375. https://doi.org/10.3390/cells10123375
Spangenberg V, Losev M, Volkhin I, Smirnova S, Nikitin P, Kolomiets O. DNA Environment of Centromeres and Non-Homologous Chromosomes Interactions in Mouse. Cells. 2021; 10(12):3375. https://doi.org/10.3390/cells10123375
Chicago/Turabian StyleSpangenberg, Victor, Mikhail Losev, Ilya Volkhin, Svetlana Smirnova, Pavel Nikitin, and Oxana Kolomiets. 2021. "DNA Environment of Centromeres and Non-Homologous Chromosomes Interactions in Mouse" Cells 10, no. 12: 3375. https://doi.org/10.3390/cells10123375
APA StyleSpangenberg, V., Losev, M., Volkhin, I., Smirnova, S., Nikitin, P., & Kolomiets, O. (2021). DNA Environment of Centromeres and Non-Homologous Chromosomes Interactions in Mouse. Cells, 10(12), 3375. https://doi.org/10.3390/cells10123375






