Positive Correlation between nNOS and Stress-Activated Bowel Motility Is Confirmed by In Vivo HiBiT System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Single Guide RNA and ssODN
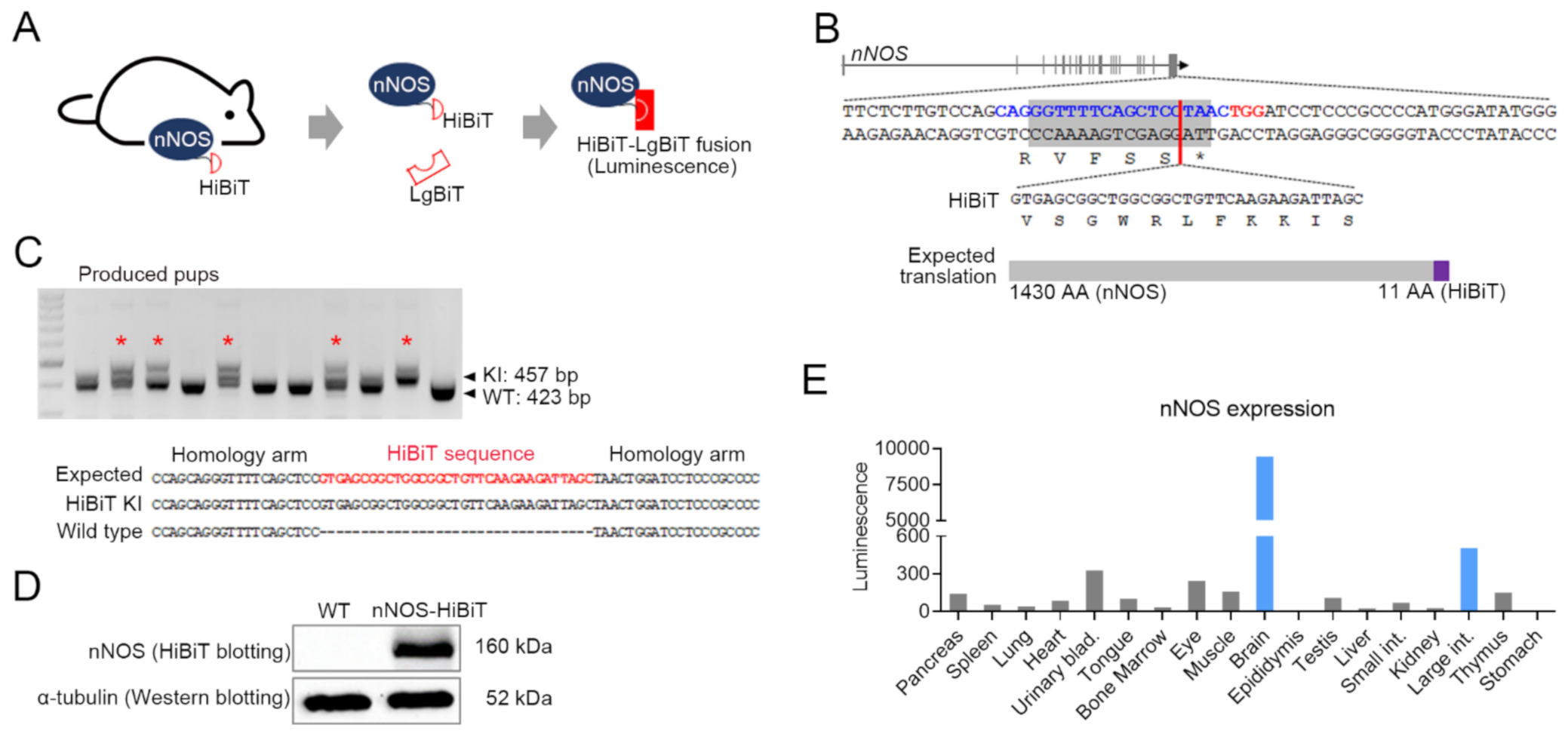
2.2. Generation of a Mouse with HiBiT Sequence KI into nNOS Gene C-Terminal End
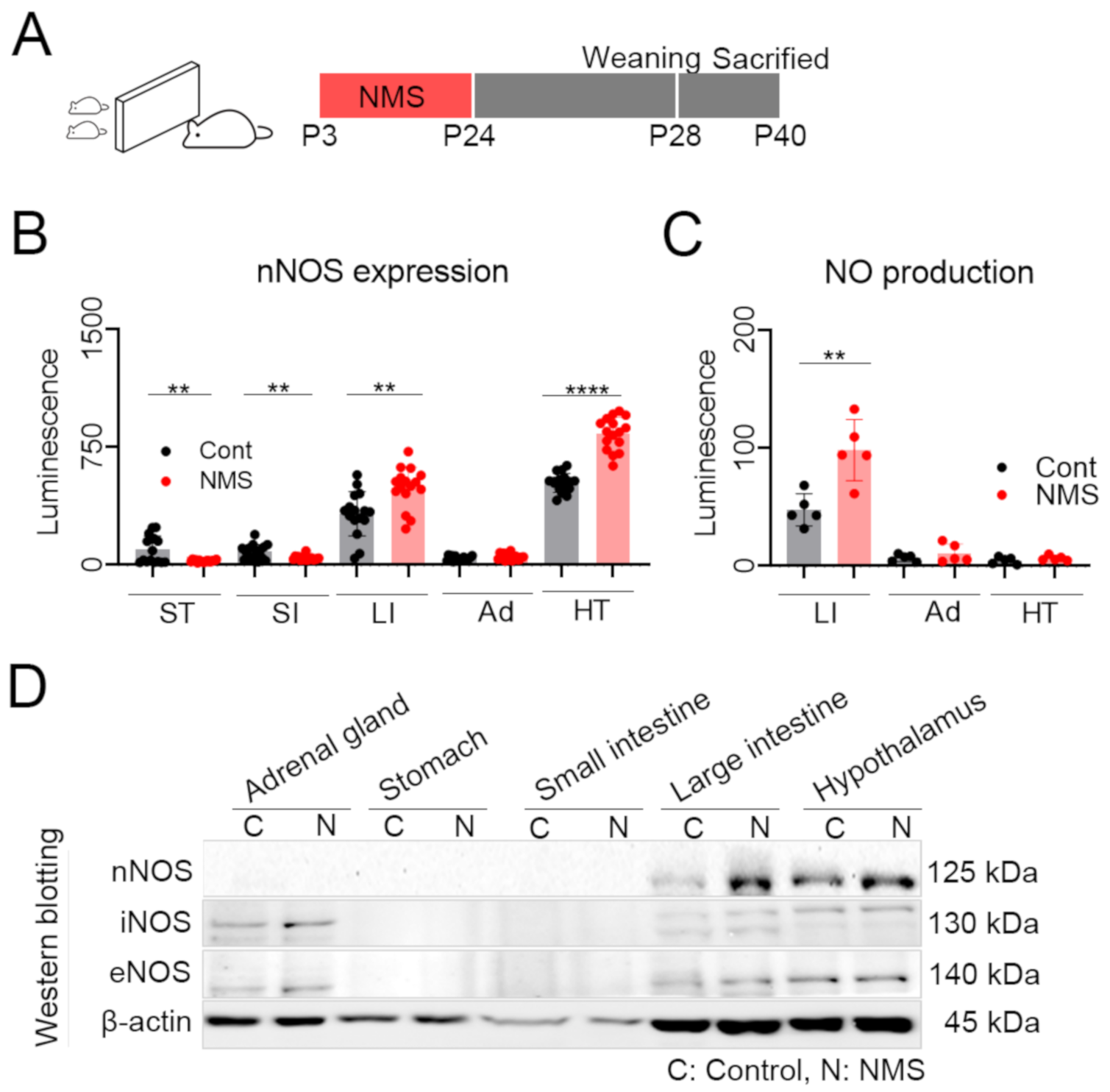
2.3. Neonatal Maternal Separation
2.4. Protein Preparation
2.5. HiBiT Blotting and HiBiT Lytic Analysis for Detecting nNOS Protein
2.6. Western Blotting
2.7. Measurement of NO Concentration
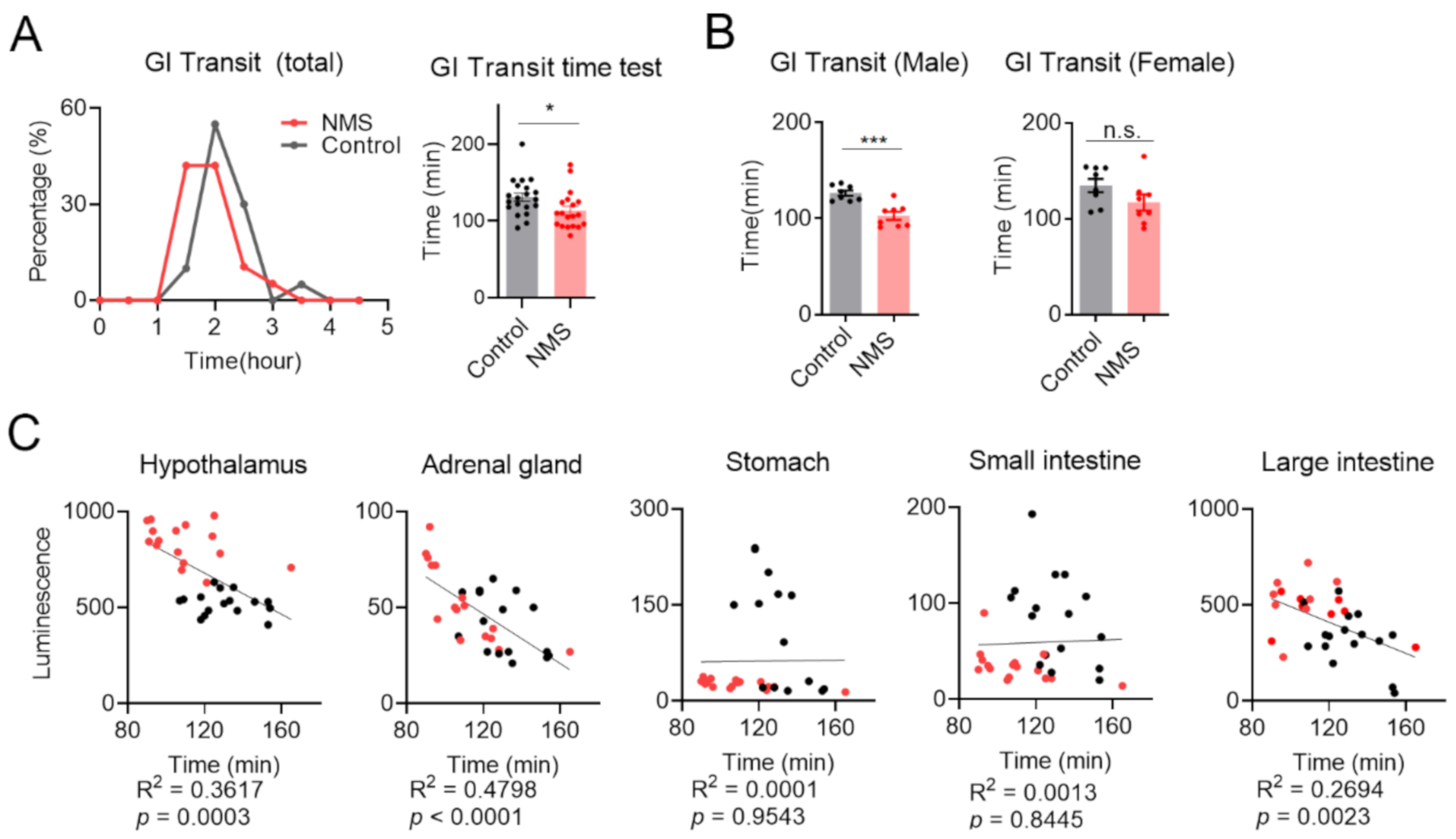
2.8. Analysis of Whole Gastrointestinal Transit Time
2.9. Statistical Analysis
3. Results
3.1. nNOS HiBiT Sequence KI Mouse Was Generated Using CRISPR/Cas9 and ssODN Template
3.2. Brain and Large Intestine Exhibited High nNOS Expression
3.3. NMS Developed a Significant Increase nNOS Expression in the Brain and Large Intestine
3.4. Whole Gastrointestinal Transit Time Was Positively Correlated with Expression Level of nNOS in the Hypothalamus-Adrenal Gland-Large Intestinal Axis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xue, Q.; Yan, Y.; Zhang, R.; Xiong, H. Regulation of iNOS on Immune Cells and Its Role in Diseases. Int. J. Mol. Sci. 2018, 19, 3805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, N.; Förstermann, U.; Li, H. Effects of resveratrol on eNOS in the endothelium and the perivascular adipose tissue. Ann. N. Y. Acad. Sci. 2017, 1403, 132–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Förstermann, U.; Sessa, W.C. Nitric oxide synthases: Regulation and function. Eur. Heart J. 2012, 33, 829–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burkard, N.; Rokita, A.G.; Kaufmann, S.G.; Hallhuber, M.; Wu, R.; Hu, K.; Hofmann, U.; Bonz, A.; Frantz, S.; Cartwright, E.J.; et al. Conditional Neuronal Nitric Oxide Synthase Overexpression Impairs Myocardial Contractility. Circ. Res. 2007, 100, e32–e44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashimo, H.; Kjellin, A.; Goyal, R.K. Gastric stasis in neuronal nitric oxide synthase–deficient knockout mice. Gastroenterology 2000, 119, 766–773. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Yu, M.; Wang, Y.; Ma, Y. An enhanced expression of hypothalamic neuronal nitric oxide synthase in a rat model of simulated transport stress. BMC Vet. Res. 2019, 15, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsigos, C.; Kyrou, I.; Kassi, E.; Chrousos, G.P. Stress, Endocrine Physiology and Pathophysiology. In Endotext; Feingold, K.R., Anawalt, B., Boyce, A., Chrousos, G., de Herder, W.W., Dungan, K., Grossman, A., Hershman, J.M., Hofland, H.J., Kaltsas, G., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2015. [Google Scholar]
- Yaribeygi, H.; Panahi, Y.; Sahraei, H.; Johnston, T.P.; Sahebkar, A. The impact of stress on body function: A review. EXCLI J. 2017, 16, 1057–1072. [Google Scholar] [PubMed]
- Moloney, R.D.; O’Leary, O.F.; Felice, D.; Bettler, B.; Dinan, T.G.; Cryan, J.F. Early-life stress induces visceral hypersensitivity in mice. Neurosci. Lett. 2012, 512, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.-Y.; Cheng, C.-W.; Tang, X.-D.; Bian, Z.-X. Impact of psychological stress on irritable bowel syndrome. World J. Gastroenterol. 2014, 20, 14126–14131. [Google Scholar] [CrossRef] [PubMed]
- Ikechi, R.; Fischer, B.D.; DeSipio, J.; Phadtare, S. Irritable Bowel Syndrome: Clinical Manifestations, Dietary Influences, and Management. Healthcare 2017, 5, 21. [Google Scholar] [CrossRef]
- Dinan, T.G.; Quigley, E.M.M.; Ahmed, S.M.M.; Scully, P.; O’Brien, S.; O’Mahony, L.; O’Mahony, S.; Shanahan, F.; Keeling, P.W.N. Hypothalamic-Pituitary-Gut Axis Dysregulation in Irritable Bowel Syndrome: Plasma Cytokines as a Potential Biomarker? Gastroenterology 2006, 130, 304–311. [Google Scholar] [CrossRef]
- Yang, M.; Fukui, H.; Eda, H.; Kitayama, Y.; Hara, K.; Kodani, M.; Tomita, T.; Oshima, T.; Watari, J.; Miwa, H. Involvement of gut microbiota in the association between gastrointestinal motility and 5-HT expression/M2 macrophage abundance in the gastrointestinal tract. Mol. Med. Rep. 2017, 16, 3482–3488. [Google Scholar] [CrossRef] [Green Version]
- Jang, D.E.; Bae, J.H.; Chang, Y.J.; Lee, Y.H.; Nam, K.T.; Kim, I.Y.; Seong, J.K.; Lee, Y.C.; Yeom, S.C. Neuronal Nitric Oxide Synthase Is a Novel Biomarker for the Interstitial Cells of Cajal in Stress-Induced Diarrhea-Dominant Irritable Bowel Syndrome. Dig. Dis. Sci. 2018, 63, 619–627. [Google Scholar] [CrossRef]
- Zhou, L.; Zhu, D.-Y. Neuronal nitric oxide synthase: Structure, subcellular localization, regulation, and clinical implications. Nitric Oxide 2009, 20, 223–230. [Google Scholar] [CrossRef]
- Spiller, R.; Lam, C. An Update on Post-infectious Irritable Bowel Syndrome: Role of Genetics, Immune Activation, Serotonin and Altered Microbiome. J. Neurogastroenterol. Motil. 2012, 18, 258–268. [Google Scholar] [CrossRef]
- Ren, T.-H.; Wu, J.; Yew, D.; Ziea, E.; Lao, L.; Leung, W.-K.; Berman, B.; Hu, P.-J.; Sung, J.J.Y. Effects of neonatal maternal separation on neurochemical and sensory response to colonic distension in a rat model of irritable bowel syndrome. Am. J. Physiol. Liver Physiol. 2007, 292, G849–G856. [Google Scholar] [CrossRef]
- Zimprich, A.; Garrett, L.; Deussing, J.M.; Wotjak, C.T.; Fuchs, H.; Gailus-Durner, V.; De Angelis, M.H.; Wurst, W.; Hölter, S.M. A robust and reliable non-invasive test for stress responsivity in mice. Front. Behav. Neurosci. 2014, 8, 125. [Google Scholar] [CrossRef] [Green Version]
- Oh-Hashi, K.; Furuta, E.; Fujimura, K.; Hirata, Y. Application of a novel HiBiT peptide tag for monitoring ATF4 protein expression in Neuro2a cells. Biochem. Biophys. Rep. 2017, 12, 40–45. [Google Scholar] [CrossRef]
- Dixon, A.S.; Schwinn, M.K.; Hall, M.P.; Zimmerman, K.; Otto, P.; Lubben, T.H.; Butler, B.L.; Binkowski, B.F.; Machleidt, T.; Kirkland, T.A.; et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 2016, 11, 400–408. [Google Scholar] [CrossRef]
- MeagherA, M.W.; Sieve, A.N.; Johnson, R.R.; Satterlee, D.; Belyavskyi, M.; Mi, W.; Prentice, T.W.; Welsh, T.H.; Welsh, C.J.R. Neonatal Maternal Separation Alters Immune, Endocrine, and Behavioral Responses to Acute Theiler’s Virus Infection in Adult Mice. Behav. Genet. 2010, 40, 233–249. [Google Scholar] [CrossRef] [Green Version]
- Vannucchi, M.G.; Evangelista, S. Experimental Models of Irritable Bowel Syndrome and the Role of the Enteric Neurotransmission. J. Clin. Med. 2018, 7, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwinn, M.K.; Machleidt, T.; Zimmerman, K.; Eggers, C.T.; Dixon, A.S.; Hurst, R.; Hall, M.P.; Encell, L.P.; Binkowski, B.F.; Wood, K.V. CRISPR-Mediated Tagging of Endogenous Proteins with a Luminescent Peptide. ACS Chem. Biol. 2017, 13, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kasper, L.H. The role of microbiome in central nervous system disorders. Brain Behav. Immun. 2014, 38, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reed, D.E.; Zhang, Y.; Beyak, M.J.; Lourenssen, S.; Blennerhassett, M.G.; Paterson, W.G.; Vanner, S.J. Stress increases descending inhibition in mouse and human colon. Neurogastroenterol. Motil. 2016, 28, 569–580. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, N. Sex-Gender Differences in Irritable Bowel Syndrome. J. Neurogastroenterol. Motil. 2018, 24, 544–558. [Google Scholar] [CrossRef] [Green Version]
- Halpin, S.J.; Ford, A.C. Prevalence of Symptoms Meeting Criteria for Irritable Bowel Syndrome in Inflammatory Bowel Disease: Systematic Review and Meta-Analysis. Am. J. Gastroenterol. 2012, 107, 1474–1482. [Google Scholar] [CrossRef]
- Yazar, A.; Büyükafpar, K.; Polat, G.; Pata, C.; Kanýk, A.; Tiftik, E.N.; Baðdatoðlu, Ö. The Urinary 5-Hydroxyindole Acetic Acid and Plasma Nitric Oxide Levels in Irritable Bowel Syndrome: A Preliminary Study. Scott. Med. J. 2005, 50, 27–29. [Google Scholar] [CrossRef]
- Reinders, C.I.; Herulf, M.; Ljung, T.; Hollenberg, J.; Weitzberg, E.; Lundberg, J.O.; Hellström, P.M. Rectal Mucosal Nitric Oxide in Differentiation of Inflammatory Bowel Disease and Irritable Bowel Syndrome. Clin. Gastroenterol. Hepatol. 2005, 3, 777–783. [Google Scholar] [CrossRef]
- Tjong, Y.-W.; Ip, S.-P.; Lao, L.; Wu, J.; Fong, H.H.S.; Sung, J.J.; Berman, B.; Che, C.-T. Role of neuronal nitric oxide synthase in colonic distension-induced hyperalgesia in distal colon of neonatal maternal separated male rats. Neurogastroenterol. Motil. 2011, 23, 666-e278. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, R.A. Pharmacological characterization of the nitrergic innervation of the stomach. Verh. K. Acad. Geneeskd. Belg. 2002, 64, 151–166. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, J.P.; Lee, J.H.; Lee, G.S.; Koo, O.J.; Yeom, S.C. Positive Correlation between nNOS and Stress-Activated Bowel Motility Is Confirmed by In Vivo HiBiT System. Cells 2021, 10, 1028. https://doi.org/10.3390/cells10051028
Han JP, Lee JH, Lee GS, Koo OJ, Yeom SC. Positive Correlation between nNOS and Stress-Activated Bowel Motility Is Confirmed by In Vivo HiBiT System. Cells. 2021; 10(5):1028. https://doi.org/10.3390/cells10051028
Chicago/Turabian StyleHan, Jeong Pil, Jeong Hyeon Lee, Geon Seong Lee, Ok Jae Koo, and Su Cheong Yeom. 2021. "Positive Correlation between nNOS and Stress-Activated Bowel Motility Is Confirmed by In Vivo HiBiT System" Cells 10, no. 5: 1028. https://doi.org/10.3390/cells10051028





