Familiarity Breeds Strategy: In Silico Untangling of the Molecular Complexity on Course of Autoimmune Liver Disease-to-Hepatocellular Carcinoma Transition Predicts Novel Transcriptional Signatures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Data Processing
2.3. Sample Clustering
2.4. Differential Gene Expression Analysis
2.5. Enrichment Analysis
2.6. Venn Diagram Construction
2.7. Gene Regulatory Network Construction
2.8. Kaplan–Meier Survival Analysis
2.9. Statistical Analysis
3. Results
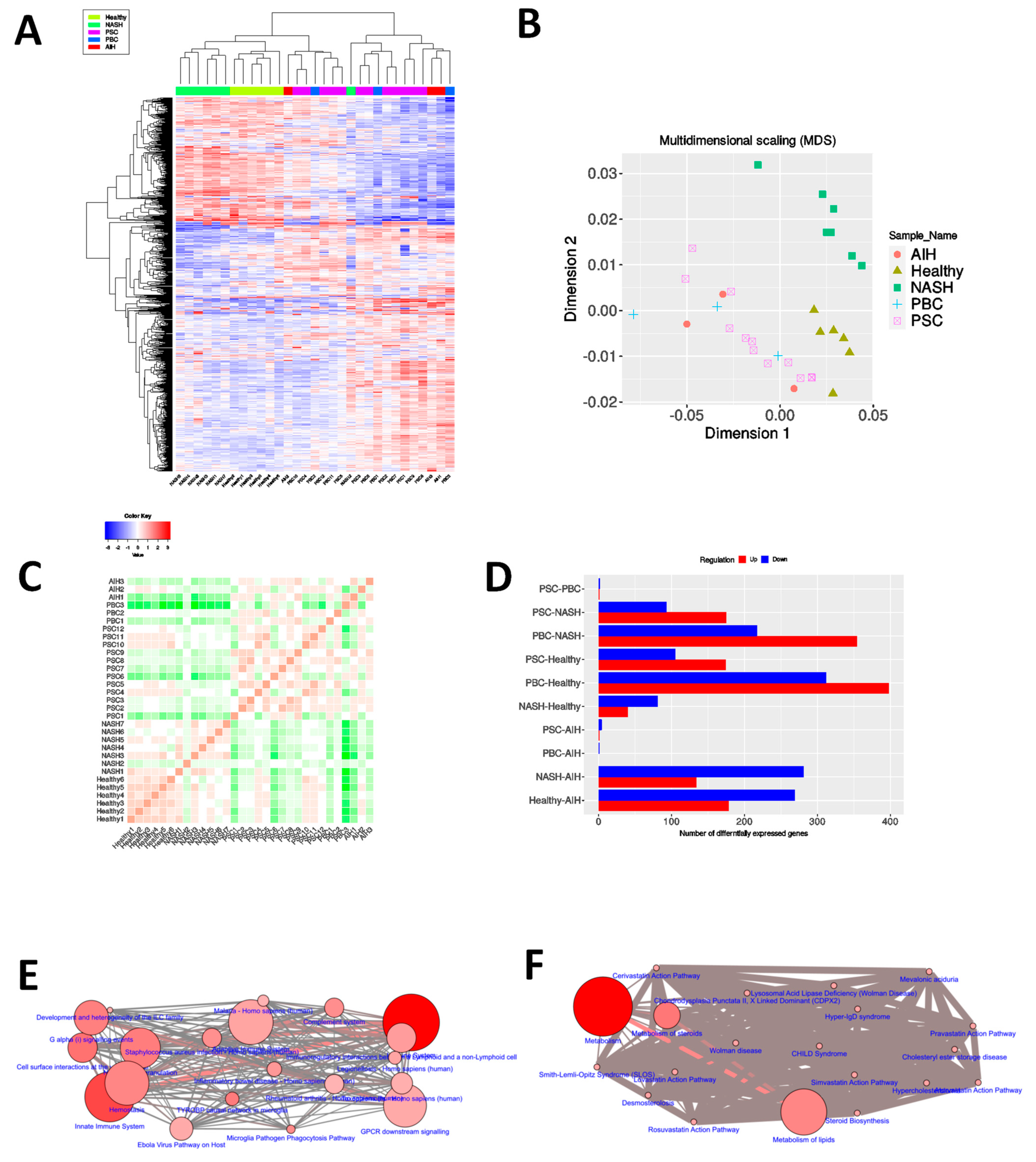
3.1. Different Autoimmune Liver Diseases Have an Overlapping Transcriptomic Signature
3.2. Progression towards Hepatocellular Carcinoma as Well as Autoimmune Liver Diseases Necessitates Large Scale Cellular Reprogramming
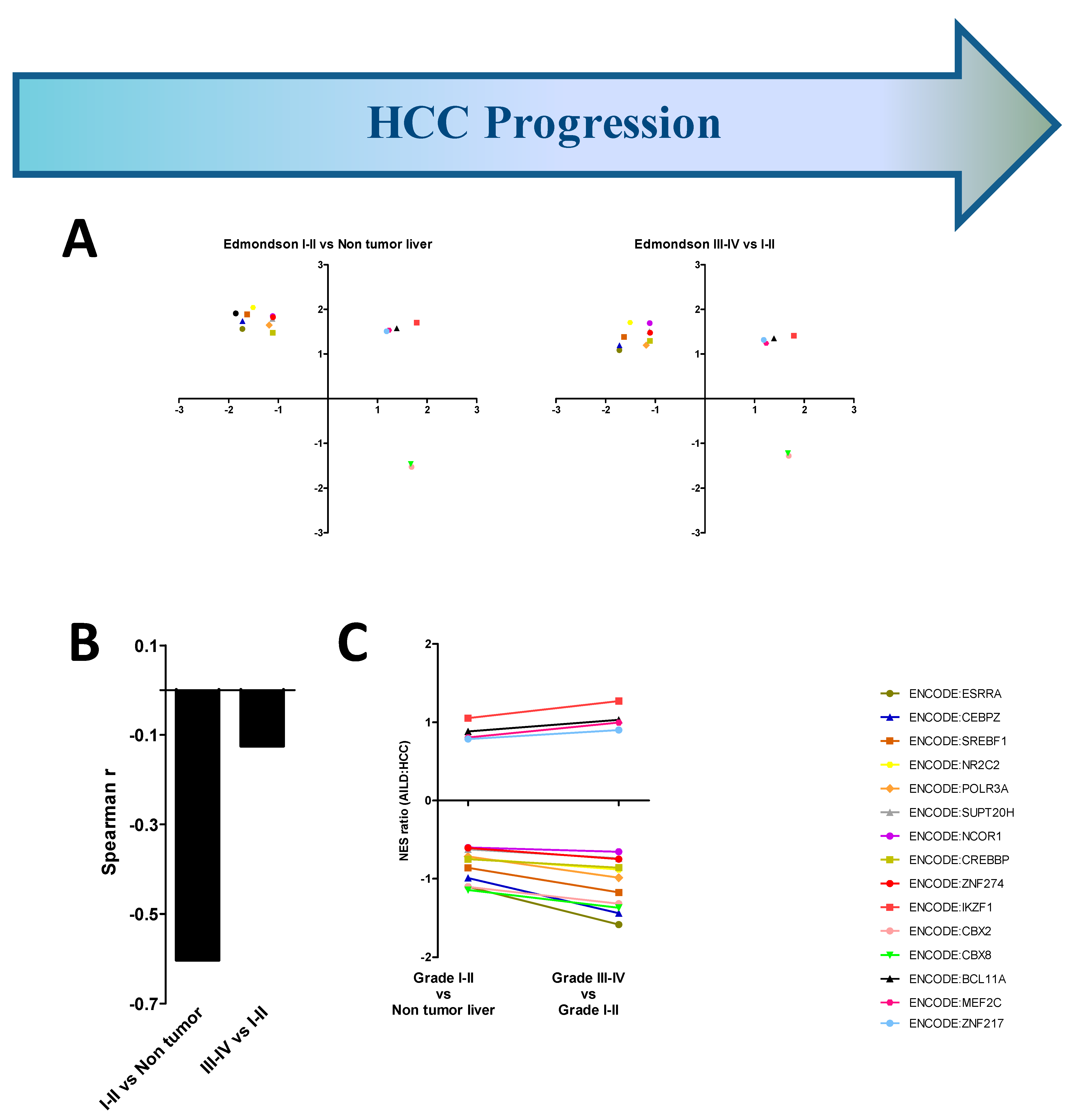
3.3. AILD-Associated HCC Signatures Develop Independent of Etiology
3.4. AILD-Associated HCC Signatures Are Weakly Associated with Histological Signs of HCC
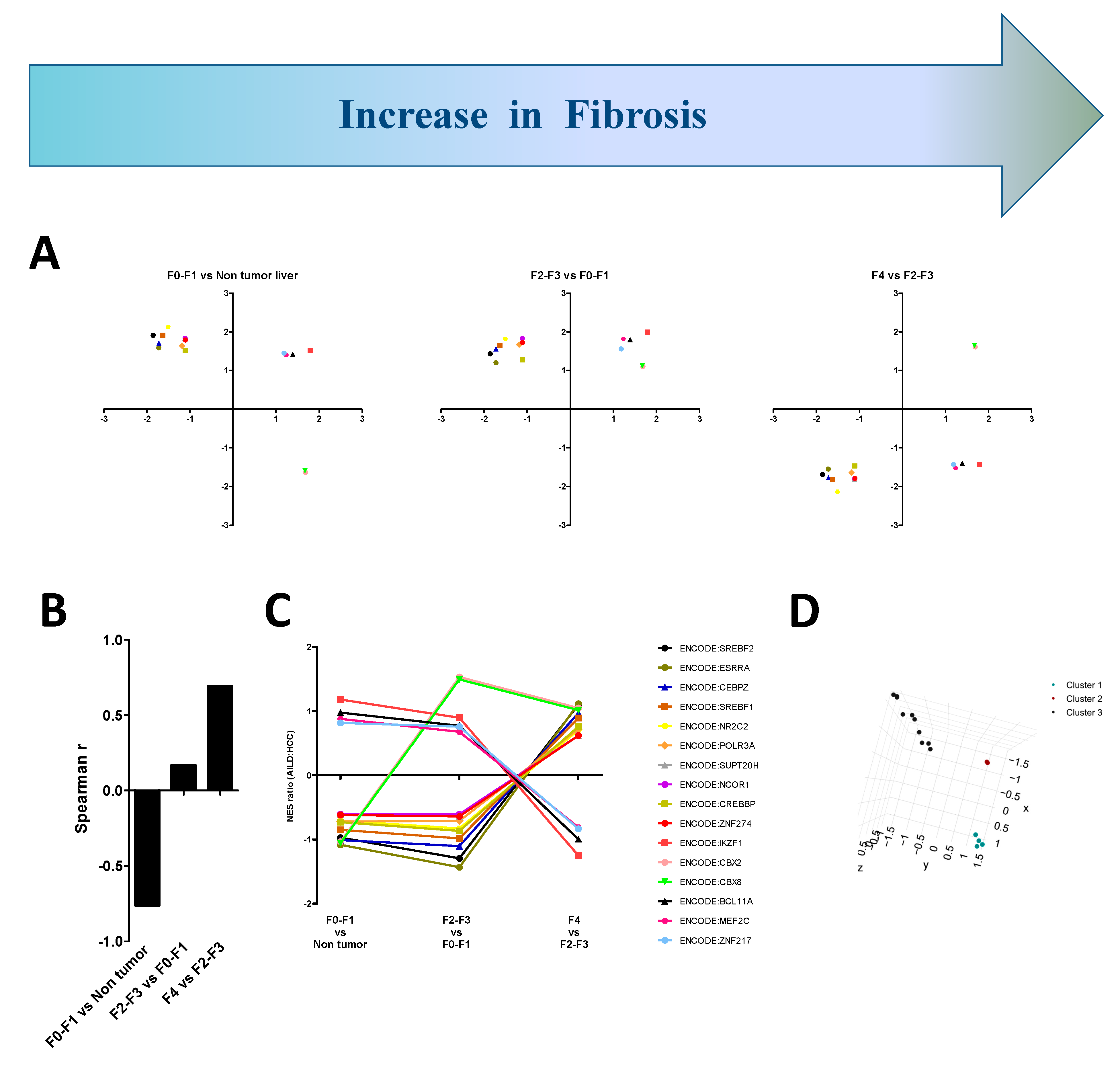
3.5. Transcriptomic Signatures of AILD Follow the Fibrotic Patterns on Carcinomatous Livers
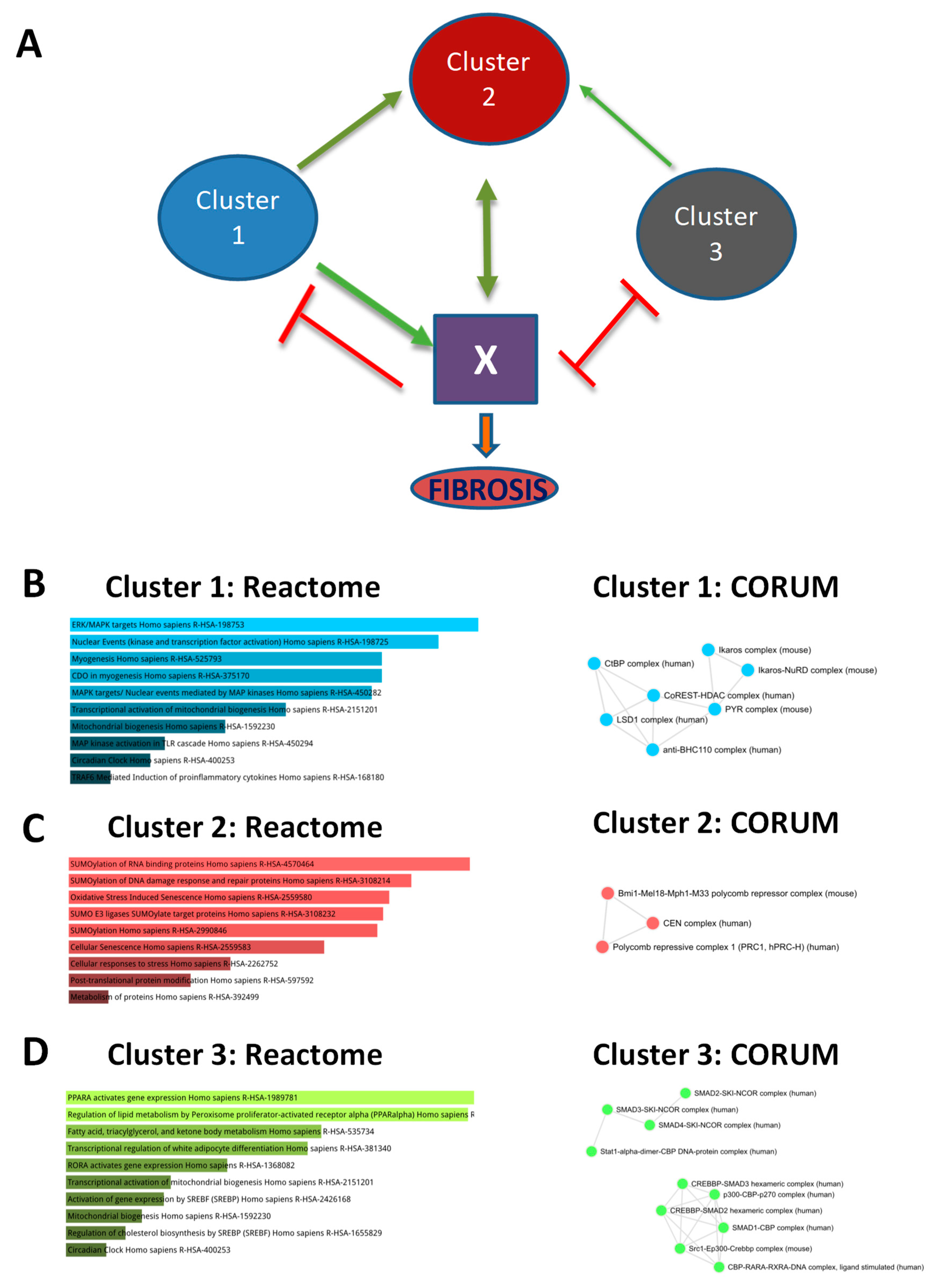
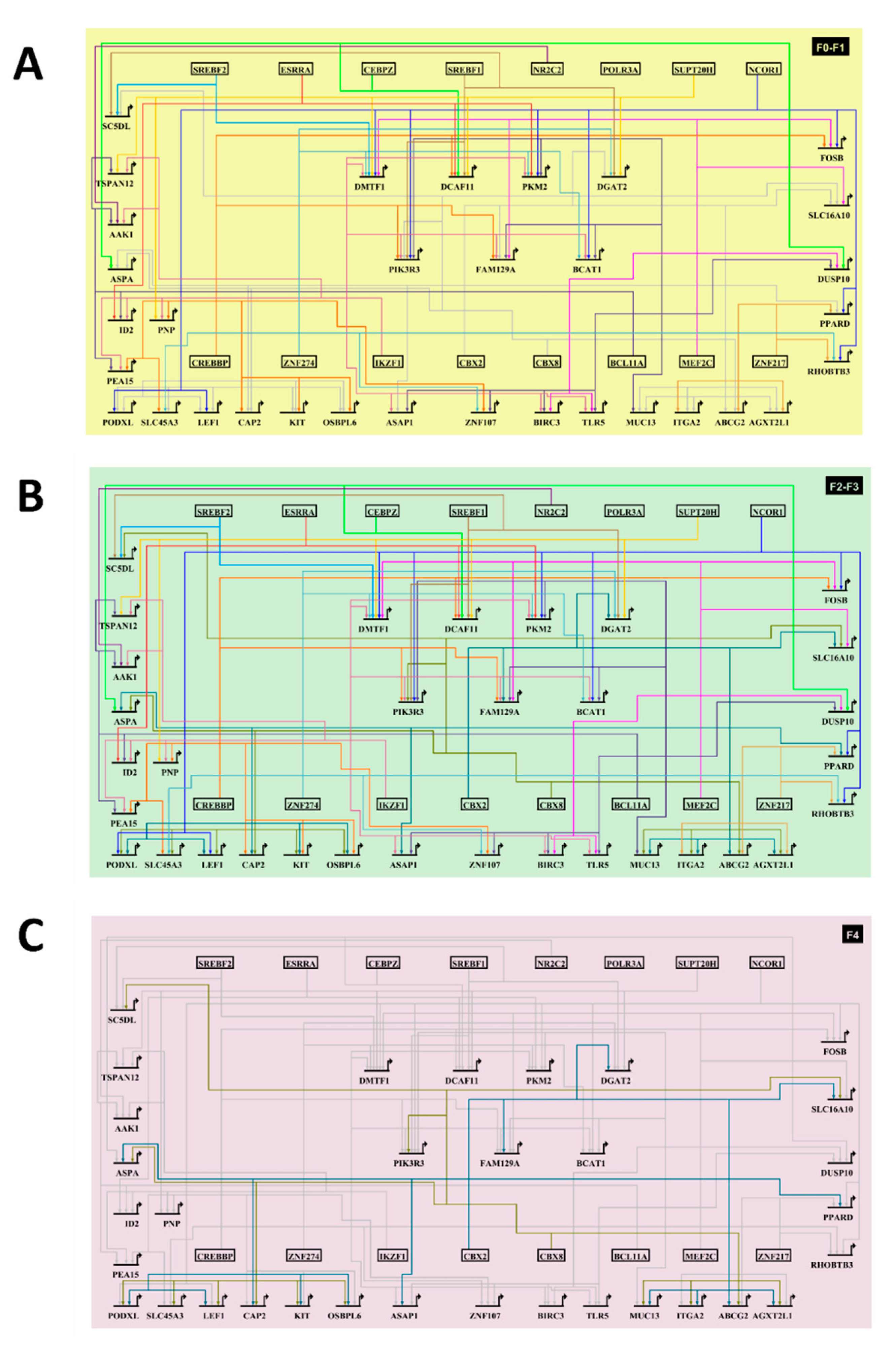
3.6. Sequential, Combinatorial Action of Different Clusters of Transcriptional Modules Promotes Fibrosis
3.7. The HCC-AILD Common Targetable Gene Regulatory Network Shifts with Progression of Fibrosis
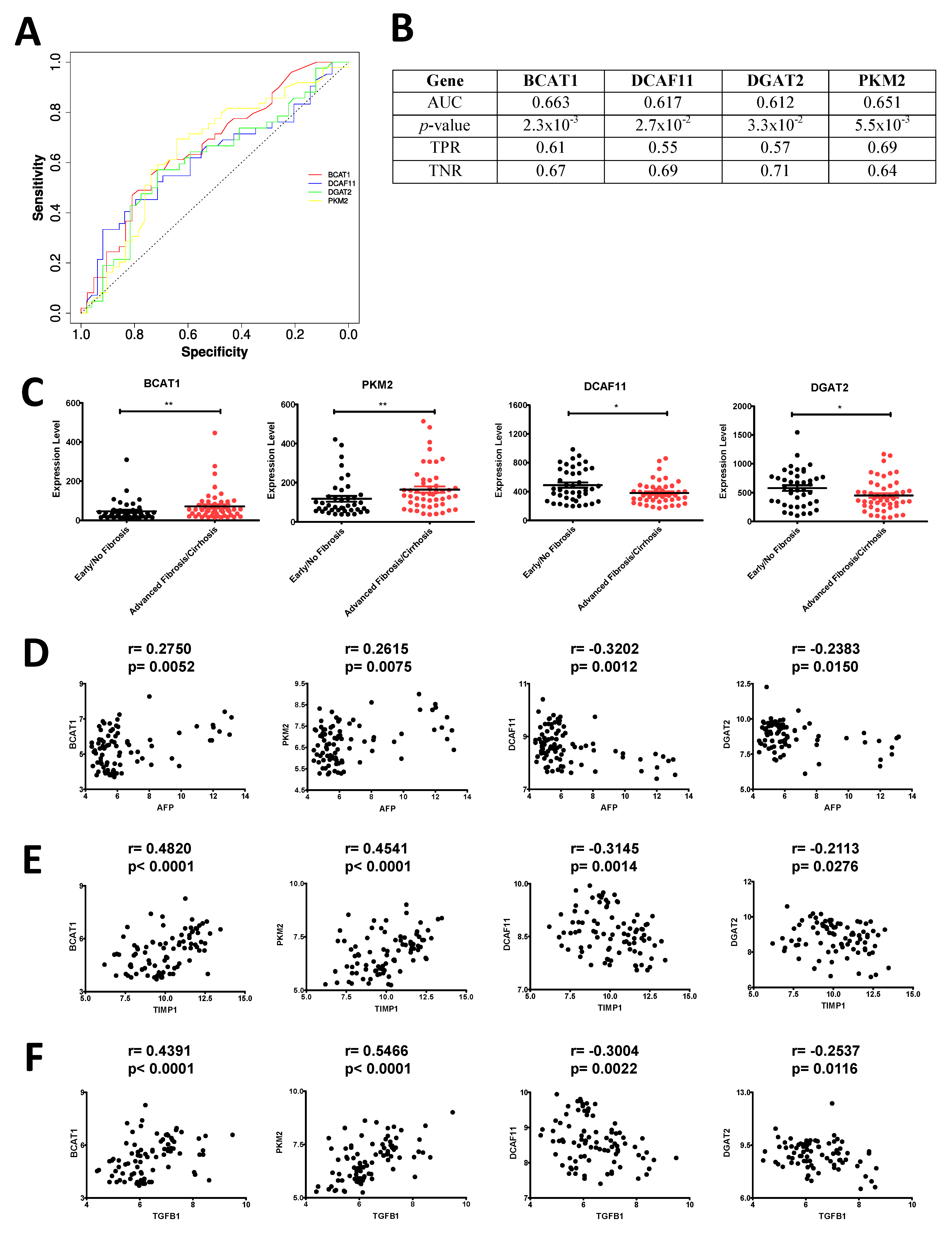
3.8. Increased Connectivity in the GRN Manifests into Better Candidacy as Predictive Marker
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arndtz, K.; Hirschfield, G.M. The Pathogenesis of Autoimmune Liver Disease. Dig. Dis. 2016, 34, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Purohit, T.; Cappell, M.S. Primary biliary cirrhosis: Pathophysiology, clinical presentation and therapy. World J. Hepatol. 2015, 7, 926–941. [Google Scholar] [CrossRef] [PubMed]
- Tabibian, J.H.; Bowlus, C.L. Primary sclerosing cholangitis: A review and update. Liver Res. 2017, 1, 221–230. [Google Scholar] [CrossRef]
- Mieli-Vergani, G.; Vergani, D.; Czaja, A.J.; Manns, M.P.; Krawitt, E.L.; Vierling, J.M.; Lohse, A.W.; Montano-Loza, A.J. Autoimmune hepatitis. Nat. Rev. Dis. Primers. 2018, 4, 18017. [Google Scholar] [CrossRef]
- Carbone, M.; Neuberger, J.M. Autoimmune liver disease, autoimmunity and liver transplantation. J. Hepatol. 2014, 60, 210–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigopoulou, E.I.; Dalekos, G.N. Current Trends and Characteristics of Hepatocellular Carcinoma in Patients with Autoimmune Liver Diseases. Cancers 2021, 13, 1023. [Google Scholar] [CrossRef]
- Lleo, A.; de Boer, Y.S.; Liberal, R.; Colombo, M. The risk of liver cancer in autoimmune liver diseases. Ther. Adv. Med. Oncol. 2019, 1, 1758835919861914. [Google Scholar] [CrossRef] [PubMed]
- Findor, J.; He, X.S.; Sord, J.; Terg, R.; Gershwin, M.E. Primary biliary cirrhosis and hepatocellular carcinoma. Autoimmun. Rev. 2002, 1, 220–225. [Google Scholar] [CrossRef]
- Tansel, A.; Katz, L.H.; El-Serag, H.B.; Thrift, A.P.; Parepally, M.; Shakhatreh, M.H.; Kanwal, F. Incidence and Determinants of Hepatocellular Carcinoma in Autoimmune Hepatitis: A Systematic Review and Meta-analysis. Clin. Gastroenterol. Hepatol. 2017, 15, 1207–1217. [Google Scholar] [CrossRef] [Green Version]
- Sucher, E.; Sucher, R.; Gradistanac, T.; Brandacher, G.; Schneeberger, S.; Berg, T. Autoimmune Hepatitis-Immunologically Triggered Liver Pathogenesis-Diagnostic and Therapeutic Strategies. J. Immunol. Res. 2019, 2019, 9437043. [Google Scholar] [CrossRef]
- Wang, M.; Xi, D.; Ning, Q. Virus-induced hepatocellular carcinoma with special emphasis on HBV. Hepatol. Int. 2017, 11, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.F.; Chang, C.W.; Wei, R.J.; Shiue, Y.L.; Wang, S.N.; Yeh, Y.T. Involvement of DNA damage response pathways in hepatocellular carcinoma. Biomed. Res. Int. 2014, 2014, 153867. [Google Scholar] [CrossRef] [PubMed]
- Bisteau, X.; Caldez, M.J.; Kaldis, P. The Complex Relationship between Liver Cancer and the Cell Cycle: A Story of Multiple Regulations. Cancers 2014, 6, 79–111. [Google Scholar] [CrossRef] [Green Version]
- Lei, Y.; Yan, W.; Lin, Z.; Liu, J.; Tian, D.; Han, P. Comprehensive analysis of partial epithelial mesenchymal transition-related genes in hepatocellular carcinoma. J. Cell. Mol. Med. 2021, 25, 448–462. [Google Scholar] [CrossRef]
- Fujiwara, N.; Friedman, S.L.; Goossens, N.; Hoshida, Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J. Hepatol. 2018, 68, 526–549. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Liu, S.; Zeng, S.; Shen, H. From bench to bed: The tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 396. [Google Scholar] [CrossRef] [Green Version]
- Refolo, M.G.; Messa, C.; Guerra, V.; Carr, B.I.; D’Alessandro, R. Inflammatory Mechanisms of HCC Development. Cancers 2020, 12, 641. [Google Scholar] [CrossRef] [Green Version]
- Czaja, A.J. Difficult treatment decisions in autoimmune hepatitis. World J. Gastroenterol. 2010, 16, 934–947. [Google Scholar] [CrossRef] [PubMed]
- Kow, A.W.C. Transplantation versus liver resection in patients with hepatocellular carcinoma. Transl. Gastroenterol. Hepatol. 2019, 4, 33. [Google Scholar] [CrossRef]
- Wong, R.J.; Gish, R.; Frederick, T.; Bzowej, N.; Frenette, C. Development of hepatocellular carcinoma in autoimmune hepatitis patients: A case series. Dig. Dis. Sci. 2011, 56, 578–585. [Google Scholar] [CrossRef]
- Watanabe, T.; Soga, K.; Hirono, H.; Hasegawa, K.; Shibasaki, K.; Kawai, H.; Aoyagi, Y. Features of hepatocellular carcinoma in cases with autoimmune hepatitis and primary biliary cirrhosis. World J. Gastroenterol. 2009, 15, 231–239. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef] [Green Version]
- Kamburov, A.; Wierling, C.; Lehrach, H.; Herwig, R. ConsensusPathDB—A database for integrating human functional interaction networks. Nucleic Acids Res. 2009, 37, D623–D628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamburov, A.; Pentchev, K.; Galicka, H.; Wierling, C.; Lehrach, H.; Herwig, R. ConsensusPathDB: Toward a more complete picture of cell biology. Nucleic Acids Res. 2011, 39, D712–D717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.J.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Longabaugh, W.J.; Davidson, E.H.; Bolouri, H. Computational representation of developmental genetic regulatory networks. Dev. Biol. 2005, 283, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Longabaugh, W.J. BioTapestry: A tool to visualize the dynamic properties of gene regulatory networks. Methods Mol. Biol. 2012, 786, 359–394. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Lánczky, A.; Menyhárt, O.; Győrffy, B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018, 8, 9227. [Google Scholar] [CrossRef] [PubMed]
- Menyhárt, O.; Nagy, Á.; Győrffy, B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R. Soc. Open Sci. 2018, 5, 181006. [Google Scholar] [CrossRef] [Green Version]
- Fekete, J.T.; Győrffy, B. ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3104 breast cancer patients. Int. J. Cancer 2019, 145, 3140–3151. [Google Scholar] [CrossRef] [PubMed]
- Casamassimi, A.; Federico, A.; Rienzo, M.; Esposito, S.; Ciccodicola, A. Transcriptome Profiling in Human Diseases: New Advances and Perspectives. Int. J. Mol. Sci. 2017, 18, 1652. [Google Scholar] [CrossRef] [Green Version]
- Park, S.Z.; Nagorney, D.M.; Czaja, A.J. Hepatocellular carcinoma in autoimmune hepatitis. Dig. Dis. Sci. 2000, 45, 1944–1948. [Google Scholar] [CrossRef]
- Schulze, K.; Imbeaud, S.; Letouzé, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015, 47, 505–511. [Google Scholar] [CrossRef]
- Edmondson, H.A.; Steiner, P.E. Primary carcinoma of the liver: A study of 100 cases among 48,900 necropsies. Cancer 1954, 7, 462–503. [Google Scholar] [CrossRef]
- Baier, J.L.; Mattner, J. Mechanisms of autoimmune liver disease. Discov. Med. 2014, 18, 255–263. [Google Scholar]
- Ramakrishna, G.; Rastogi, A.; Trehanpati, N.; Sen, B.; Khosla, R.; Sarin, S.K. From cirrhosis to hepatocellular carcinoma: New molecular insights on inflammation and cellular senescence. Liver Cancer 2013, 2, 367–383. [Google Scholar] [CrossRef]
- Bedossa, P.; Poynard, T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology 1996, 24, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Moudgil, K.D. Regulation of autoimmune inflammation by pro-inflammatory cytokines. Immunol. Lett. 2008, 120, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Franks, A.L.; Slansky, J.E. Multiple associations between a broad spectrum of autoimmune diseases, chronic inflammatory diseases and cancer. Anticancer Res. 2012, 32, 1119–1136. [Google Scholar] [PubMed]
- Tarao, K.; Nozaki, A.; Ikeda, T.; Sato, A.; Komatsu, H.; Komatsu, T.; Taguri, M.; Tanaka, K. Real impact of liver cirrhosis on the development of hepatocellular carcinoma in various liver diseases-meta-analytic assessment. Cancer Med. 2019, 8, 1054–1065. [Google Scholar] [CrossRef] [Green Version]
- Valean, S.; Acalovschi, M.; Dumitrascu, D.L.; Ciobanu, L.; Nagy, G.; Chira, R. Hepatocellular carcinoma in patients with autoimmune hepatitis—A systematic review of the literature published between 1989–2016. Med. Pharm. Rep. 2019, 92, 99–105. [Google Scholar] [CrossRef] [Green Version]
- Jeng, K.S.; Lu, S.J.; Wang, C.H.; Chang, C.F. Liver Fibrosis and Inflammation under the Control of ERK2. Int. J. Mol. Sci. 2020, 21, 3796. [Google Scholar] [CrossRef]
- Wang, Y.; Wen, H.; Fu, J.; Cai, L.; Li, P.-L.; Zhao, C.-L.; Dong, Z.-F.; Ma, J.-P.; Wang, X.; Tian, H.; et al. Hepatocyte TNF Receptor-Associated Factor 6 Aggravates Hepatic Inflammation and Fibrosis by Promoting Lysine 6-Linked Polyubiquitination of Apoptosis Signal-Regulating Kinase 1. Hepatology 2020, 71, 93–111. [Google Scholar] [CrossRef] [PubMed]
- Francque, S.; Verrijken, A.; Caron, S.; Prawitt, J.; Paumelle, R.; Derudas, B.; Lefebvre, P.; Taskinen, M.-R.; Hul, W.V.; Mertens, I.; et al. PPARα gene expression correlates with severity and histological treatment response in patients with non-alcoholic steatohepatitis. J. Hepatol. 2015, 63, 164–173. [Google Scholar] [CrossRef]
- Latella, G.; Vetuschi, A.; Sferra, R.; Catitti, V.; D’Angelo, A.; Zanninelli, G.; Flanders, K.C.; Gaudio, E. Targeted disruption of Smad3 confers resistance to the development of dimethylnitrosamine-induced hepatic fibrosis in mice. Liver Int. 2009, 29, 997–1009. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.; Stroschein, S.L.; Wang, W.; Chen, D.; Martens, E.; Zhou, S.; Zhou, Q. The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling. Genes Dev. 1999, 13, 2196–2206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, Z.; Raible, F.; Mollaaghababa, R.; Guyon, J.R.; Wu, C.-T.; Bender, W.; Kingston, R.E. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 1999, 98, 37–46. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mukherjee, S.; Kar, A.; Khatun, N.; Datta, P.; Biswas, A.; Barik, S. Familiarity Breeds Strategy: In Silico Untangling of the Molecular Complexity on Course of Autoimmune Liver Disease-to-Hepatocellular Carcinoma Transition Predicts Novel Transcriptional Signatures. Cells 2021, 10, 1917. https://doi.org/10.3390/cells10081917
Mukherjee S, Kar A, Khatun N, Datta P, Biswas A, Barik S. Familiarity Breeds Strategy: In Silico Untangling of the Molecular Complexity on Course of Autoimmune Liver Disease-to-Hepatocellular Carcinoma Transition Predicts Novel Transcriptional Signatures. Cells. 2021; 10(8):1917. https://doi.org/10.3390/cells10081917
Chicago/Turabian StyleMukherjee, Soumyadeep, Arpita Kar, Najma Khatun, Puja Datta, Avik Biswas, and Subhasis Barik. 2021. "Familiarity Breeds Strategy: In Silico Untangling of the Molecular Complexity on Course of Autoimmune Liver Disease-to-Hepatocellular Carcinoma Transition Predicts Novel Transcriptional Signatures" Cells 10, no. 8: 1917. https://doi.org/10.3390/cells10081917
APA StyleMukherjee, S., Kar, A., Khatun, N., Datta, P., Biswas, A., & Barik, S. (2021). Familiarity Breeds Strategy: In Silico Untangling of the Molecular Complexity on Course of Autoimmune Liver Disease-to-Hepatocellular Carcinoma Transition Predicts Novel Transcriptional Signatures. Cells, 10(8), 1917. https://doi.org/10.3390/cells10081917









