Tissue-Based Proteomic Profiling in Patients with Hyperplasia and Endometrial Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval and Consent to Participate
2.2. Study Design and Patient Selection
2.3. Tissue Protein Extraction
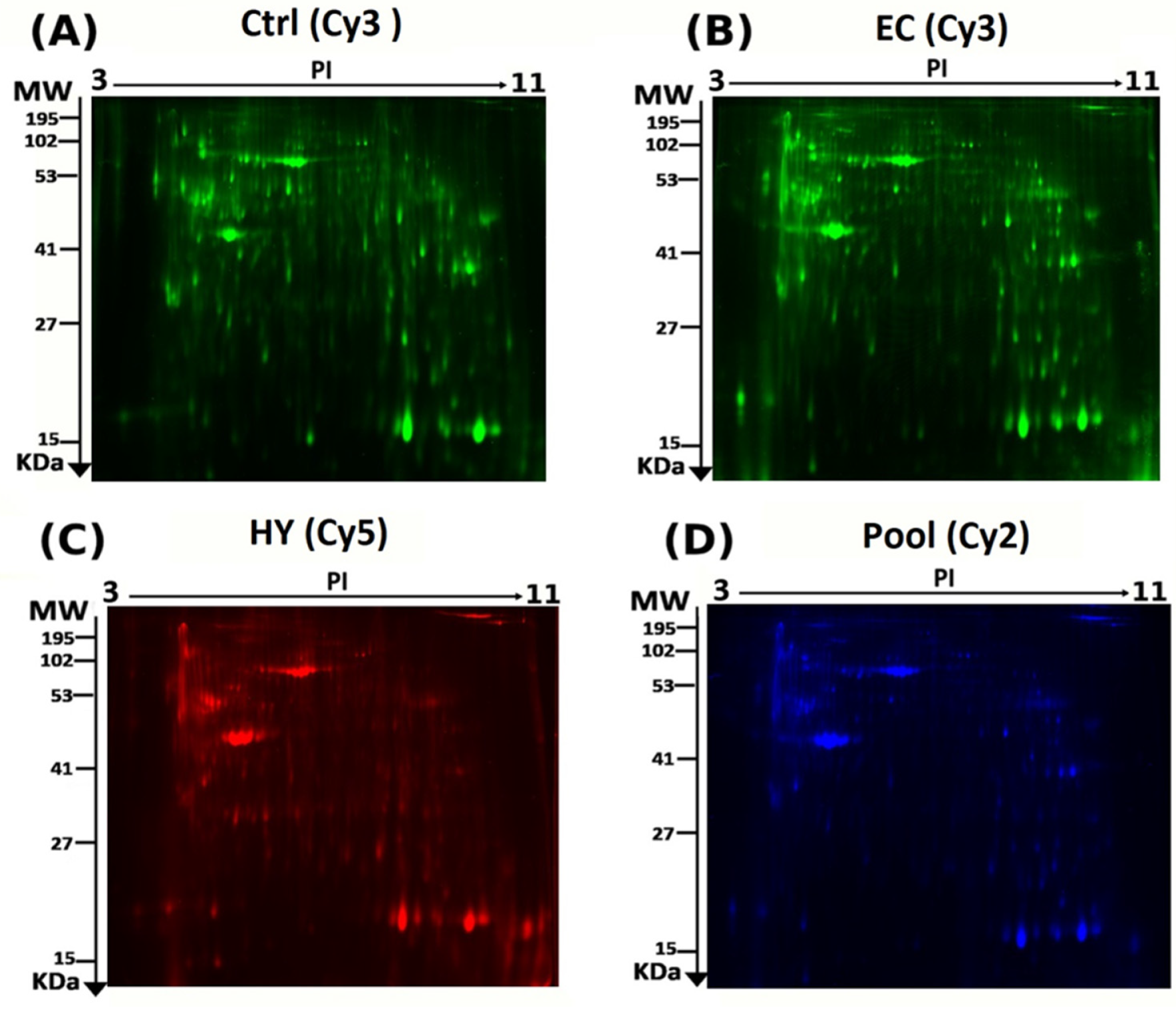
2.4. CyDye Labelling, Two-Dimensional (2D) Electrophoresis, and Image Scanning
2.5. Statistical Analysis
2.6. Protein Identification Using MALDI-TOF Mass Spectrometry
2.7. Bioinformatics Analysis
2.8. Validation of Results Using LC- MS/MS (MRM)
3. Results
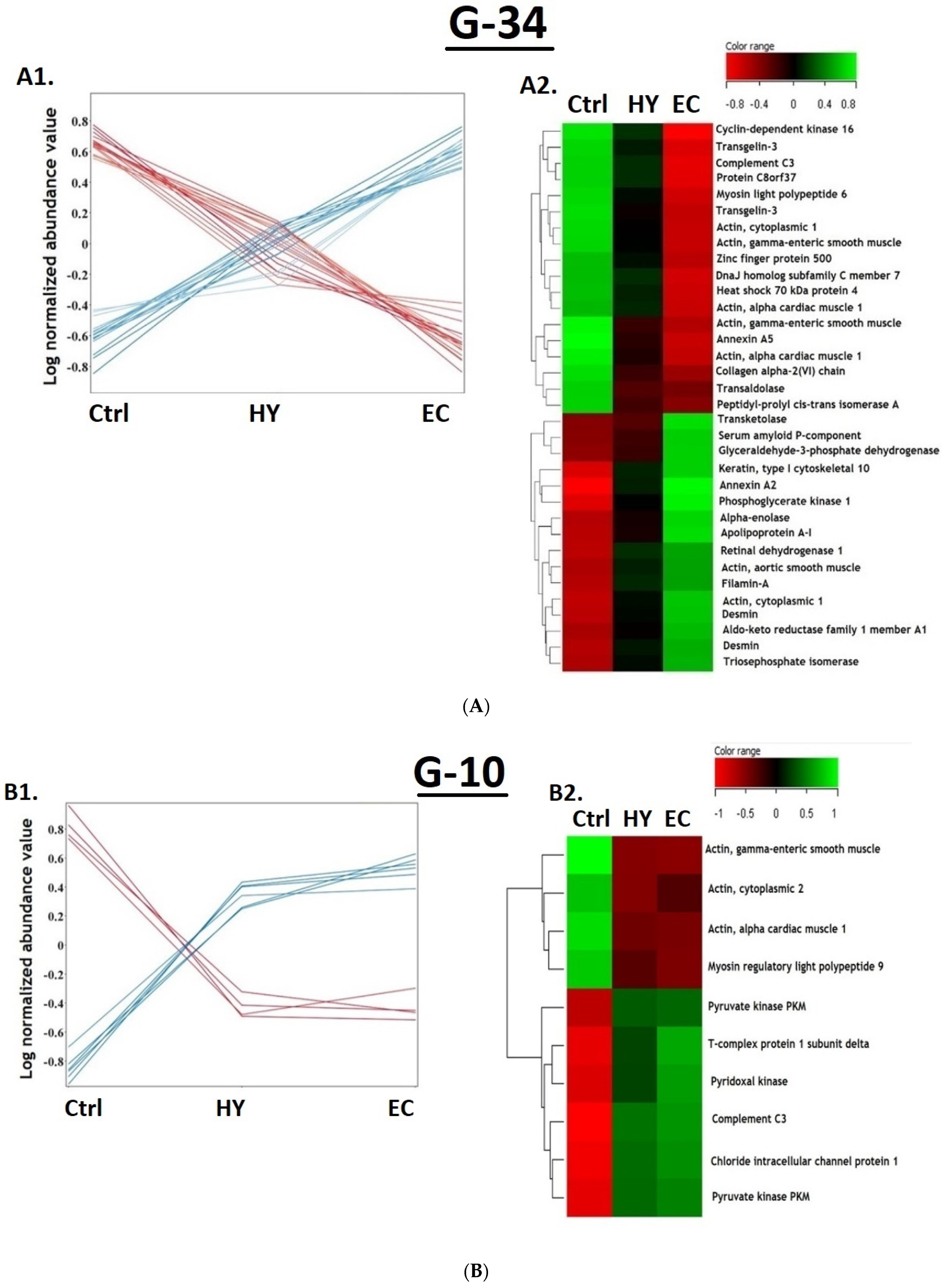
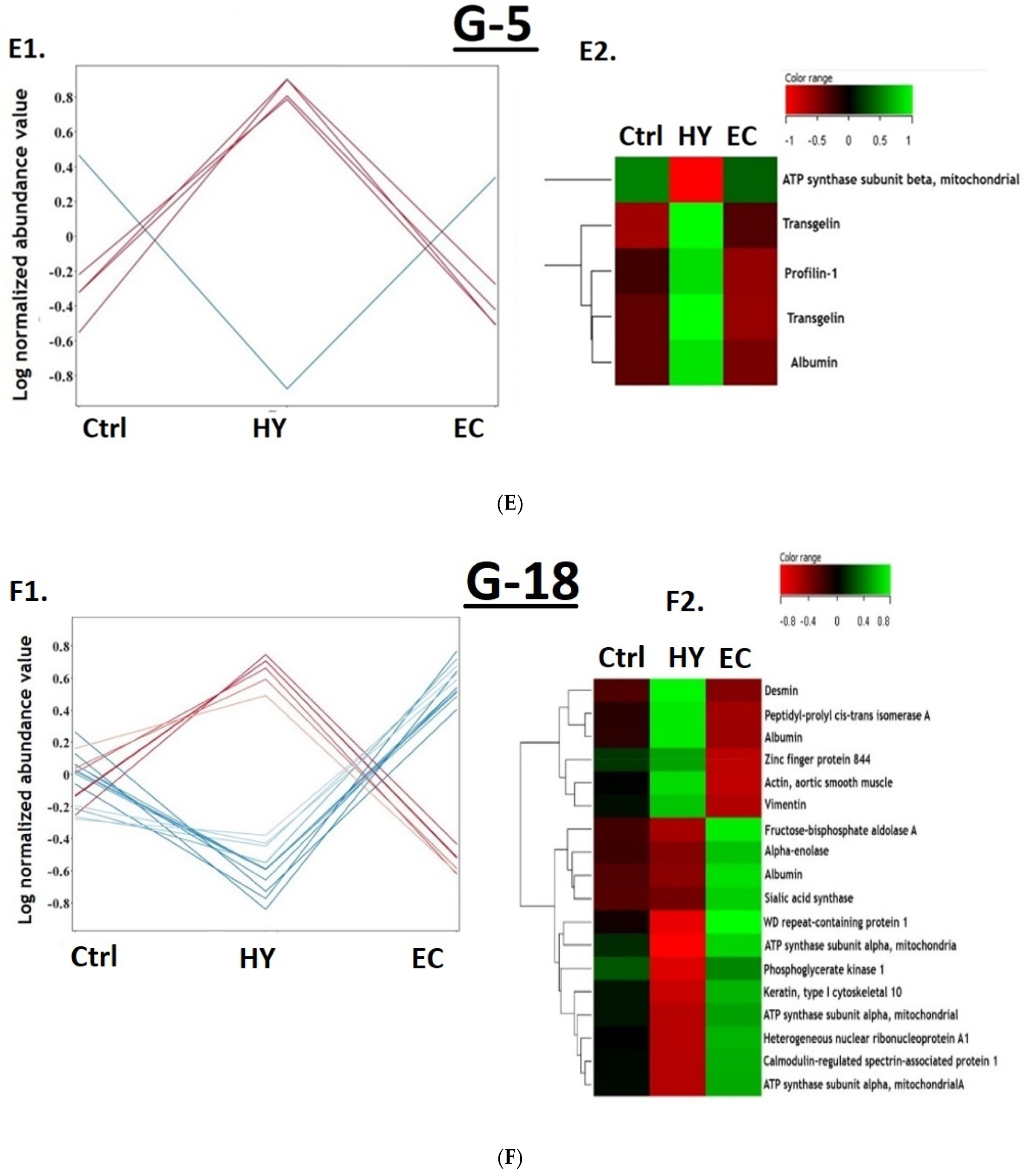
3.1. Proteomic Analysis and Identification of Differentially Expressed Proteins Using 2D-DIGE Analysis
3.2. Principal Component Analysis
3.3. The Overall Proteomic Analysis
3.4. Cancer vs. Control Proteomic Pattern
3.5. The Hyperplasia vs. Control Proteomic Pattern
3.6. The Cancer vs. Hyperplasia Proteomic Pattern
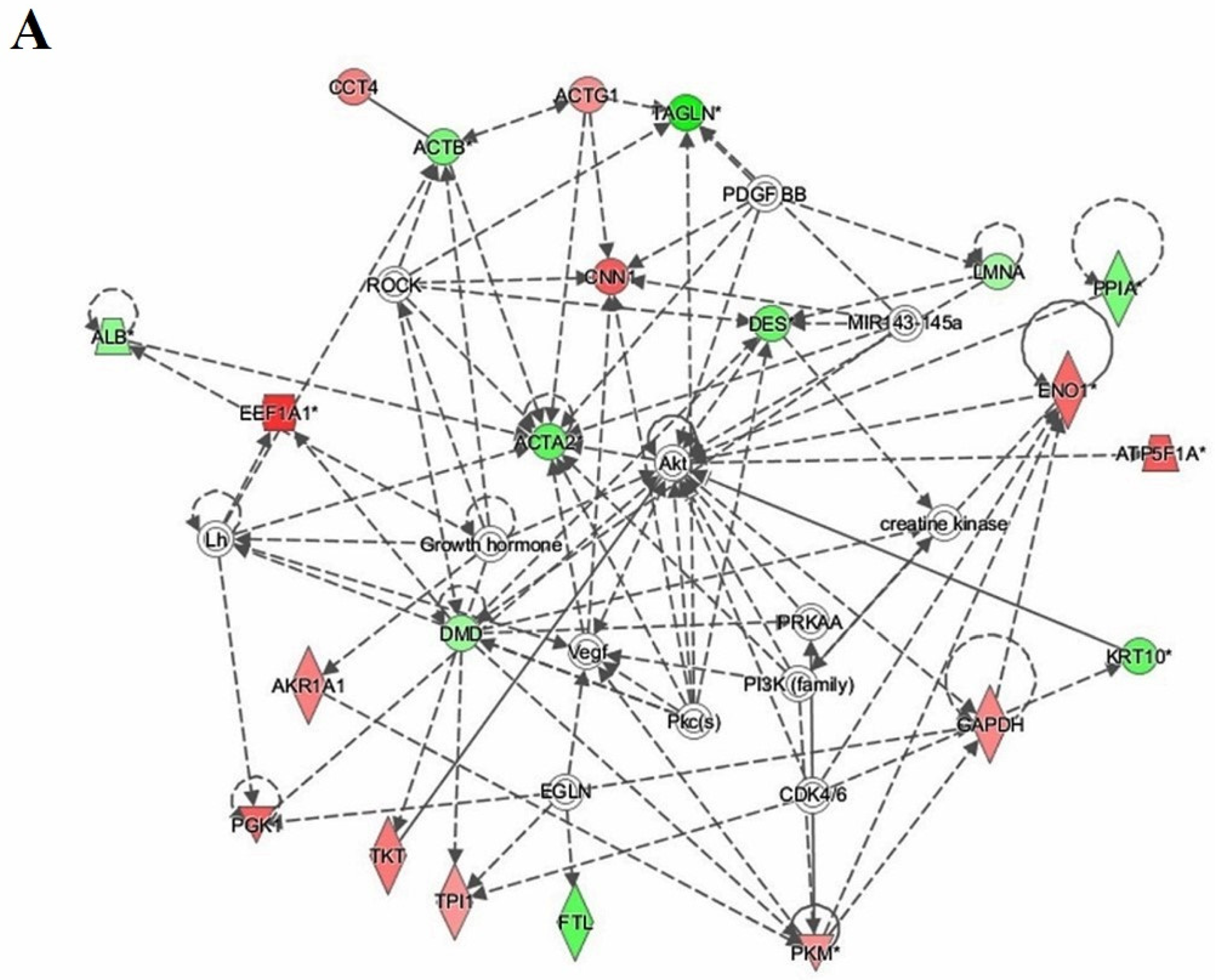
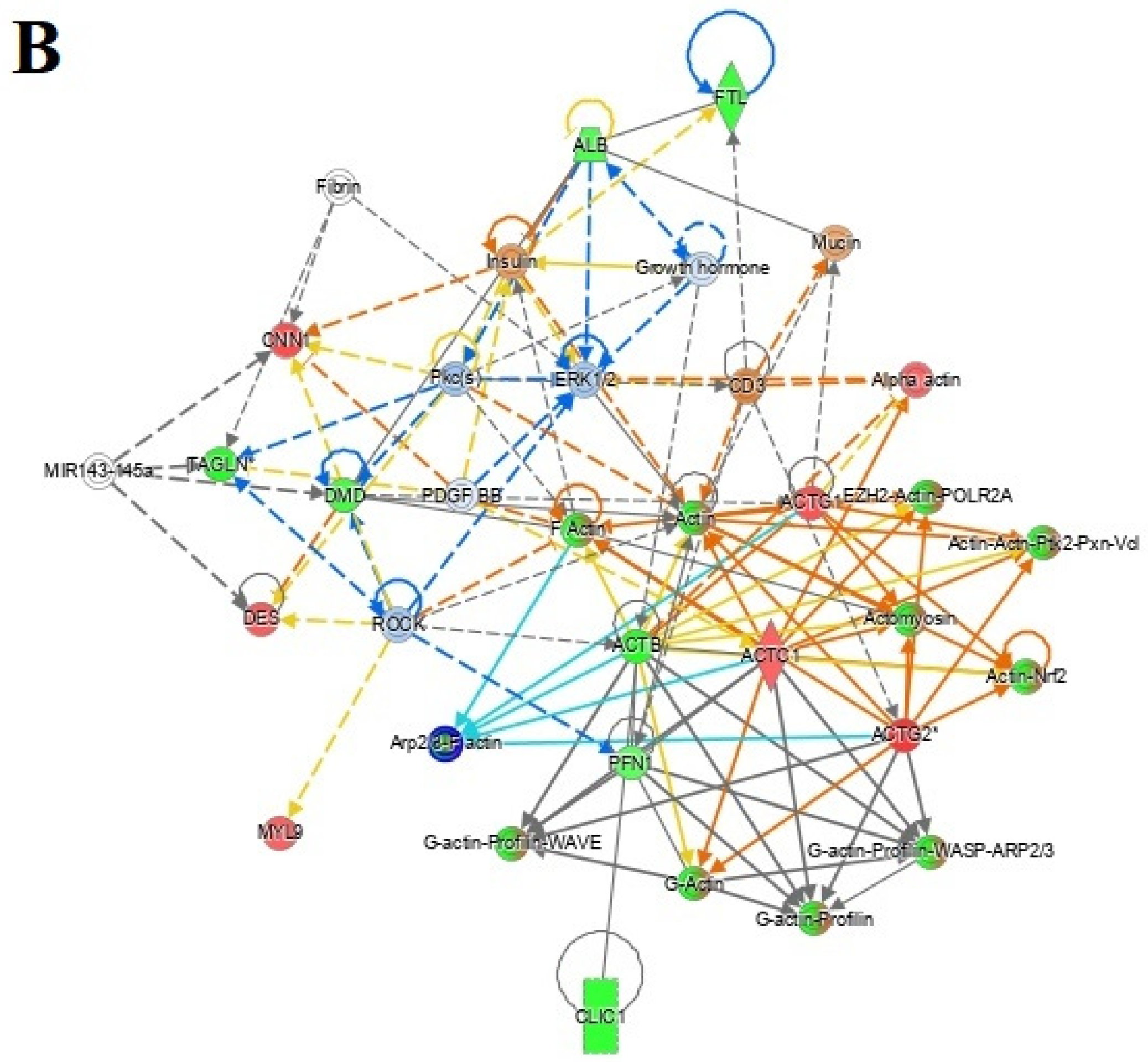
3.7. Protein–Protein Interaction Networks
3.8. Multiple Reaction Monitoring (MRM) Mass Spectrometry
4. Discussion
4.1. Proteins Differentially Expressed in EC vs. Ctrl Patients
4.2. Proteins That Are Differentially Expressed in HY Compared to Ctrl Patients
4.3. Proteins That Are Differentially Expressed in EC Compared to HY Patients
4.4. Interactions of Identified Proteins and Network Connectivity Mapping Using IPA
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felix, A.S.; Brinton, L.A. Cancer progress and priorities: Uterine cancer. Cancer Epidemiol. Prev. Biomark. 2018, 27, 985–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokhman, J.V. Two pathogenetic types of endometrial carcinoma. Gynecol. Oncol. 1983, 15, 10–17. [Google Scholar] [CrossRef]
- Suarez, A.A.; Felix, A.S.; Cohn, D.E. Bokhman Redux: Endometrial cancer “types” in the 21st century. Gynecol. Oncol. 2017, 144, 243–249. [Google Scholar] [CrossRef]
- Salvesen, H.; Carter, S.; Mannelqvist, M.; Dutt, A.; Getz, G.; Stefansson, I.; Raeder, M.; Sos, M.L.; Engelsen, I.; Trovik, J. Integrated genomic profiling of endometrial carcinoma associates aggressive tumors with indicators of PI3 kinase activation. Proc. Natl. Acad. Sci. USA 2009, 106, 4834–4839. [Google Scholar] [CrossRef] [Green Version]
- Shruthi, B.S.; Palani Vinodhkumar, S. Proteomics: A new perspective for cancer. Adv. Biomed. Res. 2016, 5, 67. [Google Scholar] [CrossRef]
- Njoku, K.; Chiasserini, D.; Whetton, A.D.; Crosbie, E.J. Proteomic biomarkers for the detection of endometrial cancer. Cancers 2019, 11, 1572. [Google Scholar] [CrossRef] [Green Version]
- Muinelo-Romay, L.; Casas-Arozamena, C.; Abal, M. Liquid biopsy in endometrial cancer: New opportunities for personalized oncology. Int. J. Mol. Sci. 2018, 19, 2311. [Google Scholar] [CrossRef] [Green Version]
- Abbott, K.L.; Lim, J.M.; Wells, L.; Benigno, B.B.; McDonald, J.F.; Pierce, M. Identification of candidate biomarkers with cancer-specific glycosylation in the tissue and serum of endometrioid ovarian cancer patients by glycoproteomic analysis. Proteomics 2010, 10, 470–481. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-S.; Cao, R.; Jin, H.; Huang, Y.-P.; Zhang, X.-Y.; Cong, Q.; He, Y.-F.; Xu, C.-J. Altered protein expression in serum from endometrial hyperplasia and carcinoma patients. J. Hematol. Oncol. 2011, 4, 15. [Google Scholar] [CrossRef] [Green Version]
- Voisin, S.N.; Krakovska, O.; Matta, A.; DeSouza, L.V.; Romaschin, A.D.; Colgan, T.J.; Siu, K.M. Identification of novel molecular targets for endometrial cancer using a drill-down LC-MS/MS approach with iTRAQ. PLoS ONE 2011, 6, e16352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxwell, G.L.; Hood, B.L.; Day, R.; Chandran, U.; Kirchner, D.; Kolli, V.K.; Bateman, N.W.; Allard, J.; Miller, C.; Sun, M. Proteomic analysis of stage I endometrial cancer tissue: Identification of proteins associated with oxidative processes and inflammation. Gynecol. Oncol. 2011, 121, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Morelli, M.; Scumaci, D.; Di Cello, A.; Venturella, R.; Donato, G.; Faniello, M.C.; Quaresima, B.; Cuda, G.; Zullo, F.; Costanzo, F. DJ-1 in endometrial cancer: A possible biomarker to improve differential diagnosis between subtypes. Int. J. Gynecol. Cancer 2014, 24, 4. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Cantor, D.I.; Huang, C.; Wang, K.; Baker, M.S.; Nice, E.C. Tissue and plasma proteomics for early stage cancer detection. Mol. Omics 2018, 14, 405–423. [Google Scholar] [CrossRef]
- Mujammami, M.; Rafiullah, M.; Alfadda, A.A.; Akkour, K.; Alanazi, I.O.; Masood, A.; Musambil, M.; Alhalal, H.; Arafah, M.; Rahman, A.M.A.; et al. Proteomic Analysis of Endometrial Cancer Tissues from Patients with Type 2 Diabetes Mellitus. Life 2022, 12, 491. [Google Scholar] [CrossRef]
- Ura, B.; Monasta, L.; Arrigoni, G.; Franchin, C.; Radillo, O.; Peterlunger, I.; Ricci, G.; Scrimin, F. A proteomic approach for the identification of biomarkers in endometrial cancer uterine aspirate. Oncotarget 2017, 8, 109536. [Google Scholar] [CrossRef]
- Ceylan, Y.; Akpınar, G.; Doger, E.; Kasap, M.; Guzel, N.; Karaosmanoglu, K.; Kopuk, S.Y.; Yucesoy, I. Proteomic analysis in endometrial cancer and endometrial hyperplasia tissues by 2D-DIGE technique. J. Gynecol. Obstet. Hum. Reprod. 2020, 49, 101652. [Google Scholar] [CrossRef]
- Byrjalsen, I.; Mose Larsen, P.; Fey, S.; Nilas, L.; Larsen, M.; Christiansen, C. Two-dimensional gel analysis of human endometrial proteins: Characterization of proteins with increased expression in hyperplasia and adenocarcinoma. Mol. Hum. Reprod. 1999, 5, 748–756. [Google Scholar] [CrossRef] [Green Version]
- Alfadda, A.A.; Benabdelkamel, H.; Masood, A.; Moustafa, A.; Sallam, R.; Bassas, A.; Duncan, M. Proteomic analysis of mature adipo cytes from obese patients in relation to aging. Exp. Gerontol. 2013, 48, 1196–1203. [Google Scholar] [CrossRef]
- Benabdelkamel, H.; Masood, A.; Almidani, G.M.; Alsadhan, A.A.; Bassas, A.F.; Duncan, M.W.; Alfadda, A.A. Mature adipocyte proteome reveals differentially altered protein abundances between lean, overweight and morbidly obese human subjects. Mol. Cell. Endocrinol. 2015, 401, 142–154. [Google Scholar] [CrossRef]
- Alfadda, A.A.; Benabdelkamel, H.; Masood, A.; Jammah, A.A.; Ekhzaimy, A.A. Differences in the plasma proteome of patients with hypothyroidism before and after thyroid hormone replacement: A proteomic analysis. Int. J. Mol. Sci. 2018, 19, 88. [Google Scholar] [CrossRef] [Green Version]
- Masood, A.; Benabdelkamel, H.; Ekhzaimy, A.A.; Alfadda, A.A. Plasma-based proteomics profiling of patients with hyperthyroidism after antithyroid treatment. Molecules 2020, 25, 2831. [Google Scholar] [CrossRef]
- Malkawi, A.K.; Masood, A.; Shinwari, Z.; Jacob, M.; Benabdelkamel, H.; Matic, G.; Almuhanna, F.; Dasouki, M.; Alaiya, A.A.; Rahman, A.M.A. Proteomic analysis of morphologically changed tissues after prolonged dexamethasone treatment. Int. J. Mol. Sci. 2019, 20, 3122. [Google Scholar] [CrossRef] [Green Version]
- Benabdelkamel, H.; Alamri, H.; Okla, M.; Masood, A.; Abdel Jabar, M.; Alanazi, I.O.; Alfadda, A.A.; Nizami, I.; Dasouki, M.; Abdel Rahman, A.M. Serum-based proteomics profiling in adult patients with cystic fibrosis. Int. J. Mol. Sci. 2020, 21, 7415. [Google Scholar] [CrossRef]
- Galal, M.A.; Jabar, M.A.; Zhra, M.; Rahman, A.M.A.; Aljada, A. Absolute quantification of senescence mediators in cells using multiple reaction monitoring liquid chromatography-Tandem mass spectrometry. Anal. Chim. Acta 2021, 1184, 339009. [Google Scholar] [CrossRef]
- Al-Qahtani, W.S.; Abduljabbar, M.; AlSuhaibani, E.S.; Abdel Rahman, A.; Aljada, A. Quantification of the lamin A/C transcript variants in cancer cell lines by targeted absolute quantitative proteomics and correlation with mRNA expression. Int. J. Mol. Sci. 2019, 20, 1902. [Google Scholar] [CrossRef] [Green Version]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [Green Version]
- Tian, W.; Zhu, Y.; Wang, Y.; Teng, F.; Zhang, H.; Liu, G.; Ma, X.; Sun, D.; Rohan, T.; Xue, F. Visfatin, a potential biomarker and prognostic factor for endometrial cancer. Gynecol. Oncol. 2013, 129, 505–512. [Google Scholar] [CrossRef]
- Yurkovetsky, Z.; Ta’asan, S.; Skates, S.; Rand, A.; Lomakin, A.; Linkov, F.; Marrangoni, A.; Velikokhatnaya, L.; Winans, M.; Gorelik, E. Development of multimarker panel for early detection of endometrial cancer. High diagnostic power of prolactin. Gynecol. Oncol. 2007, 107, 58–65. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Yu, L.; Hu, B.; Chen, L.; Jv, M.; Wang, L.; Zhou, C.; Wei, M.; Zhao, L. Advances in cancer treatment: A new therapeutic target, Annexin A2. J. Cancer 2021, 12, 3587–3596. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, H. Clinical significance of Annexin A2 expression in oral squamous cell carcinoma and its influence on cell proliferation, migration and invasion. Sci. Rep. 2021, 11, 5033. [Google Scholar] [CrossRef]
- Gabel, M.; Delavoie, F.; Royer, C.; Tahouly, T.; Gasman, S.; Bader, M.-F.; Vitale, N.; Chasserot-Golaz, S. Phosphorylation cycling of Annexin A2 Tyr23 is critical for calcium-regulated exocytosis in neuroendocrine cells. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2019, 1866, 1207–1217. [Google Scholar] [CrossRef]
- Murphy, A.G.; Foley, K.; Rucki, A.A.; Xia, T.; Jaffee, E.M.; Huang, C.-Y.; Zheng, L. Stromal Annexin A2 expression is predictive of decreased survival in pancreatic cancer. Oncotarget 2017, 8, 106405–106414. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Cui, Z.; Yang, S.; Wu, C.; Li, W.; Bao, G.; Xu, G.; Sun, Y.; Wang, L.; Zhang, J. The upregulation of annexin A2 after spinal cord injury in rats may have implication for astrocyte proliferation. Neuropeptides 2017, 61, 67–76. [Google Scholar] [CrossRef]
- Rocha, M.R.; Barcellos-de-Souza, P.; Sousa-Squiavinato, A.C.M.; Fernandes, P.V.; de Oliveira, I.M.; Boroni, M.; Morgado-Diaz, J.A. Annexin A2 overexpression associates with colorectal cancer invasiveness and TGF-ß induced epithelial mesenchymal transition via Src/ANXA2/STAT3. Sci. Rep. 2018, 8, 11285. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, H.; Katsuta, E.; Yan, L.; Dasgupta, S.; Takabe, K. High expression of Annexin A2 is associated with DNA repair, metabolic alteration, and worse survival in pancreatic ductal adenocarcinoma. Surgery 2019, 166, 150–156. [Google Scholar] [CrossRef]
- Bystricky, B.; Cierna, Z.; Sieberova, G.; Janega, P.; Karaba, M.; Minarik, G.; Benca, J.; Sedlackova, T.; Jurisova, S.; Gronesova, P.; et al. Relationship Between Circulating Tumor Cells and Annexin A2 in Early Breast Cancer Patients. Anticancer. Res. 2017, 37, 2727–2734. [Google Scholar]
- Alonso-Alconada, L.; Santacana, M.; Garcia-Sanz, P.; Muinelo-Romay, L.; Colas, E.; Mirantes, C.; Monge, M.; Cueva, J.; Oliva, E.; Soslow, R.A.; et al. Annexin-A2 as predictor biomarker of recurrent disease in endometrial cancer. Int. J. Cancer 2015, 136, 1863–1873. [Google Scholar] [CrossRef]
- Gizak, A.; Wiśniewski, J.; Heron, P.; Mamczur, P.; Sygusch, J.; Rakus, D. Targeting a moonlighting function of aldolase induces apoptosis in cancer cells. Cell Death Dis. 2019, 10, 712. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.-C.; Chan, Y.-C.; Chang, W.-M.; Lin, Y.-F.; Yang, C.-J.; Su, C.-Y.; Huang, M.-S.; Wu, A.T.H.; Hsiao, M. Feedback regulation of ALDOA activates the HIF-1α/MMP9 axis to promote lung cancer progression. Cancer Lett. 2017, 403, 28–36. [Google Scholar] [CrossRef]
- Chen, X.; Yang, T.T.; Zhou, Y.; Wang, W.; Qiu, X.C.; Gao, J.; Li, C.X.; Long, H.; Ma, B.A.; Ma, Q.; et al. Proteomic profiling of osteosarcoma cells identifies ALDOA and SULT1A3 as negative survival markers of human osteosarcoma. Mol. Carcinog. 2014, 53, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Hua, Y.; Tian, Y.; Qin, C.; Qian, J.; Bao, M.; Liu, Y.; Wang, S.; Cao, Q.; Ju, X.; et al. High expression of fructose-bisphosphate aldolase A induces progression of renal cell carcinoma. Oncol. Rep. 2018, 39, 2996–3006. [Google Scholar] [PubMed] [Green Version]
- Tang, J.; Qin, Z.; Han, P.; Wang, W.; Yang, C.; Xu, Z.; Li, R.; Liu, B.; Qin, C.; Wang, Z.; et al. High Annexin A5 expression promotes tumor progression and poor prognosis in renal cell carcinoma. Int. J. Oncol. 2017, 50, 1839–1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, G.; Hao, L.Q.; Ding, F.X.; Mei, Q.; Huang, J.J.; Fu, C.G.; Yan, H.L.; Sun, S.H. Expression of annexin a5 is associated with higher tumor stage and poor prognosis in colorectal adenocarcinomas. J. Clin. Gastroenterol. 2009, 43, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Park, N.; Chun, Y.-J. Role of Annexin A5 on Mitochondria-Dependent Apoptosis Induced by Tetramethoxystilbene in Human Breast Cancer Cells. Biomol. Ther. 2014, 22, 519–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, S.M.; Lee, C.H.; Cho, Y.L.; Nam, K.H.; Kim, Y.W.; Kim, C.K.; Han, B.D.; Lee, Y.J.; Chun, H.J.; Ahn, W.S. Two-dimensional gel analysis of protein expression profile in squamous cervical cancer patients. Gynecol. Oncol. 2005, 99, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Cappello, P.; Principe, M.; Bulfamante, S.; Novelli, F. Alpha-Enolase (ENO1), a potential target in novel immunotherapies. Front. Biosci. 2017, 22, 944–959. [Google Scholar]
- Shih, N.-Y.; Lai, H.-L.; Chang, G.-C.; Lin, H.-C.; Wu, Y.-C.; Liu, J.M.; Liu, K.-J.; Tseng, S.-W. Anti-α-enolase Autoantibodies Are Down-regulated in Advanced Cancer Patients. Jpn. J. Clin. Oncol. 2010, 40, 663–669. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Luo, Q.; Long, H.; Hu, Z.; Que, T.; Zhang, X.; Li, Z.; Wang, G.; Yi, L.; Liu, Z.; et al. Alpha-enolase as a potential cancer prognostic marker promotes cell growth, migration, and invasion in glioma. Mol. Cancer 2014, 13, 65. [Google Scholar] [CrossRef] [Green Version]
- Tsai, S.T.; Chien, I.H.; Shen, W.H.; Kuo, Y.Z.; Jin, Y.T.; Wong, T.Y.; Hsiao, J.R.; Wang, H.P.; Shih, N.Y.; Wu, L.W. ENO1, a potential prognostic head and neck cancer marker, promotes transformation partly via chemokine CCL20 induction. Eur. J. Cancer 2010, 46, 1712–1723. [Google Scholar] [CrossRef]
- Yin, H.; Wang, L.; Liu, H.-L. ENO1 Overexpression in Pancreatic Cancer Patients and Its Clinical and Diagnostic Significance. Gastroenterol. Res. Pract. 2018, 2018, 3842198. [Google Scholar] [CrossRef] [Green Version]
- Ji, M.; Wang, Z.; Chen, J.; Gu, L.; Chen, M.; Ding, Y.; Liu, T. Up-regulated ENO1 promotes the bladder cancer cell growth and proliferation via regulating β-catenin. Biosci. Rep. 2019, 39, BSR20190503. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Liu, H.; Liang, Z.; Wang, F.; Zhou, C.; Zheng, X.; Zhang, Y.; Song, Y.; Hu, J.; He, X.; et al. Tumor-intrinsic CD47 signal regulates glycolysis and promotes colorectal cancer cell growth and metastasis. Theranostics 2020, 10, 4056–4072. [Google Scholar] [CrossRef]
- Zhang, J.; Li, H.; Miao, L.; Ding, J. Silencing of ENO1 inhibits the proliferation, migration and invasion of human breast cancer cells. J. Buon. 2020, 25, 696–701. [Google Scholar]
- Liu, J.; Zhang, Y.; Li, Q.; Wang, Y. Transgelins: Cytoskeletal Associated Proteins Implicated in the Metastasis of Colorectal Cancer. Front. Cell Dev. Biol. 2020, 8, 573859. [Google Scholar] [CrossRef]
- Mo, X.; Su, Z.; Yang, B.; Zeng, Z.; Lei, S.; Qiao, H. Identification of key genes involved in the development and progression of early-onset colorectal cancer by co-expression network analysis. Oncol. Lett. 2020, 19, 177–186. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Buckhaults, P.J.; Lee, J.R.; Xiong, H.; Farrell, C.; Podolsky, R.H.; Dynan, W.S. Association of the actin-binding protein transgelin with lymph node metastasis in human colorectal cancer. Neoplasia 2009, 11, 864–873. [Google Scholar] [CrossRef] [Green Version]
- Chunhua, L.; Donglan, L.; Xiuqiong, F.; Lihua, Z.; Qin, F.; Yawei, L.; Xuegang, S. Apigenin up-regulates transgelin and inhibits invasion and migration of colorectal cancer through decreased phosphorylation of AKT. J. Nutr. Biochem. 2013, 24, 1766–1775. [Google Scholar] [CrossRef]
- Yeo, M.; Park, H.J.; Kim, D.-K.; Kim, Y.B.; Cheong, J.Y.; Lee, K.J.; Cho, S.W. Loss of SM22 is a characteristic signature of colon carcinogenesis and its restoration suppresses colon tumorigenicity in vivo and in vitro. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2010, 116, 2581–2589. [Google Scholar] [CrossRef]
- Chen, Z.; He, S.; Zhan, Y.; He, A.; Fang, D.; Gong, Y.; Li, X.; Zhou, L. TGF-β-induced transgelin promotes bladder cancer metastasis by regulating epithelial-mesenchymal transition and invadopodia formation. eBioMedicine 2019, 47, 208–220. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-L.; Chung, T.; Wu, C.-C.; Ng, K.-F.; Yu, J.-S.; Tsai, C.-H.; Chang, Y.-S.; Liang, Y.; Tsui, K.-H.; Chen, Y.-T. Comparative Tissue Proteomics of Microdissected Specimens Reveals Novel Candidate Biomarkers of Bladder Cancer. Mol. Cell Proteom. 2015, 14, 2466–2478. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Z.; Chu, Y.; Xu, B.; Wang, Q.; Jiang, M.; Li, X.; Wang, G.; Yu, P.; Liu, G.; Wang, H.; et al. Increased expression of calponin 2 is a positive prognostic factor in pancreatic ductal adenocarcinoma. Oncotarget 2017, 8, 34. [Google Scholar] [CrossRef] [Green Version]
- Ogura, T.; Kobayashi, H.; Ueoka, Y.; Okugawa, K.; Kato, K.; Hirakawa, T.; Hashimoto, S.; Taniguchi, S.; Wake, N.; Nakano, H. Adenovirus-Mediated Calponin h1 Gene Therapy Directed against Peritoneal Dissemination of Ovarian Cancer: Bifunctional Therapeutic Effects on Peritoneal Cell Layer and Cancer Cells. Clin. Cancer Res. 2006, 12, 5216–5223. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Li, T.-E.; Chen, M.; Pan, J.-J.; Shen, K.-W. miR-106b-5p contributes to the lung metastasis of breast cancer via targeting CNN1 and regulating Rho/ROCK1 pathway. Aging 2020, 12, 1867–1887. [Google Scholar] [CrossRef]
- Takeoka, M.; Ehara, T.; Sagara, J.; Hashimoto, S.; Taniguchi, S. Calponin h1 induced a flattened morphology and suppressed the growth of human fibrosarcoma HT1080 cells. Eur. J. Cancer 2002, 38, 436–442. [Google Scholar] [CrossRef]
- Mitchell, E.; Jindal, S.; Chan, T.; Narasimhan, J.; Sivagnanam, S.; Gray, E.; Chang, Y.H.; Weinmann, S.; Schedin, P. Loss of myoepithelial calponin-1 characterizes high-risk ductal carcinoma in situ cases, which are further stratified by T cell composition. Mol. Carcinog. 2020, 59, 701–712. [Google Scholar] [CrossRef]
- Yamane, T.; Asanoma, K.; Kobayashi, H.; Liu, G.; Yagi, H.; Ohgami, T.; Ichinoe, A.; Sonoda, K.; Wake, N.; Kato, K. Identification of the Critical Site of Calponin 1 for Suppression of Ovarian Cancer Properties. Anticancer. Res. 2015, 35, 5993–5999. [Google Scholar]
- Liu, H.; Li, D.; Zhou, L.; Kan, S.; He, G.; Zhou, K.; Wang, L.; Chen, M.; Shu, W. LMNA functions as an oncogene in hepatocellular carcinoma by regulating the proliferation and migration ability. J. Cell Mol. Med. 2020, 24, 12008–12019. [Google Scholar] [CrossRef]
- Wu, Z.; Wu, L.; Weng, D.; Xu, D.; Geng, J.; Zhao, F. Reduced expression of lamin A/C correlates with poor histological differentiation and prognosis in primary gastric carcinoma. J. Exp. Clin. Cancer Res. 2009, 28, 8. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wu, R.; Cho, K.R.; Thomas, D.G.; Gossner, G.; Liu, J.R.; Giordano, T.J.; Shedden, K.A.; Misek, D.E.; Lubman, D.M. Differential protein mapping of ovarian serous adenocarcinomas: Identification of potential markers for distinct tumor stage. J. Proteome Res. 2009, 8, 1452–1463. [Google Scholar] [CrossRef] [Green Version]
- Willis, N.D.; Cox, T.R.; Rahman-Casañs, S.F.; Smits, K.; Przyborski, S.A.; van den Brandt, P.; van Engeland, M.; Weijenberg, M.; Wilson, R.G.; de Bruïne, A.; et al. Lamin A/C is a risk biomarker in colorectal cancer. PLoS ONE 2008, 3, e2988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, L.; Schäfer, G.; Bu, H.; Zhang, Y.; Zhang, Y.; Klocker, H. Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis 2012, 33, 751–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cicchillitti, L.; Corrado, G.; Carosi, M.; Ewa Dabrowska, M.; Loria, R.; Falcioni, R.; Cutillo, G.; Piaggio, G.; Vizza, E. Prognostic role of NF-YA splicing isoforms and Lamin A status in low grade endometrial cancer. Oncotarget 2016, 8, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belt, E.J.; Fijneman, R.J.; van den Berg, E.G.; Bril, H.; Delis-van Diemen, P.M.; Tijssen, M.; van Essen, H.F.; de Lange-de Klerk, E.S.; Beliën, J.A.; Stockmann, H.B.; et al. Loss of lamin A/C expression in stage II and III colon cancer is associated with disease recurrence. Eur. J. Cancer 2011, 47, 1837–1845. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, A.; Hieda, M.; Yokoyama, Y.; Nishioka, Y.; Yoshidome, K.; Tsujimoto, M.; Matsuura, N. Global loss of a nuclear lamina component, lamin A/C, and LINC complex components SUN1, SUN2, and nesprin-2 in breast cancer. Cancer Med. 2015, 4, 1547–1557. [Google Scholar] [CrossRef]
- Bao, Y.; Zhao, T.L.; Zhang, Z.Q.; Liang, X.L.; Wang, Z.X.; Xiong, Y.; Lu, X.; Wang, L.H. High eukaryotic translation elongation factor 1 alpha 1 expression promotes proliferation and predicts poor prognosis in clear cell renal cell carcinoma. Neoplasma 2020, 67, 78–84. [Google Scholar] [CrossRef]
- Li, X.; Li, J.; Li, F. P21 activated kinase 4 binds translation elongation factor eEF1A1 to promote gastric cancer cell migration and invasion. Oncol. Rep. 2017, 37, 2857–2864. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.-L.; Lu, S.-X.; Liu, L.-L.; Wang, C.-H.; Yang, X.; Zhang, Z.-Y.; Zhang, H.-Z.; Yun, J.-P. eEF1A1 Overexpression Enhances Tumor Progression and Indicates Poor Prognosis in Hepatocellular Carcinoma. Transl. Oncol. 2018, 11, 125–131. [Google Scholar] [CrossRef]
- Jia, L.; Yang, T.; Gu, X.; Zhao, W.; Tang, Q.; Wang, X.; Zhu, J.; Feng, Z. Translation elongation factor eEF1Bα is identified as a novel prognostic marker of gastric cancer. Int. J. Biol. Macromol. 2019, 126, 345–351. [Google Scholar] [CrossRef]
- Pinke, D.E.; Kalloger, S.E.; Francetic, T.; Huntsman, D.G.; Lee, J.M. The prognostic significance of elongation factor eEF1A2 in ovarian cancer. Gynecol. Oncol. 2008, 108, 561–568. [Google Scholar] [CrossRef]
- Galber, C.; Acosta, M.J.; Minervini, G.; Giorgio, V. The role of mitochondrial ATP synthase in cancer. Biol. Chem. 2020, 401, 1199–1214. [Google Scholar] [CrossRef]
- Pan, J.; Sun, L.-C.; Tao, Y.-F.; Zhou, Z.; Du, X.-L.; Peng, L.; Feng, X.; Wang, J.; Li, Y.-P.; Liu, L.; et al. ATP synthase ecto-α-subunit: A novel therapeutic target for breast cancer. J. Transl Med. 2011, 9, 211. [Google Scholar] [CrossRef] [Green Version]
- Chang, F.-W.; Fan, H.-C.; Liu, J.-M.; Fan, T.-P.; Jing, J.; Yang, C.-L.; Hsu, R.-J. Estrogen Enhances the Expression of the Multidrug Transporter Gene ABCG2—Increasing Drug Resistance of Breast Cancer Cells through Estrogen Receptors. Int. J. Mol. Sci. 2017, 18, 163. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.; Chen, L.; Qian, K.; Wang, G.; Lu, M.; Qian, G.; Cao, X.; Jiang, W.; Xiao, Y.; Wang, X. A novel correlation between ATP5A1 gene expression and progression of human clear cell renal cell carcinoma identified by co-expression analysis. Oncol. Rep. 2018, 39, 525–536. [Google Scholar] [CrossRef] [Green Version]
- Feichtinger, R.G.; Schäfer, G.; Seifarth, C.; Mayr, J.A.; Kofler, B.; Klocker, H. Reduced Levels of ATP Synthase Subunit ATP5F1A Correlate with Earlier-Onset Prostate Cancer. Oxid. Med. Cell Longev. 2018, 2018, 1347174. [Google Scholar] [CrossRef] [Green Version]
- Jiang, N.; Dai, Q.; Su, X.; Fu, J.; Feng, X.; Peng, J. Role of PI3K/AKT pathway in cancer: The framework of malignant behavior. Mol. Biol. Rep. 2020, 47, 4587–4629. [Google Scholar] [CrossRef]
- Slomovitz, B.M.; Coleman, R.L. The PI3K/AKT/mTOR pathway as a therapeutic target in endometrial cancer. Clin. Cancer Res. 2012, 18, 5856–5864. [Google Scholar] [CrossRef] [Green Version]
- Lambrechts, A.; Van Troys, M.; Ampe, C. The actin cytoskeleton in normal and pathological cell motility. Int. J. Biochem. Cell Biol. 2004, 36, 1890–1909. [Google Scholar] [CrossRef]
- Jeon, M.; You, D.; Bae, S.Y.; Kim, S.W.; Nam, S.J.; Kim, H.H.; Kim, S.; Lee, J.E. Dimerization of EGFR and HER2 induces breast cancer cell motility through STAT1-dependent ACTA2 induction. Oncotarget 2016, 8, 50570–50581. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.W.; Park, Y.M.; Lee, S.J.; Cho, H.J.; Kim, D.H.; Lee, J.I.; Kang, M.S.; Seol, H.J.; Shim, Y.M.; Nam, D.H.; et al. Alpha-smooth muscle actin (ACTA2) is required for metastatic potential of human lung adenocarcinoma. Clin. Cancer Res. 2013, 19, 5879–5889. [Google Scholar] [CrossRef] [Green Version]
- Sinn, M.; Denkert, C.; Striefler, J.K.; Pelzer, U.; Stieler, J.M.; Bahra, M.; Lohneis, P.; Dörken, B.; Oettle, H.; Riess, H.; et al. α-Smooth muscle actin expression and desmoplastic stromal reaction in pancreatic cancer: Results from the CONKO-001 study. Br. J. Cancer 2014, 111, 1917–1923. [Google Scholar] [CrossRef] [Green Version]





| A: Comparison between EC and Ctrl | |||||||
|---|---|---|---|---|---|---|---|
| Sl No.: | Spot No. a | Accession No. b | Protein Name | MASCOT ID | p-Value b (ANOVA) | Ratio c EC/Ctrl | Exp d |
| 1 | 36 | A6NI72 | Putative neutrophil cytosol factor 1B | NCF1B_HUMAN | 3.74 × 10−5 | −2.9 | DOWN |
| 2 | 23 | P62736 | Actin, aortic smooth muscle | ACTA_HUMAN | 1.27 × 10−4 | −2.7 | DOWN |
| 3 | 937 | P63267 | Actin, gamma-enteric smooth muscle | ACTH_HUMAN | 1.84 × 10−4 | −2.8 | DOWN |
| 4 | 932 | P63267 | Actin, alpha cardiac muscle 1 | ACTC_HUMAN | 4.47 × 10−4 | −2.4 | DOWN |
| 5 | 1218 | O00299 | Chloride intracellular channel protein 1 | CLIC1_HUMAN | 7.66 × 10−4 | 2.7 | UP |
| 6 | 58 | Q9UI15 | Transgelin-3 | TAGL3_HUMAN | 8.94 × 10−4 | −2.5 | DOWN |
| 7 | 847 | P68104 | Elongation factor 1-alpha 1 | EF1A1_HUMAN | 9.87 × 10−4 | 2.2 | UP |
| 8 | 852 | P42331 | Rho GTPase-activating protein 25 | RHG25_HUMAN | 0.001 | 2.7 | UP |
| 9 | 1099 | P07355 | Annexin A2 | ANXA2_HUMAN | 0.001 | 3.0 | UP |
| 10 | 626 | P01024 | Complement C3 | CO3_HUMAN | 0.002 | 2.9 | UP |
| 11 | 290 | Q00536 | Cyclin-dependent kinase 16 | CDK16_HUMAN | 0.002 | −2.6 | DOWN |
| 12 | 244 | Q9UI15 | Transgelin-3 | TAGL3_HUMAN | 0.002 | −2.9 | DOWN |
| 13 | 865 | P68104 | Elongation factor 1-alpha 1 | EF1A1_HUMAN | 0.002 | 2.6 | UP |
| 14 | 1347 | P63261 | Actin, cytoplasmic 2 | ACTG_HUMAN | 0.002 | −2.0 | DOWN |
| 15 | 993 | P04075 | Fructose-bisphosphate aldolase A | ALDOA_HUMAN | 0.002 | 3.1 | UP |
| 16 | 1117 | P08758 | Annexin A5 | ANXA5_HUMAN | 0.002 | −2.7 | DOWN |
| 17 | 729 | P50991 | T-complex protein 1 subunit delta | TCPD_HUMAN | 0.002 | 2.8 | UP |
| 18 | 291 | P34932 | Heat shock 70 kDa protein 4 | HSP74_HUMAN | 0.002 | −2.4 | DOWN |
| 19 | 1593 | Q01995 | Transgelin | TAGL_HUMAN | 0.003 | −2 | DOWN |
| 20 | 1647 | P60709 | Actin, cytoplasmic 1 | ACTB_HUMAN | 0.003 | 2.80 | UP |
| 21 | 962 | P60709 | Actin, cytoplasmic 1 | ACTB_HUMAN | 0.003 | 2.41 | UP |
| 22 | 215 | Q9UI15 | Transgelin-3 | TAGL3_HUMAN | 0.003 | −2.60 | DOWN |
| 23 | 320 | Q96NL8 | Protein C8orf37 | CH037_HUMAN | 0.004 | −2.62 | DOWN |
| 24 | 1697 | P60660 | Myosin light polypeptide 6 | MYL6_HUMAN | 0.004 | −2.55 | DOWN |
| 25 | 279 | Q99615 | DnaJ homolog subfamily C member 7 | DNJC7_HUMAN | 0.004 | −2.41 | DOWN |
| 26 | 947 | P63267 | Actin, gamma-enteric smooth muscle | ACTH_HUMAN | 0.004 | −2.54 | DOWN |
| 27 | 207 | P60709 | Actin, cytoplasmic 1 | ACTB_HUMAN | 0.005 | 2.49 | UP |
| 28 | 1692 | P24844 | Myosin regulatory light polypeptide 9 | MYL9_HUMAN | 0.005 | −2.34 | DOWN |
| 29 | 950 | P13645 | Keratin, type I cytoskeletal 10 | K1C10_HUMAN | 0.006 | 2.56 | UP |
| 30 | 935 | P68032 | Actin, alpha cardiac muscle 1 | ACTC_HUMAN | 0.006 | −2.58 | DOWN |
| 31 | 761 | P14618 | Pyruvate kinase PKM | KPYM_HUMAN | 0.008 | 2.13 | UP |
| 32 | 307 | P63267 | Actin, alpha cardiac muscle 1 | ACTC_HUMAN | 0.009 | −2.32 | DOWN |
| 33 | 785 | P62736 | Actin, aortic smooth muscle | ACTA_HUMAN | 0.009 | −2.09 | DOWN |
| 34 | 723 | P14618 | Pyruvate kinase PKM | KPYM_HUMAN | 0.009 | 2.55 | UP |
| 35 | 209 | O60304 | Zinc finger protein 500 | ZN500_HUMAN | 0.01 | −2.28 | DOWN |
| 36 | 901 | P63267 | Actin, gamma-enteric smooth muscle | ACTH_HUMAN | 0.011 | −2.47 | DOWN |
| 37 | 534 | P17661 | Desmin | DESM_HUMAN | 0.012 | 2.17 | UP |
| 38 | 982 | O00764 | Pyridoxal kinase | PDXK_HUMAN | 0.012 | 2.65 | UP |
| 39 | 843 | P06733 | Alpha-enolase | ENOA_HUMAN | 0.016 | 2.37 | UP |
| 40 | 1106 | P02647 | Apolipoprotein A-I | APOA1_HUMAN | 0.02 | 2.44 | UP |
| 41 | 208 | P12110 | Collagen alpha-2(VI) chain | CO6A2_HUMAN | 0.022 | −2.26 | DOWN |
| 42 | 998 | P04075 | Fructose-bisphosphate aldolase A | ALDOA_HUMAN | 0.022 | 2.22 | UP |
| 43 | 1270 | P60174 | Triosephosphate isomerase | TPIS_HUMAN | 0.024 | 2.16 | UP |
| 44 | 775 | P00352 | Retinal dehydrogenase 1 | AL1A1_HUMAN | 0.027 | 2.18 | UP |
| 45 | 1043 | P14550 | Aldo-keto reductase family 1 member A1 | AK1A1_HUMAN | 0.029 | 2.18 | UP |
| 46 | 977 | P60709 | Actin, cytoplasmic 1 | ACTB_HUMAN | 0.032 | 2.36 | UP |
| 47 | 607 | P29401 | Transketolase | TKT_HUMAN | 0.034 | 2.17 | UP |
| 48 | 1067 | P17661 | Desmin | DESM_HUMAN | 0.038 | 2.10 | UP |
| 49 | 313 | P00966 | Argininosuccinate synthase | ASSY_HUMAN | 0.04 | −2.03 | DOWN |
| 50 | 896 | P17661 | Desmin | DESM_HUMAN | 0.04 | 2.31 | UP |
| 51 | 231 | P62937 | Peptidyl-prolyl cis-trans isomerase A | PPIA_HUMAN | 0.042 | −2.12 | DOWN |
| 52 | 905 | P04406 | Glyceraldehyde-3-phosphate dehydrogenase | G3P_HUMAN | 0.042 | 2.15 | UP |
| 53 | 1098 | P21333 | Filamin-A | FLNA_HUMAN | 0.043 | 2.12 | UP |
| B: Comparison between HY and Ctrl | |||||||
| Sl No.: | Spot No. a | Accession No. b | Protein Name | MASCOT ID | p-Value b (ANOVA) | Ratio c HY/Ctrl | Exp d |
| 1 | 937 | P63267 | Actin, gamma-enteric smooth muscle | ACTH_HUMAN | 1.84 × 10−4 | 2.74 | UP |
| 2 | 932 | P63267 | Actin, alpha cardiac muscle 1 | ACTC_HUMAN | 4.47 × 10−4 | 2.37 | UP |
| 3 | 1218 | O00299 | Chloride intracellular channel protein 1 | CLIC1_HUMAN | 7.66 × 10−4 | −2.48 | DOWN |
| 4 | 1677 | P02792 | Ferritin light chain | FRIL_HUMAN | 0.001 | −2.39 | DOWN |
| 5 | 1645 | Q01995 | Transgelin | TAGL_HUMAN | 0.001 | −2.22 | DOWN |
| 6 | 767 | P06576 | ATP synthase subunit beta, mitochondrial | ATPB_HUMAN | 0.002 | 2.53 | UP |
| 7 | 626 | P01024 | Complement C3 | CO3_HUMAN | 0.002 | −2.62 | DOWN |
| 8 | 1347 | P63261 | Actin, cytoplasmic 2 | ACTG_HUMAN | 0.002 | 2.32 | UP |
| 9 | 729 | P50991 | T-complex protein 1 subunit delta | TCPD_HUMAN | 0.002 | −2.19 | DOWN |
| 10 | 1647 | P60709 | Actin, cytoplasmic 1 | ACTB_HUMAN | 0.003 | −2.71 | DOWN |
| 11 | 1644 | Q01995 | Transgelin | TAGL_HUMAN | 0.003 | −2.34 | DOWN |
| 12 | 1639 | P13645 | Keratin, type I cytoskeletal 10 | K1C10_HUMAN | 0.004 | −2.54 | DOWN |
| 13 | 1692 | P24844 | Myosin regulatory light polypeptide 9 | MYL9_HUMAN | 0.005 | 2.12 | UP |
| 14 | 646 | P17661 | Desmin | DESM_HUMAN | 0.007 | 2.21 | UP |
| 15 | 1136 | P51911 | Calponin-1 | CNN1_HUMAN | 0.008 | 2.37 | UP |
| 16 | 761 | P14618 | Pyruvate kinase PKM | KPYM_HUMAN | 0.008 | −2.06 | DOWN |
| 17 | 723 | P14618 | Pyruvate kinase PKM | KPYM_HUMAN | 0.009 | −2.40 | DOWN |
| 18 | 1636 | Q01995 | Transgelin | TAGL_HUMAN | 0.01 | −2.74 | DOWN |
| 19 | 982 | O00764 | Pyridoxal kinase | PDXK_HUMAN | 0.012 | −2.10 | DOWN |
| 20 | 381 | P02768 | Albumin | ALBU_HUMAN | 0.015 | −2.19 | DOWN |
| 21 | 486 | Q9UBX3 | Mitochondrial dicarboxylate carrier | DIC_HUMAN | 0.018 | −2.17 | DOWN |
| 22 | 695 | P68032 | Actin, alpha cardiac muscle 1 | ACTC_HUMAN | 0.023 | 2.09 | UP |
| 23 | 1732 | P07737 | Profilin-1 | PROF1_HUMAN | 0.024 | −2.01 | DOWN |
| 24 | 369 | P02545 | Prelamin-A/C | LMNA_HUMAN | 0.026 | −2.26 | DOWN |
| 25 | 1594 | Q01995 | Transgelin | TAGL_HUMAN | 0.03 | −2.35 | DOWN |
| 26 | 1599 | P11532 | Dystrophin | DMD_HUMAN | 0.042 | −2.36 | DOWN |
| C: Comparison between EC and HY | |||||||
| Sl No.: | Spot No. a | Accession No. b | Protein Name | MASCOT ID | p-Value b(ANOVA) | Ratio c EC/HY | Exp d |
| 1 | 36 | A6NI72 | Putative neutrophil cytosol factor 1B | NCF1B_HUMAN | 3.74 × 10−5 | −2.06 | DOWN |
| 2 | 23 | P62736 | Actin, aortic smooth muscle | ACTA_HUMAN | 1.27 × 10−4 | −2.11 | DOWN |
| 3 | 54 | P07311 | Acylphosphatase-1 | ACYP1_HUMAN | 3.57 × 10−4 | −2.00 | DOWN |
| 4 | 847 | P68104 | Elongation factor 1-alpha 1 | EF1A1_HUMAN | 9.87 × 10−4 | 3.11 | UP |
| 5 | 852 | P42331 | Rho GTPase-activating protein 25 | RHG25_HUMAN | 0.001 | 2.81 | UP |
| 6 | 767 | P06576 | ATP synthase subunit beta, mitochondrial | ATPB_HUMAN | 0.002 | 2.32 | UP |
| 7 | 1613 | P62736 | Actin, aortic smooth muscle | ACTA_HUMAN | 0.002 | −2.43 | DOWN |
| 8 | 865 | P68104 | Elongation factor 1-alpha 1 | EF1A1_HUMAN | 0.002 | 2.66 | UP |
| 9 | 993 | P04075 | Fructose-bisphosphate aldolase A | ALDOA_HUMAN | 0.002 | 2.17 | UP |
| 10 | 1103 | P09651 | Heterogeneous nuclear ribonucleoprotein A1 | ROA1_HUMAN | 0.003 | 2.19 | UP |
| 11 | 962 | P60709 | Actin, cytoplasmic 1 | ACTB_HUMAN | 0.003 | 2.01 | UP |
| 12 | 1644 | Q01995 | Transgelin | TAGL_HUMAN | 0.003 | −2.66 | DOWN |
| 13 | 638 | O75083 | WD repeat-containing protein 1 | WDR1_HUMAN | 0.004 | 2.9 | UP |
| 14 | 1679 | P17661 | Desmin | DESM_HUMAN | 0.007 | −2.26 | DOWN |
| 15 | 696 | P02768 | Albumin | ALBU_HUMAN | 0.007 | 2.18 | UP |
| 16 | 1636 | Q01995 | Transgelin | TAGL_HUMAN | 0.01 | −2.26 | DOWN |
| 17 | 711 | P25705 | ATP synthase subunit alpha, mitochondria | ATPA_HUMAN | 0.01 | 2.79 | UP |
| 18 | 899 | P04075 | Fructose-bisphosphate aldolase A | ALDOA_HUMAN | 0.011 | 2.41 | UP |
| 19 | 1604 | P62937 | Peptidyl-prolyl cis-trans isomerase A | PPIA_HUMAN | 0.013 | −2.34 | DOWN |
| 20 | 381 | P02768 | Albumin | ALBU_HUMAN | 0.015 | −2.34 | DOWN |
| 21 | 1283 | P13645 | Keratin, type I cytoskeletal 10 | K1C10_HUMAN | 0.021 | 2.29 | UP |
| 22 | 1083 | Q5T5Y3 | Calmodulin-regulated spectrin-associated protein 1 | CAMP1_HUMAN | 0.022 | 2.16 | UP |
| 23 | 998 | P04075 | Fructose-bisphosphate aldolase A | ALDOA_HUMAN | 0.022 | 2.14 | UP |
| 24 | 1732 | P07737 | Profilin-1 | PROF1_HUMAN | 0.024 | −2.4 | DOWN |
| 25 | 1030 | Q9NR45 | Sialic acid synthase | SIAS_HUMAN | 0.026 | 2.00 | UP |
| 26 | 888 | P08670 | Vimentin | VIME_HUMAN | 0.027 | −2.26 | DOWN |
| 27 | 870 | P00558 | Phosphoglycerate kinase 1 | PGK1_HUMAN | 0.028 | 2.20 | UP |
| 28 | 447 | P02768 | Albumin | ALBU_HUMAN | 0.029 | −2.33 | DOWN |
| 29 | 732 | P25705 | ATP synthase subunit alpha, mitochondrial | ATPA_HUMAN | 0.036 | 2.14 | UP |
| 30 | 930 | P06733 | Alpha-enolase | ENOA_HUMAN | 0.037 | 2.03 | UP |
| 31 | 1618 | Q08AG5 | Zinc finger protein 844 | ZN844_HUMAN | 0.039 | −2.15 | DOWN |
| 32 | 1046 | P25705 | ATP synthase subunit alpha, mitochondrial | ATPA_HUMAN | 0.046 | 2.11 | UP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akkour, K.; Alanazi, I.O.; Alfadda, A.A.; Alhalal, H.; Masood, A.; Musambil, M.; Rahman, A.M.A.; Alwehaibi, M.A.; Arafah, M.; Bassi, A.; et al. Tissue-Based Proteomic Profiling in Patients with Hyperplasia and Endometrial Cancer. Cells 2022, 11, 2119. https://doi.org/10.3390/cells11132119
Akkour K, Alanazi IO, Alfadda AA, Alhalal H, Masood A, Musambil M, Rahman AMA, Alwehaibi MA, Arafah M, Bassi A, et al. Tissue-Based Proteomic Profiling in Patients with Hyperplasia and Endometrial Cancer. Cells. 2022; 11(13):2119. https://doi.org/10.3390/cells11132119
Chicago/Turabian StyleAkkour, Khalid, Ibrahim O. Alanazi, Assim A. Alfadda, Hani Alhalal, Afshan Masood, Mohthash Musambil, Anas M. Abdel Rahman, Moudi A. Alwehaibi, Maria Arafah, Ali Bassi, and et al. 2022. "Tissue-Based Proteomic Profiling in Patients with Hyperplasia and Endometrial Cancer" Cells 11, no. 13: 2119. https://doi.org/10.3390/cells11132119






