Human-T-Cell-Selective Fluorescent Probe
Abstract
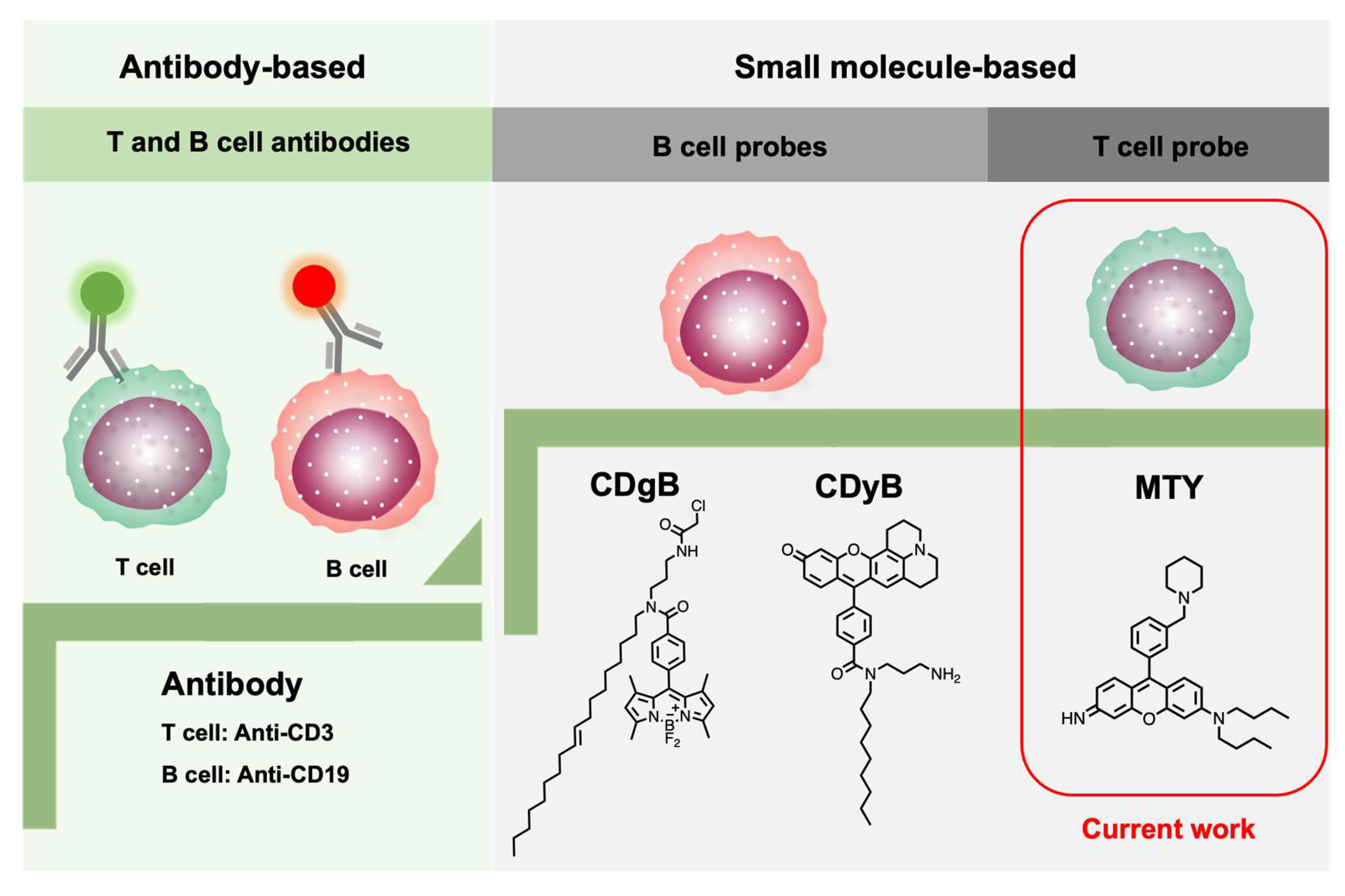
:1. Introduction
2. Materials and Methods
2.1. Compound Information
2.2. Blood
2.3. Preparation of White Blood Cell
2.4. High-Throughput Screening
2.5. Human T and B Cells’ Isolation
2.6. Fluorescence Microscopy and Flow Cytometry Analysis
3. Results and Discussion
3.1. Development of Human-T-Cell-Selective Probe
3.2. Localization of MTY
3.3. Selectivity Mechanism
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garcia, K.C. Dual Arms of Adaptive Immunity: Division of Labor and Collaboration between B and T Cells. Cell 2019, 179, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Reinherz, E.L.; Schlossman, S.F. Regulation of the Immune Response—Inducer and Suppressor T-Lymphocyte Subsets in Human Beings. N. Engl. J. Med. 1980, 303, 370–373. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.M. Overview of the avian immune system. Vet. Immunol. Immunopathol. 1991, 30, 13–17. [Google Scholar] [CrossRef]
- Reinherz, E.L.; Schlossman, S.F. The differentiation and function of human T lymphocytes. Cell 1980, 19, 821–827. [Google Scholar] [CrossRef]
- Pardoll, D.M.; Topalian, S.L. The role of CD4+ T cell responses in antitumor immunity. Curr.Opin. Immunol. 1998, 10, 588–594. [Google Scholar] [CrossRef]
- Bettelli, E.; Carrier, Y.; Gao, W.; Korn, T.; Strom, T.B.; Oukka, M.; Weiner, H.L.; Kuchroo, V.K. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 2006, 441, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Bercovici, N.; Duffour, M.T.; Agrawal, S.; Salcedo, M.; Abastado, J.P. New methods for assessing T-cell responses. Clin. Diagn. Lab Immunol. 2000, 7, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Calloni, R.; Cordero, E.A.A.; Henriques, J.A.P.; Bonatto, D. Reviewing and Updating the Major Molecular Markers for Stem Cells. Stem Cells Dev. 2013, 22, 1455–1476. [Google Scholar] [CrossRef] [PubMed]
- Ansar, W.; Ghosh, S. Monoclonal Antibodies: A Tool in Clinical Research. Indian J. Clin. Med. 2013, 4, IJCM. S11968. [Google Scholar] [CrossRef]
- Kwon, H.-Y.; Kumar Das, R.; Jung, G.T.; Lee, H.-G.; Lee, S.H.; Berry, S.N.; Tan, J.K.S.; Park, S.; Yang, J.-S.; Park, S.; et al. Lipid-Oriented Live-Cell Distinction of B and T Lymphocytes. J. Am. Chem.Soc. 2021, 143, 5836–5844. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Lee, S.H.; Das, R.K.; Kwon, H.-Y.; Kim, H.S.; Chang, Y.-T. A SLC35C2 Transporter-Targeting Fluorescent Probe for the Selective Detection of B Lymphocytes Identified by SLC-CRISPRi and Unbiased Fluorescence Library Screening. Angew.Chem.Int. Ed. 2022, 61, e202202095. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Kim, Y.K.; Vendrell, M.; Chang, Y.-T. Diversity-oriented fluorescence library approach for the discovery of sensors and probes. Mol. BioSyst. 2009, 5, 411–421. [Google Scholar] [CrossRef]
- Liu, X.; Chang, Y.-T. Fluorescent probe strategy for live cell distinction. Chem. Soc. Rev. 2022, 51, 1573–1591. [Google Scholar] [CrossRef]
- He, S.; Li, J.; Lyu, Y.; Huang, J.; Pu, K. Near-Infrared Fluorescent Macromolecular Reporters for Real-Time Imaging and Urinalysis of Cancer Immunotherapy. J. Am. Chem. Soc. 2020, 142, 7075–7082. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, S.; Chen, W.; Liu, Y.; Zhang, X.; Miao, Q.; Pu, K. Activatable Polymeric Nanoprobe for Near-Infrared Fluorescence and Photoacoustic Imaging of T Lymphocytes. Angew. Chem. Int. Ed. 2021, 60, 5921–5927. [Google Scholar] [CrossRef] [PubMed]
- Berrington, J.E.; Barge, D.; Fenton, A.C.; Cant, A.J.; Spickett, G.P. Lymphocyte subsets in term and significantly preterm UK infants in the first year of life analysed by single platform flow cytometry. Clin. Exp. Immunol. 2005, 140, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Arai, S.; Suzuki, M.; Park, S.-J.; Yoo, J.S.; Wang, L.; Kang, N.-Y.; Ha, H.-H.; Chang, Y.-T. Mitochondria-targeted fluorescent thermometer monitors intracellular temperature gradient. Chem. Commun. 2015, 51, 8044–8047. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.-W.; Borsuk, R. Shadakshari, A.; Yu, J.; Dawood, M.; Garcia, R.; Francis, L.; Tily, H.; Bartos, A.; Faraone, S.V.; et al. Mechanistic Target of Rapamycin Activation Triggers IL-4 Production and Necrotic Death of Double-Negative T Cells in Patients with Systemic Lupus Erythematosus. J. Immunol. 2013, 191, 2236. [Google Scholar] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, M.; Chang, Y.-T. Human-T-Cell-Selective Fluorescent Probe. Cells 2022, 11, 2836. https://doi.org/10.3390/cells11182836
Gao M, Chang Y-T. Human-T-Cell-Selective Fluorescent Probe. Cells. 2022; 11(18):2836. https://doi.org/10.3390/cells11182836
Chicago/Turabian StyleGao, Min, and Young-Tae Chang. 2022. "Human-T-Cell-Selective Fluorescent Probe" Cells 11, no. 18: 2836. https://doi.org/10.3390/cells11182836






