Liquid Biopsy Analysis as a Tool for TKI-Based Treatment in Non-Small Cell Lung Cancer
Abstract
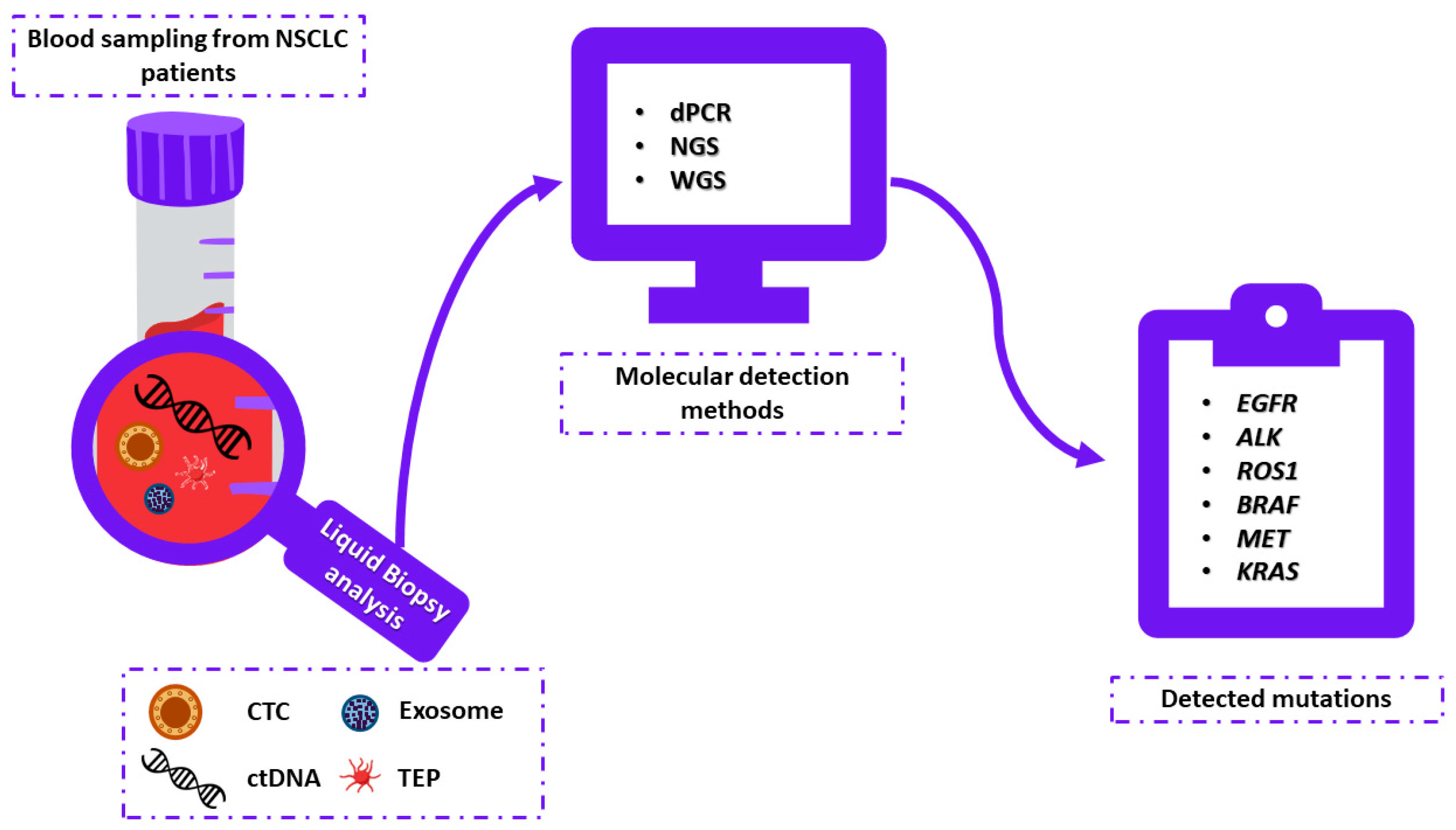
:1. Introduction
2. Therapeutic Approaches in NSCLC
2.1. EGFR-Targeted TKI Treatment
2.1.1. Major EGFR Mutations
2.1.2. Rare EGFR Mutations
2.2. ALK-Targeted TKI Treatment
2.3. ROS1-Targeted TKI Treatment
2.4. BRAF-Targeted TKI Treatment
2.5. MET-Targeted TKI Treatment
2.6. KRAS-Targeted TKI Treatment
2.7. Testing for Molecular Biomarkers
3. Liquid Biopsy
3.1. Liquid Biopsy Elements
3.1.1. CTCs
3.1.2. cfDNA
3.1.3. Exosomes
3.1.4. TEP
3.2. Cancer Screening
3.3. Prognostic Value
3.4. Therapy Selection and Monitoring of NSCLC through Liquid Biopsy
4. Liquid Biopsy Testing in NSCLC
4.1. EGFR-Mutant NSCLC
4.1.1. cfDNA Testing
4.1.2. CTCs
4.1.3. Exosomes
4.2. ALK-Rearranged NSCLC
4.2.1. ctDNA
4.2.2. CTCs
4.2.3. Exosomes and TEPs
4.3. ROS1-Positive NSCLC
4.3.1. ctDNA
4.3.2. CTCs
4.4. BRAF Mutated NSCLC
ctDNA and CTCs
4.5. MET exon14-Positive NSCLC
ctDNA
4.6. KRAS G12C Mutated NSCLC
ctDNA
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non–Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.B.; Alsubait, S. Non Small Cell Lung Cancer. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Osmani, L.; Askin, F.; Gabrielson, E.; Li, Q.K. Current WHO Guidelines and the Critical Role of Immunohistochemical Markers in the Subclassification of Non-Small Cell Lung Carcinoma (NSCLC). Moving from Targeted Therapy to Immunotherapy. Semin. Cancer Biol. 2018, 52, 103–109. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Burke, A.P.; Marx, A.; Nicholson, A.G. Introduction to The 2015 World Health Organization Classification of Tumors of the Lung, Pleura, Thymus, and Heart. J. Thorac. Oncol. 2015, 10, 1240–1242. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, A.G.; Tsao, M.S.; Beasley, M.B.; Borczuk, A.C.; Brambilla, E.; Cooper, W.A.; Dacic, S.; Jain, D.; Kerr, K.M.; Lantuejoul, S.; et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances since 2015. J. Thorac. Oncol. 2021, 17, 362–387. [Google Scholar] [CrossRef] [PubMed]
- Pignon, J.-P.; Tribodet, H.; Scagliotti, G.V.; Douillard, J.-Y.; Shepherd, F.A.; Stephens, R.J.; Dunant, A.; Torri, V.; Rosell, R.; Seymour, L.; et al. Lung Adjuvant Cisplatin Evaluation: A Pooled Analysis by the LACE Collaborative Group. J. Clin. Oncol. 2008, 26, 3552–3559. [Google Scholar] [CrossRef]
- Alexander, M.; Kim, S.Y.; Cheng, H. Update 2020: Management of Non-Small Cell Lung Cancer. Lung 2020, 198, 897–907. [Google Scholar] [CrossRef]
- Yoneda, K.; Imanishi, N.; Ichiki, Y.; Tanaka, F. Treatment of Non-Small Cell Lung Cancer with EGFR-Mutations. J. UOEH 2019, 41, 153–163. [Google Scholar] [CrossRef]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.S.; Chang, J.Y.; Chirieac, L.R.; D’Amico, T.A.; et al. Non-Small Cell Lung Cancer, Version 3.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2022, 20, 497–530. [Google Scholar] [CrossRef]
- Mazieres, J.; Drilon, A.; Lusque, A.; Mhanna, L.; Cortot, A.B.; Mezquita, L.; Thai, A.A.; Mascaux, C.; Couraud, S.; Veillon, R.; et al. Immune Checkpoint Inhibitors for Patients with Advanced Lung Cancer and Oncogenic Driver Alterations: Results from the IMMUNOTARGET Registry. Ann. Oncol. 2019, 30, 1321–1328. [Google Scholar] [CrossRef]
- Lisberg, A.; Cummings, A.; Goldman, J.W.; Bornazyan, K.; Reese, N.; Wang, T.; Coluzzi, P.; Ledezma, B.; Mendenhall, M.; Hunt, J.; et al. A Phase II Study of Pembrolizumab in EGFR-Mutant, PD-L1+, Tyrosine Kinase Inhibitor Naïve Patients with Advanced NSCLC. J. Thorac. Oncol. 2018, 13, 1138–1145. [Google Scholar] [CrossRef] [PubMed]
- Gainor, J.F.; Shaw, A.T.; Sequist, L.V.; Fu, X.; Azzoli, C.G.; Piotrowska, Z.; Huynh, T.G.; Zhao, L.; Fulton, L.; Schultz, K.R.; et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin. Cancer Res. 2016, 22, 4585–4593. [Google Scholar] [CrossRef] [PubMed]
- Naylor, E.C.; Desani, J.K.; Chung, P.K. Targeted Therapy and Immunotherapy for Lung Cancer. Surg. Oncol. Clin. N. Am. 2016, 25, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Płużański, A.; Piórek, A. Side Effects of Tyrosine Kinase Inhibitors—Management Guidelines. Oncol. Clin. Pract. 2016, 12, 113–118. [Google Scholar] [CrossRef]
- Parikh, A.R. Lung Cancer Genomics. AMA 2019, 48, 78. [Google Scholar] [CrossRef]
- Midha, A.; Dearden, S.; McCormack, R. EGFR Mutation Incidence in Non-Small-Cell Lung Cancer of Adenocarcinoma Histology: A Systematic Review and Global Map by Ethnicity (MutMapII). Am. J. Cancer Res. 2015, 5, 2892–2911. [Google Scholar]
- Lynch, T.J.; Bell, D.W.; Sordella, R.; Gurubhagavatula, S.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Haserlat, S.M.; Supko, J.G.; Haluska, F.G.; et al. Activating Mutations in the Epidermal Growth Factor Receptor Underlying Responsiveness of Non-Small-Cell Lung Cancer to Gefitinib. N. Engl. J. Med. 2004, 350, 2129–2139. [Google Scholar] [CrossRef]
- Castellanos, E.; Feld, E.; Horn, L. Driven by Mutations: The Predictive Value of Mutation Subtype in EGFR -Mutated Non–Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 612–623. [Google Scholar] [CrossRef]
- Li, W.-Q.; Cui, J.-W. Non-Small Cell Lung Cancer Patients with Ex19del or Exon 21 L858R Mutation: Distinct Mechanisms, Different Efficacies to Treatments. J. Cancer Res. Clin. Oncol. 2020, 146, 2329–2338. [Google Scholar] [CrossRef]
- Ramalingam, S.S.; Vansteenkiste, J.; Planchard, D.; Cho, B.C.; Gray, J.E.; Ohe, Y.; Zhou, C.; Reungwetwattana, T.; Cheng, Y.; Chewaskulyong, B.; et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N. Engl. J. Med. 2020, 382, 41–50. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Tsuboi, M.; He, J.; John, T.; Grohe, C.; Majem, M.; Goldman, J.W.; Laktionov, K.; Kim, S.-W.; Kato, T.; et al. Osimertinib in Resected EGFR-Mutated Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Beau-Faller, M.; Prim, N.; Ruppert, A.-M.; Nanni-Metéllus, I.; Lacave, R.; Lacroix, L.; Escande, F.; Lizard, S.; Pretet, J.-L.; Rouquette, I.; et al. Rare EGFR Exon 18 and Exon 20 Mutations in Non-Small-Cell Lung Cancer on 10 117 Patients: A Multicentre Observational Study by the French ERMETIC-IFCT Network. Ann. Oncol. 2014, 25, 126–131. [Google Scholar] [CrossRef]
- Cheng, C.; Wang, R.; Li, Y.; Pan, Y.; Zhang, Y.; Li, H.; Zheng, D.; Zheng, S.; Shen, X.; Sun, Y.; et al. EGFR Exon 18 Mutations in East Asian Patients with Lung Adenocarcinomas: A Comprehensive Investigation of Prevalence, Clinicopathologic Characteristics and Prognosis. Sci. Rep. 2015, 5, 13959. [Google Scholar] [CrossRef] [PubMed]
- Kancha, R.K.; von Bubnoff, N.; Peschel, C.; Duyster, J. Functional Analysis of Epidermal Growth Factor Receptor (EGFR) Mutations and Potential Implications for EGFR Targeted Therapy. Clin. Cancer Res. 2009, 15, 460–467. [Google Scholar] [CrossRef]
- Baek, J.H.; Sun, J.-M.; Min, Y.J.; Cho, E.K.; Cho, B.C.; Kim, J.-H.; Ahn, M.-J.; Park, K. Efficacy of EGFR Tyrosine Kinase Inhibitors in Patients with EGFR-Mutated Non-Small Cell Lung Cancer except Both Exon 19 Deletion and Exon 21 L858R: A Retrospective Analysis in Korea. Lung Cancer 2015, 87, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Lohinai, Z.; Hoda, M.A.; Fabian, K.; Ostoros, G.; Raso, E.; Barbai, T.; Timar, J.; Kovalszky, I.; Cserepes, M.; Rozsas, A.; et al. Distinct Epidemiology and Clinical Consequence of Classic Versus Rare EGFR Mutations in Lung Adenocarcinoma. J. Thorac. Oncol. 2015, 10, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.-H.; Sequist, L.V.; Geater, S.L.; Tsai, C.-M.; Mok, T.S.K.; Schuler, M.; Yamamoto, N.; Yu, C.-J.; Ou, S.-H.I.; Zhou, C.; et al. Clinical Activity of Afatinib in Patients with Advanced Non-Small-Cell Lung Cancer Harbouring Uncommon EGFR Mutations: A Combined Post-Hoc Analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015, 16, 830–838. [Google Scholar] [CrossRef]
- Asahina, H.; Yamazaki, K.; Kinoshita, I.; Yokouchi, H.; Dosaka-Akita, H.; Nishimura, M. Non-Responsiveness to Gefitinib in a Patient with Lung Adenocarcinoma Having Rare EGFR Mutations S768I and V769L. Lung Cancer 2006, 54, 419–422. [Google Scholar] [CrossRef]
- Siegelin, M.D.; Borczuk, A.C. Epidermal Growth Factor Receptor Mutations in Lung Adenocarcinoma. Lab Investig. 2014, 94, 129–137. [Google Scholar] [CrossRef]
- Camidge, D.R.; Dziadziuszko, R.; Peters, S.; Mok, T.; Noe, J.; Nowicka, M.; Gadgeel, S.M.; Cheema, P.; Pavlakis, N.; de Marinis, F.; et al. Updated Efficacy and Safety Data and Impact of the EML4-ALK Fusion Variant on the Efficacy of Alectinib in Untreated ALK-Positive Advanced Non-Small Cell Lung Cancer in the Global Phase III ALEX Study. J. Thorac. Oncol. 2019, 14, 1233–1243. [Google Scholar] [CrossRef]
- Shaw, A.T.; Solomon, B. Targeting Anaplastic Lymphoma Kinase in Lung Cancer. Clin. Cancer Res. 2011, 17, 2081–2086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, X.; Shao, Y.; Qin, H.-F.; Tai, Y.-H.; Gao, H.-J. ALK-Rearrangement in Non-Small-Cell Lung Cancer (NSCLC). Thorac. Cancer 2018, 9, 423–430. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, A.; Sobhani, N.; Chapman, R.; Bagby, S.; Bortoletti, C.; Traversini, M.; Ferrari, K.; Voltolini, L.; Darlow, J.; Roviello, G. Focus on ROS1-Positive Non-Small Cell Lung Cancer (NSCLC): Crizotinib, Resistance Mechanisms and the Newer Generation of Targeted Therapies. Cancers 2020, 12, E3293. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.T.; Ou, S.-H.I.; Bang, Y.-J.; Camidge, D.R.; Solomon, B.J.; Salgia, R.; Riely, G.J.; Varella-Garcia, M.; Shapiro, G.I.; Costa, D.B.; et al. Crizotinib in ROS1-Rearranged Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2014, 371, 1963–1971. [Google Scholar] [CrossRef]
- Marchetti, A.; Felicioni, L.; Malatesta, S.; Grazia Sciarrotta, M.; Guetti, L.; Chella, A.; Viola, P.; Pullara, C.; Mucilli, F.; Buttitta, F. Clinical Features and Outcome of Patients with Non-Small-Cell Lung Cancer Harboring BRAF Mutations. J. Clin. Oncol. 2011, 29, 3574–3579. [Google Scholar] [CrossRef]
- O’Leary, C.G.; Andelkovic, V.; Ladwa, R.; Pavlakis, N.; Zhou, C.; Hirsch, F.; Richard, D.; O’Byrne, K. Targeting BRAF Mutations in Non-Small Cell Lung Cancer. Transl. Lung Cancer Res. 2019, 8, 1119–1124. [Google Scholar] [CrossRef]
- Han, J.; Liu, Y.; Yang, S.; Wu, X.; Li, H.; Wang, Q. MEK Inhibitors for the Treatment of Non-Small Cell Lung Cancer. J. Hematol. Oncol. 2021, 14, 1. [Google Scholar] [CrossRef]
- Fujino, T.; Suda, K.; Mitsudomi, T. Lung Cancer with MET Exon 14 Skipping Mutation: Genetic Feature, Current Treatments, and Future Challenges. Lung Cancer (Auckl.) 2021, 12, 35–50. [Google Scholar] [CrossRef]
- Paik, P.K.; Felip, E.; Veillon, R.; Sakai, H.; Cortot, A.B.; Garassino, M.C.; Mazieres, J.; Viteri, S.; Senellart, H.; Van Meerbeeck, J.; et al. Tepotinib in Non-Small-Cell Lung Cancer with MET Exon 14 Skipping Mutations. N. Engl. J. Med. 2020, 383, 931–943. [Google Scholar] [CrossRef]
- Mo, H.-N.; Liu, P. Targeting MET in Cancer Therapy. Chronic Dis. Transl. Med. 2017, 3, 148–153. [Google Scholar] [CrossRef]
- Cortot, A.B.; Kherrouche, Z.; Descarpentries, C.; Wislez, M.; Baldacci, S.; Furlan, A.; Tulasne, D. Exon 14 Deleted MET Receptor as a New Biomarker and Target in Cancers. J. Natl. Cancer Inst. 2017, 109, djw262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frampton, G.M.; Ali, S.M.; Rosenzweig, M.; Chmielecki, J.; Lu, X.; Bauer, T.M.; Akimov, M.; Bufill, J.A.; Lee, C.; Jentz, D.; et al. Activation of MET via Diverse Exon 14 Splicing Alterations Occurs in Multiple Tumor Types and Confers Clinical Sensitivity to MET Inhibitors. Cancer Discov. 2015, 5, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Awad, M.M.; Oxnard, G.R.; Jackman, D.M.; Savukoski, D.O.; Hall, D.; Shivdasani, P.; Heng, J.C.; Dahlberg, S.E.; Jänne, P.A.; Verma, S.; et al. MET Exon 14 Mutations in Non-Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J. Clin. Oncol. 2016, 34, 721–730. [Google Scholar] [CrossRef]
- Paik, P.K.; Drilon, A.; Fan, P.-D.; Yu, H.; Rekhtman, N.; Ginsberg, M.S.; Borsu, L.; Schultz, N.; Berger, M.F.; Rudin, C.M.; et al. Response to MET Inhibitors in Patients with Stage IV Lung Adenocarcinomas Harboring MET Mutations Causing Exon 14 Skipping. Cancer Discov. 2015, 5, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.H.; Yeung, S.F.; Chan, A.W.H.; Chung, L.Y.; Chau, S.L.; Lung, R.W.M.; Tong, C.Y.; Chow, C.; Tin, E.K.Y.; Yu, Y.H.; et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non-Small Cell Lung Carcinoma with Poor Prognosis. Clin. Cancer Res. 2016, 22, 3048–3056. [Google Scholar] [CrossRef] [PubMed]
- Pruis, M.A.; Geurts-Giele, W.R.R.; von der, T.J.H.; Meijssen, I.C.; Dinjens, W.N.M.; Aerts, J.G.J.V.; Dingemans, A.M.C.; Lolkema, M.P.; Paats, M.S.; Dubbink, H.J. Highly Accurate DNA-Based Detection and Treatment Results of MET Exon 14 Skipping Mutations in Lung Cancer. Lung Cancer 2020, 140, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Bladt, F.; Friese-Hamim, M.; Ihling, C.; Wilm, C.; Blaukat, A. The C-Met Inhibitor MSC2156119J Effectively Inhibits Tumor Growth in Liver Cancer Models. Cancers 2014, 6, 1736–1752. [Google Scholar] [CrossRef]
- Bladt, F.; Faden, B.; Friese-Hamim, M.; Knuehl, C.; Wilm, C.; Fittschen, C.; Grädler, U.; Meyring, M.; Dorsch, D.; Jaehrling, F.; et al. EMD 1214063 and EMD 1204831 Constitute a New Class of Potent and Highly Selective C-Met Inhibitors. Clin. Cancer Res. 2013, 19, 2941–2951. [Google Scholar] [CrossRef]
- Friese-Hamim, M.; Bladt, F.; Locatelli, G.; Stammberger, U.; Blaukat, A. The Selective C-Met Inhibitor Tepotinib Can Overcome Epidermal Growth Factor Receptor Inhibitor Resistance Mediated by Aberrant c-Met Activation in NSCLC Models. Am. J. Cancer Res 2017, 7, 962–972. [Google Scholar]
- Mikami, K.; Medová, M.; Nisa, L.; Francica, P.; Glück, A.A.; Tschan, M.P.; Blaukat, A.; Bladt, F.; Aebersold, D.M.; Zimmer, Y. Impact of P53 Status on Radiosensitization of Tumor Cells by MET Inhibition-Associated Checkpoint Abrogation. Mol. Cancer Res. 2015, 13, 1544–1553. [Google Scholar] [CrossRef]
- Falchook, G.S.; Kurzrock, R.; Amin, H.M.; Xiong, W.; Fu, S.; Piha-Paul, S.A.; Janku, F.; Eskandari, G.; Catenacci, D.V.; Klevesath, M.; et al. First-in-Man Phase I Trial of the Selective MET Inhibitor Tepotinib in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2020, 26, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Clark, J.W.; Weiss, J.; Ou, S.-H.I.; Camidge, D.R.; Solomon, B.J.; Otterson, G.A.; Villaruz, L.C.; Riely, G.J.; Heist, R.S.; et al. Antitumor Activity of Crizotinib in Lung Cancers Harboring a MET Exon 14 Alteration. Nat. Med. 2020, 26, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Pakkala, S.; Ramalingam, S.S. Personalized Therapy for Lung Cancer: Striking a Moving Target. JCI Insight 2018, 3, 120858. [Google Scholar] [CrossRef] [PubMed]
- Janes, M.R.; Zhang, J.; Li, L.-S.; Hansen, R.; Peters, U.; Guo, X.; Chen, Y.; Babbar, A.; Firdaus, S.J.; Darjania, L.; et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018, 172, 578–589.e17. [Google Scholar] [CrossRef]
- Heo, S.J.; Jung, I.; Lee, C.-K.; Kim, J.H.; Lim, S.M.; Moon, Y.W.; Shim, H.S.; Jeong, J.; Kim, J.-H.; Kim, H.R.; et al. A Randomized Phase II Trial of ERCC1 and RRM1 MRNA Expression-Based Chemotherapy versus Docetaxel/Carboplatin in Advanced Non-Small Cell Lung Cancer. Cancer Chemother. Pharm. 2016, 77, 539–548. [Google Scholar] [CrossRef]
- Riely, G.J.; Marks, J.; Pao, W. KRAS Mutations in Non-Small Cell Lung Cancer. Proc. Am. Thorac. Soc. 2009, 6, 201–205. [Google Scholar] [CrossRef]
- Mao, C.; Qiu, L.-X.; Liao, R.-Y.; Du, F.-B.; Ding, H.; Yang, W.-C.; Li, J.; Chen, Q. KRAS Mutations and Resistance to EGFR-TKIs Treatment in Patients with Non-Small Cell Lung Cancer: A Meta-Analysis of 22 Studies. Lung Cancer 2010, 69, 272–278. [Google Scholar] [CrossRef]
- Del Re, M.; Rofi, E.; Restante, G.; Crucitta, S.; Arrigoni, E.; Fogli, S.; Di Maio, M.; Petrini, I.; Danesi, R. Implications of KRAS Mutations in Acquired Resistance to Treatment in NSCLC. Oncotarget 2018, 9, 6630–6643. [Google Scholar] [CrossRef]
- Shigematsu, H.; Gazdar, A.F. Somatic Mutations of Epidermal Growth Factor Receptor Signaling Pathway in Lung Cancers. Int. J. Cancer 2006, 118, 257–262. [Google Scholar] [CrossRef]
- Schmid, S.; Gautschi, O.; Rothschild, S.; Mark, M.; Froesch, P.; Klingbiel, D.; Reichegger, H.; Jochum, W.; Diebold, J.; Früh, M. Clinical Outcome of ALK-Positive Non-Small Cell Lung Cancer (NSCLC) Patients with De Novo EGFR or KRAS Co-Mutations Receiving Tyrosine Kinase Inhibitors (TKIs). J. Thorac. Oncol. 2017, 12, 681–688. [Google Scholar] [CrossRef]
- Sahnane, N.; Frattini, M.; Bernasconi, B.; Zappa, F.; Schiavone, G.; Wannesson, L.; Antonelli, P.; Balzarini, P.; Sessa, F.; Mazzucchelli, L.; et al. EGFR and KRAS Mutations in ALK-Positive Lung Adenocarcinomas: Biological and Clinical Effect. Clin. Lung Cancer 2016, 17, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Guibert, N.; Barlesi, F.; Descourt, R.; Léna, H.; Besse, B.; Beau-Faller, M.; Mosser, J.; Pichon, E.; Merlio, J.-P.; Ouafik, L.; et al. Characteristics and Outcomes of Patients with Lung Cancer Harboring Multiple Molecular Alterations: Results from the IFCT Study Biomarkers France. J. Thorac. Oncol. 2017, 12, 963–973. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, X.; Zhao, C.; Li, J.; Su, C.; Chen, X.; Ren, S.; Li, X.; Zhou, C. Clinical Features and Therapeutic Options in Non-Small Cell Lung Cancer Patients with Concomitant Mutations of EGFR, ALK, ROS1, KRAS or BRAF. Cancer Med. 2019, 8, 2858–2866. [Google Scholar] [CrossRef] [PubMed]
- Canon, J.; Rex, K.; Saiki, A.Y.; Mohr, C.; Cooke, K.; Bagal, D.; Gaida, K.; Holt, T.; Knutson, C.G.; Koppada, N.; et al. The Clinical KRAS(G12C) Inhibitor AMG 510 Drives Anti-Tumour Immunity. Nature 2019, 575, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Hallin, J.; Engstrom, L.D.; Hargis, L.; Calinisan, A.; Aranda, R.; Briere, D.M.; Sudhakar, N.; Bowcut, V.; Baer, B.R.; Ballard, J.A.; et al. The KRASG12C Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov. 2020, 10, 54–71. [Google Scholar] [CrossRef]
- Sankar, K.; Gadgeel, S.M.; Qin, A. Molecular Therapeutic Targets in Non-Small Cell Lung Cancer. Expert Rev. Anticancer 2020, 20, 647–661. [Google Scholar] [CrossRef]
- Jennings, L.J.; Arcila, M.E.; Corless, C.; Kamel-Reid, S.; Lubin, I.M.; Pfeifer, J.; Temple-Smolkin, R.L.; Voelkerding, K.V.; Nikiforova, M.N. Guidelines for Validation of Next-Generation Sequencing-Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2017, 19, 341–365. [Google Scholar] [CrossRef]
- Aziz, N.; Zhao, Q.; Bry, L.; Driscoll, D.K.; Funke, B.; Gibson, J.S.; Grody, W.W.; Hegde, M.R.; Hoeltge, G.A.; Leonard, D.G.B.; et al. College of American Pathologists’ Laboratory Standards for next-Generation Sequencing Clinical Tests. Arch. Pathol. Lab. Med. 2015, 139, 481–493. [Google Scholar] [CrossRef]
- Luthra, R.; Chen, H.; Roy-Chowdhuri, S.; Singh, R.R. Next-Generation Sequencing in Clinical Molecular Diagnostics of Cancer: Advantages and Challenges. Cancers 2015, 7, 2023–2036. [Google Scholar] [CrossRef]
- Drilon, A.; Wang, L.; Arcila, M.E.; Balasubramanian, S.; Greenbowe, J.R.; Ross, J.S.; Stephens, P.; Lipson, D.; Miller, V.A.; Kris, M.G.; et al. Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin. Cancer Res. 2015, 21, 3631–3639. [Google Scholar] [CrossRef]
- Robson, M.E.; Bradbury, A.R.; Arun, B.; Domchek, S.M.; Ford, J.M.; Hampel, H.L.; Lipkin, S.M.; Syngal, S.; Wollins, D.S.; Lindor, N.M. American Society of Clinical Oncology Policy Statement Update: Genetic and Genomic Testing for Cancer Susceptibility. J. Clin. Oncol. 2015, 33, 3660–3667. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.P.; Vose, J.M.; Hayes, D.F. Genetic Cancer Susceptibility Testing: Increased Technology, Increased Complexity. J. Clin. Oncol. 2015, 33, 3533–3534. [Google Scholar] [CrossRef] [PubMed]
- Cardarella, S.; Ortiz, T.M.; Joshi, V.A.; Butaney, M.; Jackman, D.M.; Kwiatkowski, D.J.; Yeap, B.Y.; Jänne, P.A.; Lindeman, N.I.; Johnson, B.E. The Introduction of Systematic Genomic Testing for Patients with Non-Small-Cell Lung Cancer. J. Thorac. Oncol. 2012, 7, 1767–1774. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Kung, H.-J.; Mack, P.C.; Gandara, D.R. Genotyping and Genomic Profiling of Non-Small-Cell Lung Cancer: Implications for Current and Future Therapies. J. Clin. Oncol. 2013, 31, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Planchard, D. Identification of Driver Mutations in Lung Cancer: First Step in Personalized Cancer. Target Oncol. 2013, 8, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Soria, J.-C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and Localization of Surgically Resectable Cancers with a Multi-Analyte Blood Test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Klein, E.A.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical Validation of a Targeted Methylation-Based Multi-Cancer Early Detection Test Using an Independent Validation Set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef]
- Zhang, T.; Armstrong, A.J. Clinical Utility of Circulating Tumor Cells in Advanced Prostate Cancer. Curr. Oncol. Rep. 2016, 18, 3. [Google Scholar] [CrossRef]
- Cabel, L.; Proudhon, C.; Gortais, H.; Loirat, D.; Coussy, F.; Pierga, J.-Y.; Bidard, F.-C. Circulating Tumor Cells: Clinical Validity and Utility. Int. J. Clin. Oncol. 2017, 22, 421–430. [Google Scholar] [CrossRef]
- Vasseur, A.; Kiavue, N.; Bidard, F.; Pierga, J.; Cabel, L. Clinical Utility of Circulating Tumor Cells: An Update. Mol. Oncol. 2021, 15, 1647–1666. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, F.A.; Papadimitrakopoulou, V.; Mok, T.; Wu, Y.-L.; Han, J.-Y.; Ahn, M.-J.; Ramalingam, S.S.; John, T.; Sebastian, M.; Theelen, W.; et al. Early Clearance of Plasma EGFR Mutations as a Predictor of Response to Osimertinib in the AURA3 Trial. J. Clin. Oncol. 2018, 36, 9027. [Google Scholar] [CrossRef]
- Castellanos-Rizaldos, E.; Zhang, X.; Tadigotla, V.R.; Grimm, D.G.; Karlovich, C.; Raez, L.E.; Skog, J.K. Exosome-Based Detection of Activating and Resistance EGFR Mutations from Plasma of Non-Small Cell Lung Cancer Patients. Oncotarget 2019, 10, 2911–2920. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; van den Broek, D.; Denis, M.G.; Hofman, P.; Hubank, M.; Mouliere, F.; Paz-Ares, L.; Schuuring, E.; Sültmann, H.; Vainer, G.; et al. Recommendations for a Practical Implementation of Circulating Tumor DNA Mutation Testing in Metastatic Non-Small-Cell Lung Cancer. ESMO Open 2022, 7, 100399. [Google Scholar] [CrossRef]
- Bonanno, L.; Dal Maso, A.; Pavan, A.; Zulato, E.; Calvetti, L.; Pasello, G.; Guarneri, V.; Conte, P.; Indraccolo, S. Liquid Biopsy and Non-Small Cell Lung Cancer: Are We Looking at the Tip of the Iceberg? Br. J. Cancer 2022, 127, 383–393. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced NSCLC: A Consensus Statement From the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2021, 16, 1647–1662. [Google Scholar] [CrossRef]
- Xu, M.J.; Dorsey, J.F.; Amaravadi, R.; Karakousis, G.; Simone, C.B.; Xu, X.; Xu, W.; Carpenter, E.L.; Schuchter, L.; Kao, G.D. Circulating Tumor Cells, DNA, and MRNA: Potential for Clinical Utility in Patients With Melanoma. Oncologist 2016, 21, 84–94. [Google Scholar] [CrossRef]
- Kang, B.J.; Ra, S.W.; Lee, K.; Lim, S.; Son, S.H.; Ahn, J.J.; Kim, B.C. Circulating Tumor Cell Number Is Associated with Primary Tumor Volume in Patients with Lung Adenocarcinoma. Tuberc. Respir. Dis. 2020, 83, 61–70. [Google Scholar] [CrossRef]
- Kim, H.; Heo, C.M.; Oh, J.; Chung, H.H.; Lee, E.M.; Park, J.; Lee, S.-H.; Lee, K.H.; Lee, K.T.; Lee, J.K.; et al. Clinical Significance of Circulating Tumor Cells after Chemotherapy in Unresectable Pancreatic Ductal Adenocarcinoma. Transl. Oncol. 2022, 16, 101321. [Google Scholar] [CrossRef]
- Lindsay, C.R.; Faugeroux, V.; Michiels, S.; Pailler, E.; Facchinetti, F.; Ou, D.; Bluthgen, M.V.; Pannet, C.; Ngo-Camus, M.; Bescher, G.; et al. A Prospective Examination of Circulating Tumor Cell Profiles in Non-Small-Cell Lung Cancer Molecular Subgroups. Ann. Oncol. 2017, 28, 1523–1531. [Google Scholar] [CrossRef]
- Buszka, K.; Kamińska, P.; Budna-Tukan, J. Płynna Biopsja Jako Nieinwazyjna Metoda Oceny Wybranych Mutacji Genowych w Niedrobnokomórkowym Raku Płuca. PBK 2020, 47, 413–424. [Google Scholar]
- Hochmair, M.J.; Buder, A.; Schwab, S.; Burghuber, O.C.; Prosch, H.; Hilbe, W.; Cseh, A.; Fritz, R.; Filipits, M. Liquid-Biopsy-Based Identification of EGFR T790M Mutation-Mediated Resistance to Afatinib Treatment in Patients with Advanced EGFR Mutation-Positive NSCLC, and Subsequent Response to Osimertinib. Target. Oncol. 2019, 14, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Zhang, Y.; Xu, J.; He, J.; Gao, W. Progress and Application of Circulating Tumor Cells in Non-Small Cell Lung Cancer. Mol. Oncolytics 2021, 22, 72–84. [Google Scholar] [CrossRef]
- Zhang, Q.; Nong, J.; Wang, J.; Yan, Z.; Yi, L.; Gao, X.; Liu, Z.; Zhang, H.; Zhang, S. Isolation of Circulating Tumor Cells and Detection of EGFR Mutations in Patients with Non-Small-Cell Lung Cancer. Oncol Lett 2019, 17, 3799–3807. [Google Scholar] [CrossRef] [PubMed]
- Hong, B.; Zu, Y. Detecting Circulating Tumor Cells: Current Challenges and New Trends. Theranostics 2013, 3, 377–394. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabières, C. Cell Lines from Circulating Tumor Cells. Oncoscience 2015, 2, 815–816. [Google Scholar] [CrossRef]
- Martignano, F. Cell-Free DNA: An Overview of Sample Types and Isolation Procedures. Methods Mol. Biol. 2019, 1909, 13–27. [Google Scholar] [CrossRef]
- Yao, W.; Mei, C.; Nan, X.; Hui, L. Evaluation and Comparison of in Vitro Degradation Kinetics of DNA in Serum, Urine and Saliva: A Qualitative Study. Gene 2016, 590, 142–148. [Google Scholar] [CrossRef]
- Kamat, A.A.; Bischoff, F.Z.; Dang, D.; Baldwin, M.F.; Han, L.Y.; Lin, Y.G.; Merritt, W.M.; Landen, C.N.; Lu, C.; Gershenson, D.M.; et al. Circulating Cell-Free DNA: A Novel Biomarker for Response to Therapy in Ovarian Carcinoma. Cancer Biol. 2006, 5, 1369–1374. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Parkinson, C.A.; Gale, D.; Piskorz, A.M.; Biggs, H.; Hodgkin, C.; Addley, H.; Freeman, S.; Moyle, P.; Sala, E.; Sayal, K.; et al. Exploratory Analysis of TP53 Mutations in Circulating Tumour DNA as Biomarkers of Treatment Response for Patients with Relapsed High-Grade Serous Ovarian Carcinoma: A Retrospective Study. PLoS Med. 2016, 13, e1002198. [Google Scholar] [CrossRef] [PubMed]
- Bu, H.; He, D.; He, X.; Wang, K. Exosomes: Isolation, Analysis, and Applications in Cancer Detection and Therapy. Chembiochem 2019, 20, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Calverley, D.C.; Phang, T.L.; Choudhury, Q.G.; Gao, B.; Oton, A.B.; Weyant, M.J.; Geraci, M.W. Significant Downregulation of Platelet Gene Expression in Metastatic Lung Cancer. Clin. Transl. Sci. 2010, 3, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Haemmerle, M.; Taylor, M.L.; Gutschner, T.; Pradeep, S.; Cho, M.S.; Sheng, J.; Lyons, Y.M.; Nagaraja, A.S.; Dood, R.L.; Wen, Y.; et al. Platelets Reduce Anoikis and Promote Metastasis by Activating YAP1 Signaling. Nat. Commun. 2017, 8, 310. [Google Scholar] [CrossRef]
- Labelle, M.; Begum, S.; Hynes, R.O. Direct Signaling between Platelets and Cancer Cells Induces an Epithelial-Mesenchymal-like Transition and Promotes Metastasis. Cancer Cell 2011, 20, 576–590. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, H. Next-Generation Sequencing in Liquid Biopsy: Cancer Screening and Early Detection. Hum. Genom. 2019, 13, 34. [Google Scholar] [CrossRef]
- Lozar, T.; Gersak, K.; Cemazar, M.; Kuhar, C.G.; Jesenko, T. The Biology and Clinical Potential of Circulating Tumor Cells. Radiol. Oncol. 2019, 53, 131–147. [Google Scholar] [CrossRef]
- Ilie, M.; Hofman, V.; Long-Mira, E.; Selva, E.; Vignaud, J.-M.; Padovani, B.; Mouroux, J.; Marquette, C.-H.; Hofman, P. “Sentinel” Circulating Tumor Cells Allow Early Diagnosis of Lung Cancer in Patients with Chronic Obstructive Pulmonary Disease. PLoS ONE 2014, 9, e111597. [Google Scholar] [CrossRef]
- Fiorelli, A.; Accardo, M.; Carelli, E.; Angioletti, D.; Santini, M.; Di Domenico, M. Circulating Tumor Cells in Diagnosing Lung Cancer: Clinical and Morphologic Analysis. Ann. Thorac. Surg. 2015, 99, 1899–1905. [Google Scholar] [CrossRef]
- Marquette, C.-H.; Boutros, J.; Benzaquen, J.; Ferreira, M.; Pastre, J.; Pison, C.; Padovani, B.; Bettayeb, F.; Fallet, V.; Guibert, N.; et al. Circulating Tumour Cells as a Potential Biomarker for Lung Cancer Screening: A Prospective Cohort Study. Lancet Respir. Med. 2020, 8, 709–716. [Google Scholar] [CrossRef]
- Jia, S.; Xie, L.; Li, L.; Qian, Y.; Wang, J.; Wang, S.; Zhang, W.; Qian, B. Values of Liquid Biopsy in Early Detection of Cancer: Results from Meta-Analysis. Expert Rev. Mol. Diagn. 2021, 21, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Rapanotti, M.C.; Campione, E.; Spallone, G.; Orlandi, A.; Bernardini, S.; Bianchi, L. Minimal Residual Disease in Melanoma: Circulating Melanoma Cells and Predictive Role of MCAM/MUC18/MelCAM/CD146. Cell Death Discov. 2017, 3, 17005. [Google Scholar] [CrossRef] [PubMed]
- Wislez, M.; Domblides, C.; Greillier, L.; Mazières, J.; Monnet, I.; Kiakouama-Maleka, L.; Quantin, X.; Spano, J.P.; Ricordel, C.; Fraisse, P.; et al. Circulating Tumor DNA in Advanced Non-Small-Cell Lung Cancer Patients with HIV Is Associated with Shorter Overall Survival: Results from a Phase II Trial (IFCT-1001 CHIVA). Lung Cancer 2021, 157, 124–130. [Google Scholar] [CrossRef]
- Dziadziuszko, R.; Peters, S.; Mok, T.; Camidge, D.R.; Gadgeel, S.M.; Ou, S.-H.I.; Konopa, K.; Noé, J.; Nowicka, M.; Bordogna, W.; et al. Circulating Cell-Free DNA as a Prognostic Biomarker in Patients with Advanced ALK+ Non-Small Cell Lung Cancer in the Global Phase III ALEX Trial. Clin. Cancer Res. 2022, 28, 1800–1808. [Google Scholar] [CrossRef] [PubMed]
- de Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.W.M.; Pienta, K.J.; Raghavan, D. Circulating Tumor Cells Predict Survival Benefit from Treatment in Metastatic Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef]
- Liu, W.; Yin, B.; Wang, X.; Yu, P.; Duan, X.; Liu, C.; Wang, B.; Tao, Z. Circulating Tumor Cells in Prostate Cancer: Precision Diagnosis and Therapy. Oncol. Lett. 2017, 14, 1223–1232. [Google Scholar] [CrossRef] [PubMed]
- Broncy, L.; Paterlini-Bréchot, P. Clinical Impact of Circulating Tumor Cells in Patients with Localized Prostate Cancer. Cells 2019, 8, 676. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.; Chu, C.; Gui, X.; Li, J.; Chen, Q. The Prognostic Value of Circulating Cell-Free DNA in Breast Cancer: A Meta-Analysis. Medicine 2018, 97, e0197. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Cheng, L.; Zhang, J.; Chen, L.; Wang, D.; Guo, X.; Ma, X. Predictive Value of Circulating Cell-Free DNA in the Survival of Breast Cancer Patients: A Systemic Review and Meta-Analysis. Medicine 2018, 97, e11417. [Google Scholar] [CrossRef]
- Cavallone, L.; Aguilar-Mahecha, A.; Lafleur, J.; Brousse, S.; Aldamry, M.; Roseshter, T.; Lan, C.; Alirezaie, N.; Bareke, E.; Majewski, J.; et al. Prognostic and Predictive Value of Circulating Tumor DNA during Neoadjuvant Chemotherapy for Triple Negative Breast Cancer. Sci. Rep. 2020, 10, 14704. [Google Scholar] [CrossRef]
- Papakonstantinou, A.; Gonzalez, N.S.; Pimentel, I.; Suñol, A.; Zamora, E.; Ortiz, C.; Espinosa-Bravo, M.; Peg, V.; Vivancos, A.; Saura, C.; et al. Prognostic Value of CtDNA Detection in Patients with Early Breast Cancer Undergoing Neoadjuvant Therapy: A Systematic Review and Meta-Analysis. Cancer Treat. Rev. 2022, 104, 102362. [Google Scholar] [CrossRef] [PubMed]
- Hamfjord, J.; Guren, T.K.; Dajani, O.; Johansen, J.S.; Glimelius, B.; Sorbye, H.; Pfeiffer, P.; Lingjærde, O.C.; Tveit, K.M.; Kure, E.H.; et al. Total Circulating Cell-Free DNA as a Prognostic Biomarker in Metastatic Colorectal Cancer before First-Line Oxaliplatin-Based Chemotherapy. Ann. Oncol. 2019, 30, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.P.; Pugh, S.A.; Graham, J.; Primrose, J.N.; Barriuso, J. Circulating Tumour DNA as a Biomarker in Resectable and Irresectable Stage IV Colorectal Cancer; a Systematic Review and Meta-Analysis. Eur. J. Cancer 2021, 144, 368–381. [Google Scholar] [CrossRef]
- Taieb, J.; Taly, V.; Henriques, J.; Bourreau, C.; Mineur, L.; Bennouna, J.; Desrame, J.; Louvet, C.; Lepere, C.; Mabro, M.; et al. Prognostic Value and Relation with Adjuvant Treatment Duration of CtDNA in Stage III Colon Cancer: A Post Hoc Analysis of the PRODIGE-GERCOR IDEA-France Trial. Clin. Cancer Res. 2021, 27, 5638–5646. [Google Scholar] [CrossRef] [PubMed]
- Hiltermann, T.J.N.; Pore, M.M.; van den Berg, A.; Timens, W.; Boezen, H.M.; Liesker, J.J.W.; Schouwink, J.H.; Wijnands, W.J.A.; Kerner, G.S.M.A.; Kruyt, F.a.E.; et al. Circulating Tumor Cells in Small-Cell Lung Cancer: A Predictive and Prognostic Factor. Ann. Oncol. 2012, 23, 2937–2942. [Google Scholar] [CrossRef]
- Hou, J.-M.; Krebs, M.G.; Lancashire, L.; Sloane, R.; Backen, A.; Swain, R.K.; Priest, L.J.C.; Greystoke, A.; Zhou, C.; Morris, K.; et al. Clinical Significance and Molecular Characteristics of Circulating Tumor Cells and Circulating Tumor Microemboli in Patients with Small-Cell Lung Cancer. J. Clin. Oncol. 2012, 30, 525–532. [Google Scholar] [CrossRef]
- Jansson, S.; Bendahl, P.-O.; Larsson, A.-M.; Aaltonen, K.E.; Rydén, L. Prognostic Impact of Circulating Tumor Cell Apoptosis and Clusters in Serial Blood Samples from Patients with Metastatic Breast Cancer in a Prospective Observational Cohort. BMC Cancer 2016, 16, 433. [Google Scholar] [CrossRef]
- Lim, M.; Park, S.; Jeong, H.-O.; Park, S.H.; Kumar, S.; Jang, A.; Lee, S.; Kim, D.U.; Cho, Y.-K. Circulating Tumor Cell Clusters Are Cloaked with Platelets and Correlate with Poor Prognosis in Unresectable Pancreatic Cancer. Cancers 2021, 13, 5272. [Google Scholar] [CrossRef]
- Zhang, L.; Riethdorf, S.; Wu, G.; Wang, T.; Yang, K.; Peng, G.; Liu, J.; Pantel, K. Meta-Analysis of the Prognostic Value of Circulating Tumor Cells in Breast Cancer. Clin. Cancer Res. 2012, 18, 5701–5710. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.-L.; Christie, M.; et al. Circulating Tumor DNA Analysis Detects Minimal Residual Disease and Predicts Recurrence in Patients with Stage II Colon Cancer. Sci. Transl. Med. 2016, 8, 346ra92. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid Biopsy and Minimal Residual Disease-Latest Advances and Implications for Cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Kilgour, E.; Rothwell, D.G.; Brady, G.; Dive, C. Liquid Biopsy-Based Biomarkers of Treatment Response and Resistance. Cancer Cell 2020, 37, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.M.M.; et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [Green Version]
- Tol, J.; Koopman, M.; Miller, M.C.; Tibbe, A.; Cats, A.; Creemers, G.J.M.; Vos, A.H.; Nagtegaal, I.D.; Terstappen, L.W.M.M.; Punt, C.J.A. Circulating Tumour Cells Early Predict Progression-Free and Overall Survival in Advanced Colorectal Cancer Patients Treated with Chemotherapy and Targeted Agents. Ann. Oncol. 2010, 21, 1006–1012. [Google Scholar] [CrossRef]
- Goldberg, S.B.; Narayan, A.; Kole, A.J.; Decker, R.H.; Teysir, J.; Carriero, N.J.; Lee, A.; Nemati, R.; Nath, S.K.; Mane, S.M.; et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin. Cancer Res. 2018, 24, 1872–1880. [Google Scholar] [CrossRef] [PubMed]
- Alama, A.; Coco, S.; Genova, C.; Rossi, G.; Fontana, V.; Tagliamento, M.; Giovanna Dal Bello, M.; Rosa, A.; Boccardo, S.; Rijavec, E.; et al. Prognostic Relevance of Circulating Tumor Cells and Circulating Cell-Free DNA Association in Metastatic Non-Small Cell Lung Cancer Treated with Nivolumab. J. Clin. Med. 2019, 8, E1011. [Google Scholar] [CrossRef] [PubMed]
- Brozos-Vázquez, E.M.; Díaz-Peña, R.; García-González, J.; León-Mateos, L.; Mondelo-Macía, P.; Peña-Chilet, M.; López-López, R. Immunotherapy in Nonsmall-Cell Lung Cancer: Current Status and Future Prospects for Liquid Biopsy. Cancer Immunol. Immunother. 2021, 70, 1177–1188. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Thorac. Oncol. 2018, 13, 323–358. [Google Scholar] [CrossRef]
- Rolfo, C.; Cardona, A.F.; Cristofanilli, M.; Paz-Ares, L.; Diaz Mochon, J.J.; Duran, I.; Raez, L.E.; Russo, A.; Lorente, J.A.; Malapelle, U.; et al. Challenges and Opportunities of CfDNA Analysis Implementation in Clinical Practice: Perspective of the International Society of Liquid Biopsy (ISLB). Crit. Rev. Oncol./Hematol. 2020, 151, 102978. [Google Scholar] [CrossRef]
- Gray, J.; Thompson, J.C.; Carpenter, E.L.; Elkhouly, E.A.C. Plasma Cell-Free DNA Genotyping : From an Emerging Concept to a Standard-of-Care Tool in Metastatic Non-Small Cell Lung Cancer. Oncologist 2021, 26, e1812–e1821. [Google Scholar] [CrossRef] [PubMed]
- Douillard, J.; Ostoros, G.; Cobo, M.; Ciuleanu, T.; Mccormack, R.; Webster, A.; Milenkova, T. First-Line Gefitinib in Caucasian EGFR Mutation-Positive NSCLC Patients: A Phase-IV, Open-Label, Single-Arm Study. Br. J. Cancer 2013, 110, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Douillard, J.Y.; Ostoros, G.; Cobo, M.; Ciuleanu, T.; Cole, R.; McWalter, G.; Walker, J.; Dearden, S.; Webster, A.; Milenkova, T.; et al. Treatment in EGFR Mutated Caucasian NSCLC: Circulating-Free Tumor DNA as a Surrogate for Determination of EGFR Status. J. Thorac. Oncol. 2014, 9, 1345–1353. [Google Scholar] [CrossRef] [Green Version]
- Morales-espinosa, D.; Viteri, S.; Drozdowskyj, A.; Jordana-ariza, N.; Ramirez-serrano, J.L.; Molina-vila, M.A.; Rosell, R. Association OfEGFRL858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol. 2015, 1, 149. [Google Scholar] [CrossRef]
- Zhou, C.; Wu, G.; Liu, X.; Zhong, Z.; Lu, S.; Cheng, Y.; Han, B.; Chen, L.; Huang, C.; Qin, S.; et al. First-Line Erlotinib versus Gemcitabine/Cisplatin in Patients with Advanced EGFR Mutation-Positive Non-Small-Cell Lung Cancer: Analyses from the Phase III. Ann. Oncol. 2015, 26, 54–59. [Google Scholar] [CrossRef] [PubMed]
- cobas EGFR Mutation Test v2. Available online: https://www.Fda.Gov/Drugs/Resources-Information-Approved-Drugs/Cobas-Egfr-Mutation-Test-V2 (accessed on 6 February 2016).
- Yu, H.A.; Arcila, M.E.; Rekhtman, N.; Sima, C.S.; Maureen, F.; Pao, W.; Kris, M.G.; Miller, V.A.; Ladanyi, M.; Riely, G.J. Analysis of Tumor Specimens at the Time of Acquired Resistance to EGFR-TKI Therapy in 155 Patients with EGFR-Mutant Lung Cancers. Clin. Cancer Res. 2013, 19, 2240–2247. [Google Scholar] [CrossRef] [PubMed]
- Goss, G.; Tsai, C.M.; Shepherd, F.A.; Bazhenova, L.; Lee, J.S.; Chang, G.C.; Crino, L.; Satouchi, M.; Chu, Q.; Hida, T.; et al. Osimertinib for Pretreated EGFR Thr790Met-Positive Advanced Non-Small-Cell Lung Cancer (AURA2): A Multicentre, Open-Label, Single-Arm, Phase 2 Study. Lancet Oncol. 2016, 17, 1643–1652. [Google Scholar] [CrossRef]
- Jenkins, S.; Yang, J.C.H.; Ramalingam, S.S.; Yu, K.; Patel, S.; Weston, S.; Hodge, R.; Cantarini, M.; Jänne, P.A.; Mitsudomi, T.; et al. Plasma CtDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non–Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 1061–1070. [Google Scholar] [CrossRef]
- Papadimitrakopoulou, V.A.; Han, J.Y.; Ahn, M.J.; Ramalingam, S.S.; Delmonte, A.; Hsia, T.C.; Laskin, J.; Kim, S.W.; He, Y.; Tsai, C.M.; et al. Epidermal Growth Factor Receptor Mutation Analysis in Tissue and Plasma from the AURA3 Trial: Osimertinib versus Platinum-Pemetrexed for T790M Mutation-Positive Advanced Non–Small Cell Lung Cancer. Cancer 2020, 126, 373–380. [Google Scholar] [CrossRef]
- Karlovich, C.; Goldman, J.W.; Sun, J.M.; Mann, E.; Sequist, L.V.; Konopa, K.; Wen, W.; Angenendt, P.; Horn, L.; Spigel, D.; et al. Assessment of EGFR Mutation Status in Matched Plasma and Tumor Tissue of NSCLC Patients from a Phase I Study of Rociletinib (CO-1686). Clin. Cancer Res. 2016, 22, 2386–2395. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Thress, K.S.; Alden, R.S.; Lawrance, R.; Paweletz, C.P.; Cantarini, M.; Yang, J.C.H.; Barrett, J.C.; Jänne, P.A. Association between Plasma Genotyping and Outcomes of Treatment with Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2016, 34, 3375–3382. [Google Scholar] [CrossRef]
- Gray, J.E.; Okamoto, I.; Sriuranpong, V.; Vansteenkiste, J.; Imamura, F.; Lee, J.S.; Pang, Y.K.; Cobo, M.; Kasahara, K.; Cheng, Y.; et al. Tissue and Plasma EGFR Mutation Analysis in the FLAURA Trial: Osimertinib versus Comparator EGFR Tyrosine Kinase Inhibitor as First-Line Treatment in Patients with EGFR-Mutated Advanced Non–Small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 6644–6652. [Google Scholar] [CrossRef] [PubMed]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance Mechanisms to Osimertinib in EGFR-Mutated Non-Small Cell Lung Cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Thress, K.S.; Paweletz, C.P.; Felip, E.; Cho, B.C.; Stetson, D.; Dougherty, B.; Lai, Z.; Markovets, A.; Vivancos, A.; Kuang, Y.; et al. Acquired EGFR C797S Mediates Resistance to AZD9291 in Advanced Non-Small Cell Lung Cancer Harboring EGFR T790M. Nat. Med. 2015, 21, 560–562. [Google Scholar] [CrossRef] [PubMed]
- Oxnard, G.R.; Hu, Y.; Mileham, K.F.; Husain, H.; Costa, D.B.; Tracy, P.; Feeney, N.; Sholl, L.M.; Dahlberg, S.E.; Redig, A.J.; et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients with EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol. 2018, 4, 1527–1534. [Google Scholar] [CrossRef]
- Papadimitrakopoulou, V.A.; Wu, Y.-L.; Han, J.-Y.; Ahn, M.-J.; Ramalingam, S.S.; John, T.; Okamoto, I.; Yang, J.C.-H.; Bulusu, K.C.; Laus, G.; et al. Analysis of Resistance Mechanisms to Osimertinib in Patients with EGFR T790M Advanced NSCLC from the AURA3 Study. Ann. Oncol. 2018, 29, viii741. [Google Scholar] [CrossRef]
- Ramalingam, S.S.; Cheng, Y.; Zhou, C.; Ohe, Y.; Imamura, F.; Cho, B.C.; Lin, M.; Majem, M.; Shah, R.; Rukazenkov, Y.; et al. Mechanisms of Acquired Resistance to First-Line Osimertinib: Preliminary Data from the Phase III FLAURA Study. Ann. Oncol. 2018, 29, viii740. [Google Scholar] [CrossRef]
- Buder, A.; Hochmair, M.J.; Filipits, M. The Allele Frequency of EGFR Mutations Predicts Survival in Advanced EGFR T790M-Positive Non-Small Cell Lung Cancer Patients Treated with Osimertinib. Target. Oncol. 2021, 16, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.; Wu, Y.; Lee, J.S.; Yu, C.; Sriuranpong, V.; Sandoval-tan, J.; Ladrera, G.; Thongprasert, S.; Srimuninnimit, V. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-Line Intercalated Erlotinib and Chemotherapy. Clin. Cancer Res. 2015, 21, 3196–3203. [Google Scholar] [CrossRef]
- Zhou, C.; Imamura, F.; Cheng, Y.; Okamoto, I.; Cho, B.C.; Lin, M.C.; Majem, M.; Gautschi, O.; Gray, J.E.; Boyer, M.J.; et al. Early Clearance of Plasma EGFR Mutations as a Predictor of Response to Osimertinib and Comparator EGFR-TKIs in the FLAURA Trial. J. Clin. Oncol. 2019, 37, 9020. [Google Scholar] [CrossRef]
- Zheng, D.; Ye, X.; Zhang, M.Z.; Sun, Y.; Wang, J.Y.; Ni, J.; Zhang, H.P.; Zhang, L. Plasma EGFR T790M CtDNA Status Is Associated with Clinical Outcome in Advanced NSCLC Patients with Acquired EGFR-TKI Resistance. Sci. Rep. 2016, 12, 20913. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, B.S.; Wu, L.; Wei, W.; Tsai, J.; Weber, B. Monitoring of Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor-Sensitizing and Resistance Mutations in the Plasma DNA of Patients With Advanced Non-Small Cell Lung Cancer During Treatment With Erlotinib. Cancer 2014, 120, 3896–3901. [Google Scholar] [CrossRef] [PubMed]
- Chabon, J.J.; Simmons, A.D.; Lovejoy, A.F.; Esfahani, M.S.; Newman, A.M.; Haringsma, H.J.; Kurtz, D.M.; Stehr, H.; Scherer, F.; Karlovich, C.A.; et al. Circulating Tumour DNA Profiling Reveals Heterogeneity of EGFR Inhibitor Resistance Mechanisms in Lung Cancer Patients. Nat. Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef]
- Cheng, M.L.; Lau, C.J.; Milan, M.S.D.; Supplee, J.G.; Riess, J.W.; Bradbury, P.A.; Jänne, P.A.; Oxnard, G.R.; Paweletz, C.P. Plasma CtDNA Response Is an Early Marker of Treatment Effect in Advanced NSCLC. JCO Precis. Oncol. 2021, 5, 393–402. [Google Scholar] [CrossRef]
- Punnoose, E.A.; Atwal, S.; Liu, W.; Raja, R.; Fine, B.M.; Hughes, B.G.M.; Hicks, R.J.; Hampton, G.M.; Amler, L.C.; Pirzkall, A.; et al. Evaluation of Circulating Tumor Cells and Circulating Tumor DNA in Non-Small Cell Lung Cancer: Association with Clinical Endpoints in a Phase II Clinical Trial of Pertuzumab and Erlotinib. Clin. Cancer Res. 2012, 18, 2391–2401. [Google Scholar] [CrossRef]
- Sundaresan, T.K.; Sequist, L.V.; Heymach, J.V.; Riely, G.J.; Jänne, P.A.; Koch, W.H.; Sullivan, J.P.; Fox, D.B.; Maher, R.; Muzikansky, A.; et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin. Cancer Res. 2016, 22, 1103–1110. [Google Scholar] [CrossRef]
- Ntzifa, A.; Kotsakis, A.; Georgoulias, V.; Lianidou, E. Detection of EGFR Mutations in Plasma CfDNA and Paired CTCs of NSCLC Patients before and after Osimertinib Therapy Using Crystal Digital PCR. Cancers 2021, 13, 2736. [Google Scholar] [CrossRef]
- Liu, H.E.; Vuppalapaty, M.; Wilkerson, C.; Renier, C.; Chiu, M.; Lemaire, C.; Che, J.; Matsumoto, M.; Carroll, J.; Crouse, S.; et al. Detection of EGFR Mutations in CfDNA and CTCs, and Comparison to Tumor Tissue in Non-Small-Cell-Lung-Cancer (NSCLC) Patients. Front. Oncol. 2020, 10, 572895. [Google Scholar] [CrossRef]
- Owen, S.; Lo, T.W.; Fouladdel, S.; Zeinali, M.; Keller, E.; Azizi, E.; Ramnath, N.; Simultaneous, N.S. Single Cell Gene Expression and EGFR Mutation Analysis of Circulating Tumor Cells Reveals Distinct Phenotypes in NSCLC. Adv. Biosyst. 2020, 4, e2000. [Google Scholar] [CrossRef]
- Maheswaran, S.; Sequist, L.V.; Nagrath, S.; Ulkus, L.; Brannigan, B.; Collura, C.V.; Inserra, E.; Diederichs, S.; Iafrate, A.J.; Bell, D.W.; et al. Detection of Mutations in EGFR in Circulating Lung-Cancer Cells. N. Engl. J. Med. 2008, 24, 366–377. [Google Scholar] [CrossRef]
- Breitenbuecher, F.; Hoffarth, S.; Worm, K.; Cortes-Incio, D.; Gauler, T.C.; Köhler, J.; Herold, T.; Schmid, K.W.; Freitag, L.; Kasper, S.; et al. Development of a Highly Sensitive and Specific Method for Detection of Circulating Tumor Cells Harboring Somatic Mutations in Non-Small-Cell Lung Cancer Patients. PLoS ONE 2014, 9, e85350. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, A.; Del Grammastro, M.; Felicioni, L.; Malatesta, S.; Filice, G.; Centi, I.; De Pas, T.; Santoro, A.; Chella, A.; Brandes, A.A.; et al. Assessment of EGFR Mutations in Circulating Tumor Cell Preparations from NSCLC Patients by next Generation Sequencing: Toward a Real-Time Liquid Biopsy for Treatment. PLoS ONE 2014, 9, e103833. [Google Scholar] [CrossRef] [PubMed]
- Gorges, T.M.; Penkalla, N.; Schalk, T.; Joosse, S.A.; Riethdorf, S.; Wikman, H.; Jackson, S.; Brychta, N.; Tucholski, J.; Klaus, L. Enumeration and Molecular Characterization of Tumor Cells in Lung Cancer Patients Using a Novel In Vivo Device for Capturing Circulating Tumor Cells. Clin. Cancer Res. 2016, 22, 2197–2206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castellanos-Rizaldos, E.; Grimm, D.G.; Tadigotla, V.; Hurley, J.; Healy, J.; Neal, P.L.; Sher, M.; Venkatesan, R.; Karlovich, C.; Raponi, M.; et al. Exosome-Based Detection of EGFR T790M in Plasma from Non–Small Cell Lung Cancer Patients. Clin. Cancer Res. 2018, 24, 2944–2950. [Google Scholar] [CrossRef] [PubMed]
- Möhrmann, L.; Huang, H.J.; Hong, D.S.; Tsimberidou, A.M.; Fu, S.; Piha-Paul, S.A.; Subbiah, V.; Karp, D.D.; Naing, A.; Krug, A.; et al. Liquid Biopsies Using Plasma Exosomal Nucleic Acids and Plasma Cell-Free DNA Compared with Clinical Outcomes of Patients with Advanced Cancers. Clin. Cancer Res. 2018, 24, 181–188. [Google Scholar] [CrossRef]
- Krug, A.K.; Enderle, D.; Karlovich, C.; Priewasser, T.; Bentink, S.; Spiel, A.; Brinkmann, K. Improved EGFR Mutation Detection Using Combined Exosomal RNA and Circulating Tumor DNA in NSCLC Patient Plasma. Ann. Oncol. 2018, 29, 700–706. [Google Scholar] [CrossRef]
- Kim, Y.; Shin, S.; Lee, K.A. Exosome-Based Detection of EGFR T790M in Plasma and Pleural Fluid of Prospectively Enrolled Non-Small Cell Lung Cancer Patients after First-Line Tyrosine Kinase Inhibitor Therapy. Cancer Cell Int. 2021, 21, 1–8. [Google Scholar] [CrossRef]
- Park, J.; Lee, C.; Eom, J.S.; Kim, M.H.; Cho, Y.K. Detection of Egfr Mutations Using Bronchial Washing-Derived Extracellular Vesicles in Patients with Non-Small-Cell Lung Carcinoma. Cancers 2020, 12, 2822. [Google Scholar] [CrossRef]
- Purcell, E.; Owen, S.; Prantzalos, E.; Radomski, A.; Carman, N.; Lo, T.W.; Zeinali, M.; Subramanian, C.; Ramnath, N.; Nagrath, S. Epidermal Growth Factor Receptor Mutations Carried in Extracellular Vesicle-Derived Cargo Mirror Disease Status in Metastatic Non-Small Cell Lung Cancer. Front. Cell Dev. Biol. 2021, 9, 1–12. [Google Scholar] [CrossRef]
- Jouida, A.; McCarthy, C.; Fabre, A.; Keane, M.P. Exosomes: A New Perspective in EGFR-Mutated Lung Cancer. Cancer Metastasis Rev. 2021, 40, 589–601. [Google Scholar] [CrossRef]
- Guibert, N.; Hu, Y.; Feeney, N.; Kuang, Y.; Plagnol, V.; Jones, G.; Howarth, K.; Beeler, J.F.; Paweletz, C.P.; Oxnard, G.R. Amplicon-Based next-Generation Sequencing of Plasma Cell-Free DNA for Detection of Driver and Resistance Mutations in Advanced Non-Small Cell Lung Cancer. Ann. Oncol. 2018, 29, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Mezquita, L.; Swalduz, A.; Jovelet, C.; Ortiz-Cuaran, S.; Howarth, K.; Planchard, D.; Avrillon, V.; Recondo, G.; Marteau, S.; Benitez, J.C.; et al. Clinical Relevance of an Amplicon-Based Liquid Biopsy for Detecting ALK and ROS1 Fusion and Resistance Mutations in Patients With Non–Small-Cell Lung Cancer. JCO Precis. Oncol. 2020, 4, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Brannon, A.R.; Ferris, L.A.; Campbell, C.D.; Lin, J.J.; Schultz, K.R.; Ackil, J.; Stevens, S.; Dardaei, L.; Yoda, S.; et al. Tracking the Evolution of Resistance to ALK Tyrosine Kinase Inhibitors Through Longitudinal Analysis of Circulating Tumor DNA. JCO Precis. Oncol. 2018, 2018, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Horn, L.; Whisenant, J.G.; Wakelee, H.; Reckamp, K.L.; Qiao, H.; Leal, T.A.; Du, L.; Hernandez, J.; Huang, V.; Blumenschein, G.R.; et al. Monitoring Therapeutic Response and Resistance: Analysis of Circulating Tumor DNA in Patients With ALK+ Lung Cancer. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2019, 14, 1901–1911. [Google Scholar] [CrossRef] [PubMed]
- Dziadziuszko, R.; Mok, T.; Peters, S.; Han, J.Y.; Alatorre-Alexander, J.; Leighl, N.; Sriuranpong, V.; Pérol, M.; de Castro Junior, G.; Nadal, E.; et al. Blood First Assay Screening Trial (BFAST) in Treatment-Naive Advanced or Metastatic NSCLC: Initial Results of the Phase 2 ALK-Positive Cohort. J. Thorac. Oncol. 2021, 16, 2040–2050. [Google Scholar] [CrossRef]
- McCoach, C.E.; Blakely, C.M.; Banks, K.C.; Levy, B.; Chue, B.M.; Raymond, V.M.; Le, A.T.; Lee, C.E.; Diaz, J.; Waqar, S.N.; et al. Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non–Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 2758–2770. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Zhang, W.; Xiong, L.; Pan, F.; Niu, Y.; Chu, T.; Wang, H.; Zhao, Y.; Jiang, L. Use of Capture-Based next-Generation Sequencing to Detect ALK Fusion in Plasma Cell-Free DNA of Patients with Non-Small-Cell Lung Cancer. Oncotarget 2017, 8, 2771–2780. [Google Scholar] [CrossRef]
- Wang, Y.; Tian, P.W.; Wang, W.Y.; Wang, K.; Zhang, Z.; Chen, B.J.; He, Y.Q.; Li, L.; Liu, H.; Chuai, S.; et al. Noninvasive Genotyping and Monitoring of Anaplastic Lymphoma Kinase (ALK) Rearranged Non-Small Cell Lung Cancer by Capturebased next-Generation Sequencing. Oncotarget 2016, 7, 65208–65217. [Google Scholar] [CrossRef]
- Dietz, S.; Christopoulos, P.; Yuan, Z.; Angeles, A.K.; Gu, L.; Volckmar, A.L.; Ogrodnik, S.J.; Janke, F.; Fratte, C.D.; Zemojtel, T.; et al. Longitudinal Therapy Monitoring of ALK-Positive Lung Cancer by Combined Copy Number and Targeted Mutation Profiling of Cell-Free DNA. EBioMedicine 2020, 62, 103103. [Google Scholar] [CrossRef]
- Hua, G.; Zhang, X.; Zhang, M.; Wang, Q.; Chen, X.; Yu, R.; Bao, H.; Liu, J.; Wu, X.; Shao, Y.; et al. Real-World Circulating Tumor DNA Analysis Depicts Resistance Mechanism and Clonal Evolution in ALK Inhibitor-Treated Lung Adenocarcinoma Patients. ESMO Open 2022, 7, 100337. [Google Scholar] [CrossRef]
- Mondaca, S.; Lebow, E.S.; Namakydoust, A.; Razavi, P.; Reis-Filho, J.S.; Shen, R.; Offin, M.; Tu, H.Y.; Murciano-Goroff, Y.; Xu, C.; et al. Clinical Utility of Next-Generation Sequencing-Based CtDNA Testing for Common and Novel ALK Fusions. Lung Cancer 2021, 159, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.N.; Tamen, R.M.; Martinez, P.; Sable-Hunt, A.; Addario, T.; Barbour, P.; Shaffer, T.; Hosseini, S.A.; Bertucci, C.; Lim, L.P.; et al. SPACEWALK: A Remote Participation Study of ALK Resistance Leveraging Plasma Cell-Free DNA Genotyping. JTO Clin. Res. Rep. 2021, 2, 100151. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.T.; Chiang, C.L.; Hung, J.Y.; Lee, M.H.; Su, W.C.; Wu, S.Y.; Wei, Y.F.; Lee, K.Y.; Tseng, Y.H.; Su, J.; et al. Resistance Profiles of Anaplastic Lymphoma Kinase Tyrosine Kinase Inhibitors in Advanced Non–Small-Cell Lung Cancer: A Multicenter Study Using Targeted next-Generation Sequencing. Eur. J. Cancer 2021, 156, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bordi, P.; Tiseo, M.; Rofi, E.; Petrini, I.; Restante, G.; Danesi, R.; Del Re, M. Detection of ALK and KRAS Mutations in Circulating Tumor DNA of Patients With Advanced ALK-Positive NSCLC With Disease Progression During Crizotinib Treatment. Clin. Lung Cancer 2017, 18, 692–697. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, J.; Wang, T.; Zhou, J.; Zheng, J.; Feng, J.; Zhuang, W.; Chen, J.; Zhao, J.; Zhong, W.; et al. Decoding the Evolutionary Response to Ensartinib in Patients With ALK-Positive NSCLC by Dynamic Circulating Tumor DNA Sequencing. J. Thorac. Oncol. 2021, 16, 827–839. [Google Scholar] [CrossRef]
- Shaw, A.T.; Solomon, B.J.; Besse, B.; Bauer, T.M.; Lin, C.C.; Soo, R.A.; Riely, G.J.; Ignatius Ou, S.H.; Clancy, J.S.; Li, S.; et al. ALK Resistance Mutations and Efficacy of Lorlatinib in Advanced Anaplastic Lymphoma Kinase-Positive Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2019, 37, 1370–1379. [Google Scholar] [CrossRef]
- Soo, R.A.; Martini, J.-F.; van der Wekken, A.J.; Teraoka, S.; Shaw, A.T.; Shepard, D.; Calella, A.M.; Polli, A.; Toffalorio, F.; Tomasini, P.; et al. Early Circulating Tumor (Ct) DNA Dynamics and Efficacy of Lorlatinib: Analysis from the CROWN Study. J. Clin. Oncol. 2021, 39, 9011. [Google Scholar] [CrossRef]
- Aldea, M.; Hendriks, L.; Mezquita, L.; Jovelet, C.; Planchard, D.; Auclin, E.; Remon, J.; Howarth, K.; Benitez, J.C.; Gazzah, A.; et al. Circulating Tumor DNA Analysis for Patients with Oncogene-Addicted NSCLC With Isolated Central Nervous System Progression. J. Thorac. Oncol. 2019, 15, 383–391. [Google Scholar] [CrossRef]
- Zheng, M.M.; Li, Y.S.; Jiang, B.Y.; Tu, H.Y.; Tang, W.F.; Yang, J.J.; Zhang, X.C.; Ye, J.Y.; Yan, H.H.; Su, J.; et al. Clinical Utility of Cerebrospinal Fluid Cell-Free DNA as Liquid Biopsy for Leptomeningeal Metastases in ALK-Rearranged NSCLC. J. Thorac. Oncol. 2019, 14, 924–932. [Google Scholar] [CrossRef]
- Ilié, M.; Long, E.; Butori, C.; Hofman, V.; Coelle, C.; Mauro, V.; Zahaf, K.; Marquette, C.H.; Mouroux, J.; Paterlini-Bréchot, P.; et al. ALK-Gene Rearrangement: A Comparative Analysis on Circulating Tumour Cells and Tumour Tissue from Patients with Lung Adenocarcinoma. Ann. Oncol. 2012, 23, 2907–2913. [Google Scholar] [CrossRef]
- Pailler, E.; Adam, J.; Barthélémy, A.; Oulhen, M.; Auger, N.; Valent, A.; Borget, I.; Planchard, D.; Taylor, M.; André, F.; et al. Detection of Circulating Tumor Cells Harboring a Unique ALK Rearrangement in ALK-Positive Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2013, 31, 2273–2281. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.L.; Lim, T.H.; Lim, T.K.H.; Tan, D.S.W.; Chua, Y.W.; Ang, M.K.; Pang, B.; Lim, C.T.; Takano, A.; Lim, A.S.T.; et al. Concordance of Anaplastic Lymphoma Kinase (ALK) Gene Rearrangements between Circulating Tumor Cells and Tumor in Non-Small Cell Lung Cancer. Oncotarget 2016, 7, 23251–23262. [Google Scholar] [CrossRef] [PubMed]
- Pailler, E.; Oulhen, M.; Borget, I.; Remon, J.; Ross, K.; Auger, N.; Billiot, F.; Ngo Camus, M.; Commo, F.; Lindsay, C.R.; et al. Circulating Tumor Cells with Aberrant ALK Copy Number Predict Progression-Free Survival during Crizotinib Treatment in ALK-Rearranged Non-Small Cell Lung Cancer Patients. Cancer Res. 2017, 77, 2222–2230. [Google Scholar] [CrossRef] [Green Version]
- Aieta, M.; Facchinetti, A.; De Faveri, S.; Manicone, M.; Tartarone, A.; Possidente, L.; Lerose, R.; Mambella, G.; Calderone, G.; Zamarchi, R.; et al. Monitoring and Characterization of Circulating Tumor Cells (CTCs) in a Patient With EML4-ALK–Positive Non–Small Cell Lung Cancer (NSCLC). Clin. Lung Cancer 2016, 17, e173–e177. [Google Scholar] [CrossRef]
- Provencio, M.; Pérez-Callejo, D.; Torrente, M.; Martin, P.; Calvo, V.; Gutiérrez, L.; Franco, F.; Coronado, M.J.; Cruz-Bermúdez, J.L.; Ruiz-Valdepeñas, A.M.; et al. Concordance between Circulating Tumor Cells and Clinical Status during Follow-up in Anaplastic Lymphoma Kinase (ALK) Nonsmall- Cell Lung Cancer Patients. Oncotarget 2017, 8, 59408–59416. [Google Scholar] [CrossRef] [PubMed]
- Ilié, M.; Mazières, J.; Chamorey, E.; Heeke, S.; Benzaquen, J.; Thamphya, B.; Boutros, J.; Tiotiu, A.; Fayada, J.; Cadranel, J.; et al. Prospective Multicenter Validation of the Detection of ALK Rearrangements of Circulating Tumor Cells for Noninvasive Longitudinal Management of Patients With Advanced NSCLC. J. Thorac. Oncol. 2021, 16, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Pailler, E.; Faugeroux, V.; Oulhen, M.; Mezquita, L.; Laporte, M.; Honore, A.; Lecluse, Y.; Queffelec, P.; NgoCamus, M.; Nicotra, C.; et al. Acquired Resistance Mutations to ALK Inhibitors Identified by Single Circulating Tumor Cell Sequencing in ALK-Rearranged Non–Small-Cell Lung Cancer. Clin. Cancer Res. 2021, 5, 67. [Google Scholar] [CrossRef]
- Oulhen, M.; Pawlikowska, P.; Tayoun, T.; Garonzi, M.; Buson, G.; Forcato, C.; Manaresi, N.; Aberlenc, A.; Mezquita, L.; Lecluse, Y.; et al. Circulating Tumor Cell Copy-Number Heterogeneity in ALK-Rearranged Non-Small-Cell Lung Cancer Resistant to ALK Inhibitors. NPJ Precis. Oncol. 2021, 5, 67. [Google Scholar] [CrossRef]
- Brinkmann, K.; Enderle, D.; Flinspach, C.; Meyer, L.; Skog, J.; Noerholm, M. Exosome Liquid Biopsies of NSCLC Patients for Longitudinal Monitoring of ALK Fusions and Resistance Mutations. J. Clin. Oncol. 2018, 36, e24090. [Google Scholar] [CrossRef]
- Reclusa, P.; Laes, J.; Malapelle, U.; Valentino, A.; Rocco, D.; Gil-bazo, I.; Rolfo, C. EML4-ALK Translocation Identification in RNA Exosomal Cargo (ExoALK) in NSCLC Patients: A Novel Role for Liquid Biopsy. Transl. Cancer Res. 2019, 8, 76–78. [Google Scholar] [CrossRef]
- Nilsson, R.J.A.; Karachaliou, N.; Berenguer, J.; Gimenez-Capitan, A.; Schellen, P.; Teixido, C.; Tannous, J.; Kuiper, J.L.; Drees, E.; Grabowska, M.; et al. Rearranged EML4-ALK Fusion Transcripts Sequester in Circulating Blood Platelets and Enable Blood-Based Crizotinib Response Monitoring in Non-Small-Cell Lung Cancer. Oncotarget 2016, 7, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Park, C.K.; Kim, J.E.; Kim, M.S.; Kho, B.G.; Park, H.Y.; Kim, T.O.; Shin, H.J.; Cho, H.J.; Choi, Y.D.; Oh, I.J.; et al. Feasibility of Liquid Biopsy Using Plasma and Platelets for Detection of Anaplastic Lymphoma Kinase Rearrangements in Non - Small Cell Lung Cancer. J. Cancer Res. Clin. Oncol. 2019, 145, 2071–2082. [Google Scholar] [CrossRef]
- Aggarwal, C.; Thompson, J.C.; Black, T.A.; Katz, S.I.; Fan, R.; Yee, S.S.; Chien, A.L.; Evans, T.L.; Bauml, J.M.; Alley, E.W.; et al. Clinical Implications of Plasma-Based Genotyping with the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol. 2019, 5, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Han, Y.; Li, P.; Zhang, J.; Ou, Q.; Tong, X.; Zhao, R.; Dong, N.; Wu, X.; Li, W.; et al. Molecular and Clinicopathological Characteristics of ROS1-Rearranged Non-Small-Cell Lung Cancers Identified by next-Generation Sequencing. Mol. Oncol. 2020, 14, 2787–2795. [Google Scholar] [CrossRef] [PubMed]
- Zugazagoitia, J.; Gómez-Rueda, A.; Jantus-Lewintre, E.; Isla, D.; Camps, C.; Ramos, I.; Trigo, J.M.; Bernabé, R.; Juan-Vidal, O.; Sanchez-Torres, J.M.; et al. Clinical Utility of Plasma-Based Digital next-Generation Sequencing in Oncogene-Driven Non-Small-Cell Lung Cancer Patients with Tyrosine Kinase Inhibitor Resistance. Lung Cancer 2019, 134, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Page, R.D.; Drusbosky, L.M.; Dada, H.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; et al. Clinical Outcomes for Plasma-Based Comprehensive Genomic Profiling Versus Standard-of-Care Tissue Testing in Advanced Non–Small Cell Lung Cancer. Clin. Lung Cancer 2022, 23, 72–81. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Rooney, M.; Nagy, R.J.; Lin, J.J.; Chin, E.; Ferris, L.A.; Ackil, J.; Lennerz, J.K.; Lanman, R.B.; Gainor, J.F. Molecular Analysis of Plasma From Patients With ROS1-Positive NSCLC. J. Thorac. Oncol. 2019, 14, 816–824. [Google Scholar] [CrossRef]
- Shaw, A.T.; Solomon, B.J.; Chiari, R.; Riely, G.J.; Besse, B.; Soo, R.A.; Kao, S.; Lin, C.C.; Bauer, T.M.; Clancy, J.S.; et al. Lorlatinib in Advanced ROS1-Positive Non-Small-Cell Lung Cancer: A Multicentre, Open-Label, Single-Arm, Phase 1–2 Trial. Lancet Oncol. 2019, 20, 1691–1701. [Google Scholar] [CrossRef]
- Dziadziuszko, R.; Hung, T.; Wang, K.; Choeurng, V.; Drilon, A.; Doebele, R.C.; Barlesi, F.; Wu, C.; Dennis, L.; Skoletsky, J.; et al. Pre- and Post-treatment Blood-based Genomic Landscape of Patients with ROS1 or NTRK Fusion-positive Solid Tumors Treated with Entrectinib. Mol. Oncol. 2022, 16, 2000–2014. [Google Scholar] [CrossRef]
- Lee, J.K.; Hazar-Rethinam, M.; Decker, B.; Gjoerup, O.; Madison, R.W.; Lieber, D.S.; Chung, J.H.; Schrock, A.B.; Creeden, J.; Venstrom, J.; et al. The Pan-Tumor Landscape of Targetable Kinase Fusions in Circulating Tumor DNA. Clin. Cancer Res. 2022, 28, 728–737. [Google Scholar] [CrossRef]
- Pailler, E.; Auger, N.; Lindsay, C.R.; Vielh, P.; Islas-Morris-Hernandez, A.; Borget, I.; Ngo-Camus, M.; Planchard, D.; Soria, J.C.; Besse, B.; et al. High Level of Chromosomal Instability in Circulating Tumor Cells of ROS1-Rearranged Non-Small-Cell Lung Cancer. Ann. Oncol. 2015, 26, 1408–1415. [Google Scholar] [CrossRef] [PubMed]
- Leong, M.C.; Lim, T.H.; Chua, Y.W.; Tan, C.L.; Tan, D.S.-W.; Wu, A.; Takano, A.; Lim, K.H.; Lim, A.S.T.; Lim, W.-T. Molecular Detection of ROS1-Rearranged Circulating Tumor Cells (CTC) in Non-Small Cell Lung Cancer (NSCLC) Patients. J. Clin. Oncol. 2016, 34, e23086. [Google Scholar] [CrossRef]
- Raez, L.E.; Manca, P.; Rolfo, C.; Singh, V. ROS-1 Rearrangements in Circulating Tumor Cells. J. Thorac. Oncol. 2018, 13, e71–e72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical Utility of Comprehensive Cell-Free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-Small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 4691–4700. [Google Scholar] [CrossRef]
- Iaccarino, A.; Pisapia, P.; Pepe, F.; Sgariglia, R.; Nacchio, M.; Russo, G.; Gragnano, G.; De Luca, C.; Troncone, G.; Malapelle, U. Liquid Biopsy for BRAF Mutations Testing in Non-Small Cell Lung Cancer: A Retrospective Study. J. Clin. Pathol. 2022, 75, 58–60. [Google Scholar] [CrossRef]
- Ortiz-Cuaran, S.; Mezquita, L.; Swalduz, A.; Aldea, M.; Mazieres, J.; Leonce, C.; Jovelet, C.; Pradines, A.; Avrillon, V.; Chumbi Flores, W.R.; et al. Circulating Tumor DNA Genomics Reveal Potential Mechanisms of Resistance to BRAF-Targeted Therapies in Patients with BRAF-Mutant Metastatic Non-Small Cell Lung Cancer. Clin. Cancer Res. 2020, 26, 6242–6253. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, X.; Li, R.; Shen, J.; Zhang, H.; Yu, L.; Liu, B.; Wang, L. The Detection and Significance of EGFR and BRAF in Cell-Free DNA of Peripheral Blood in NSCLC. Oncotarget 2017, 8, 49773–49782. [Google Scholar] [CrossRef]
- Solassol, J.; Vendrell, J.A.; Senal, R.; Audran, P.; Leenhardt, F.; Quantin, X. Challenging BRAF/EGFR Co-Inhibition in NSCLC Using Sequential Liquid Biopsies. Lung Cancer 2019, 133, 45–47. [Google Scholar] [CrossRef]
- Ding, H.; Zhuang, Z.; Xie, J.; Huang, H.; Tao, Z.; Liu, Z. Durable Clinical Response of Advanced Lung Adenocarcinoma Harboring Egfr-19del/T790m/Brafv600e Mutations after Treating with Osimertinib and Dabrafenib plus Trametinib: A Case Report. OncoTargets Ther. 2020, 13, 7933–7939. [Google Scholar] [CrossRef]
- Zhou, F.; Zhao, W.; Chen, X.; Zhang, J.; Zhou, C. Response to the Combination of Dabrafenib, Trametinib and Osimertinib in a Patient with EGFR-Mutant NSCLC Harboring an Acquired BRAFV600E Mutation. Lung Cancer 2020, 139, 219–220. [Google Scholar] [CrossRef]
- Bracht, J.W.P.; Karachaliou, N.; Bivona, T.; Lanman, R.B.; Faull, I.; Nagy, R.J.; Drozdowskyj, A.; Berenguer, J.; Fernandez-Bruno, M.; Molina-Vila, M.A.; et al. BRAF Mutations Classes I, II, and III in NSCLC Patients Included in the SLLIP Trial: The Need for a New Pre-Clinical Treatment Rationale. Cancers 2019, 11, 1381. [Google Scholar] [CrossRef]
- Guibert, N.; Pradines, A.; Casanova, A.; Farella, M.; Keller, L.; Soria, J.C.; Favre, G.; Mazières, J. Detection and Monitoring of the BRAF Mutation in Circulating Tumor Cells and Circulating Tumor DNA in BRAF-Mutated Lung Adenocarcinoma. J. Thorac. Oncol. 2016, 11, e109–e112. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.-H.I.; Young, L.; Schrock, A.B.; Johnson, A.; Klempner, S.J.; Zhu, V.W.; Miller, V.A.; Ali, S.M. Emergence of Preexisting MET Y1230C Mutation as a Resistance Mechanism to Crizotinib in NSCLC with MET Exon 14 Skipping. J. Thorac. Oncol. 2017, 12, 137–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nacchio, M.; Sgariglia, R.; Gristina, V.; Pisapia, P.; Pepe, F.; De Luca, C.; Migliatico, I.; Clery, E.; Greco, L.; Vigliar, E.; et al. KRAS Mutations Testing in Non-Small Cell Lung Cancer: The Role of Liquid Biopsy in the Basal Setting. J. Thorac. Dis. 2020, 12, 3836–3843. [Google Scholar] [CrossRef] [PubMed]
- Nicolazzo, C.; Gelibter, A.; Bottillo, I.; Belardinilli, F.; Pisegna, S.; De Renzi, G.; Marinelli, D.; Grammatico, P.; Cortesi, E.; Giannini, G.; et al. Comparison of Two Blood-Based Genotyping Tests to Investigate the KRAS G12C Mutation in Patients with Non-Small-Cell Lung Cancer at Failure of First-Line Treatments. Diagnostics 2021, 11, 2196. [Google Scholar] [CrossRef] [PubMed]
- Thein, K.Z.; Biter, A.B.; Banks, K.C.; Duda, A.W.; Saam, J.; Roszik, J.; Janku, F.; Skoulidis, F.; Heymach, J.V.; Kopetz, S.; et al. Identification of KRASG12C Mutations in Circulating Tumor DNA in Patients With Cancer. JCO Precis. Oncol. 2022, 6, e2100547. [Google Scholar] [CrossRef]
- Sama, S.; Le, T.; Ullah, A.; Elhelf, I.A.; Kavuri, S.K.; Karim, N.A. The Role of Serial Liquid Biopsy in the Management of Metastatic Non-Small Cell Lung Cancer (NSCLC). Clin. Pract. 2022, 12, 419–424. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buszka, K.; Ntzifa, A.; Owecka, B.; Kamińska, P.; Kolecka-Bednarczyk, A.; Zabel, M.; Nowicki, M.; Lianidou, E.; Budna-Tukan, J. Liquid Biopsy Analysis as a Tool for TKI-Based Treatment in Non-Small Cell Lung Cancer. Cells 2022, 11, 2871. https://doi.org/10.3390/cells11182871
Buszka K, Ntzifa A, Owecka B, Kamińska P, Kolecka-Bednarczyk A, Zabel M, Nowicki M, Lianidou E, Budna-Tukan J. Liquid Biopsy Analysis as a Tool for TKI-Based Treatment in Non-Small Cell Lung Cancer. Cells. 2022; 11(18):2871. https://doi.org/10.3390/cells11182871
Chicago/Turabian StyleBuszka, Karolina, Aliki Ntzifa, Barbara Owecka, Paula Kamińska, Agata Kolecka-Bednarczyk, Maciej Zabel, Michał Nowicki, Evi Lianidou, and Joanna Budna-Tukan. 2022. "Liquid Biopsy Analysis as a Tool for TKI-Based Treatment in Non-Small Cell Lung Cancer" Cells 11, no. 18: 2871. https://doi.org/10.3390/cells11182871
APA StyleBuszka, K., Ntzifa, A., Owecka, B., Kamińska, P., Kolecka-Bednarczyk, A., Zabel, M., Nowicki, M., Lianidou, E., & Budna-Tukan, J. (2022). Liquid Biopsy Analysis as a Tool for TKI-Based Treatment in Non-Small Cell Lung Cancer. Cells, 11(18), 2871. https://doi.org/10.3390/cells11182871







