Multiple Light-Dark Signals Regulate Expression of the DEAD-Box RNA Helicase CrhR in Synechocystis PCC 6803
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Stress and Inhibitor Conditions
2.3. Protein Abundance
2.4. RNA Sampling and Extraction
2.5. Quantification of RNA and Quality Confirmation
2.6. RNA Abundance
2.7. cDNA Generation
2.8. Primer Design and Selection of Endogenous Control Gene
2.9. qPCR Amplification
2.10. Data Manipulation
3. Results
3.1. CrhR Protein Accumulation Is Regulated by Light Driven Reduction of the PQ Pool
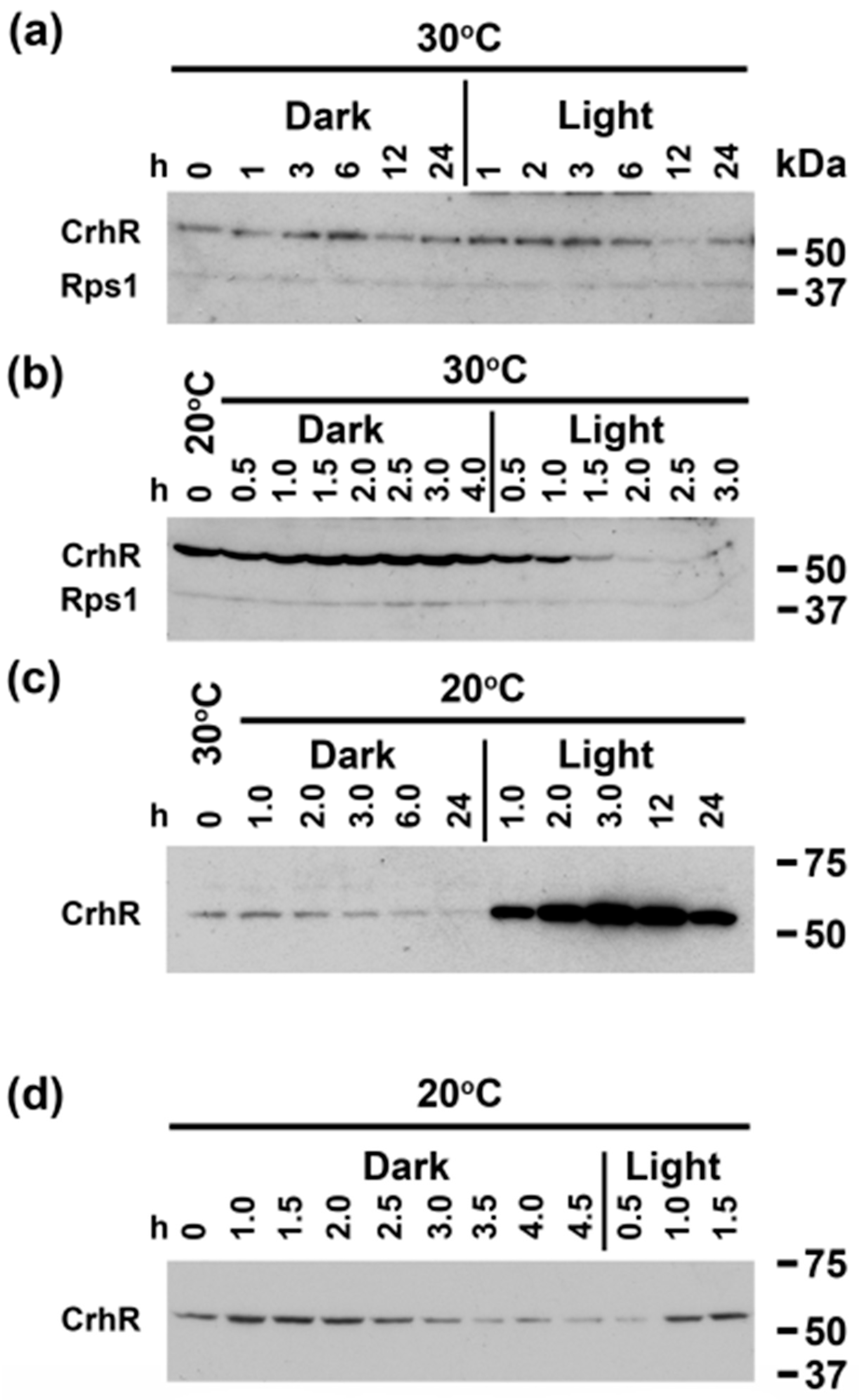
3.2. Light Quantity and Light-Dark Transition Regulates CrhR Expression
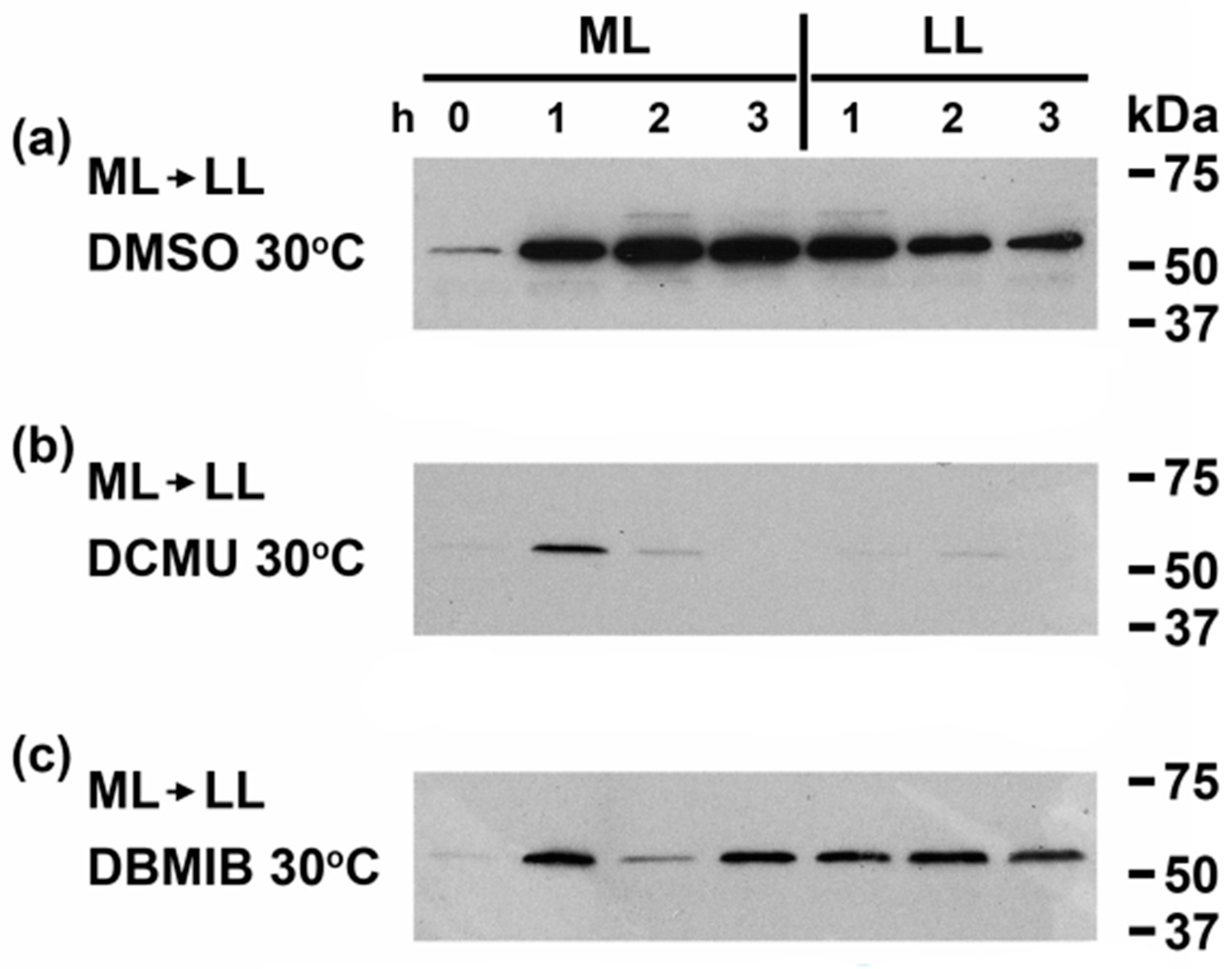
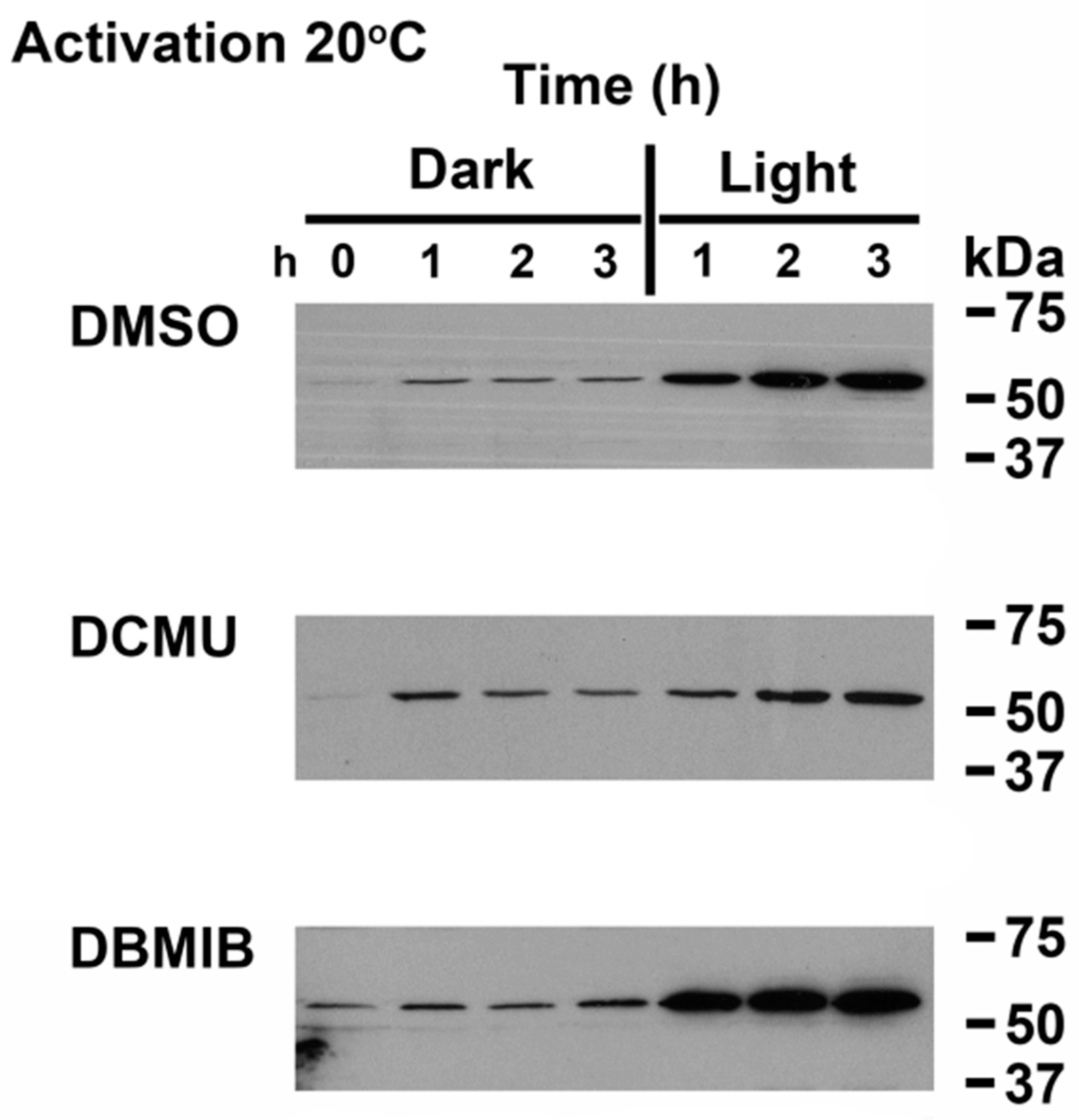
3.3. Linking Light Quantity and PQ Redox Poise to Regulation of CrhR Expression
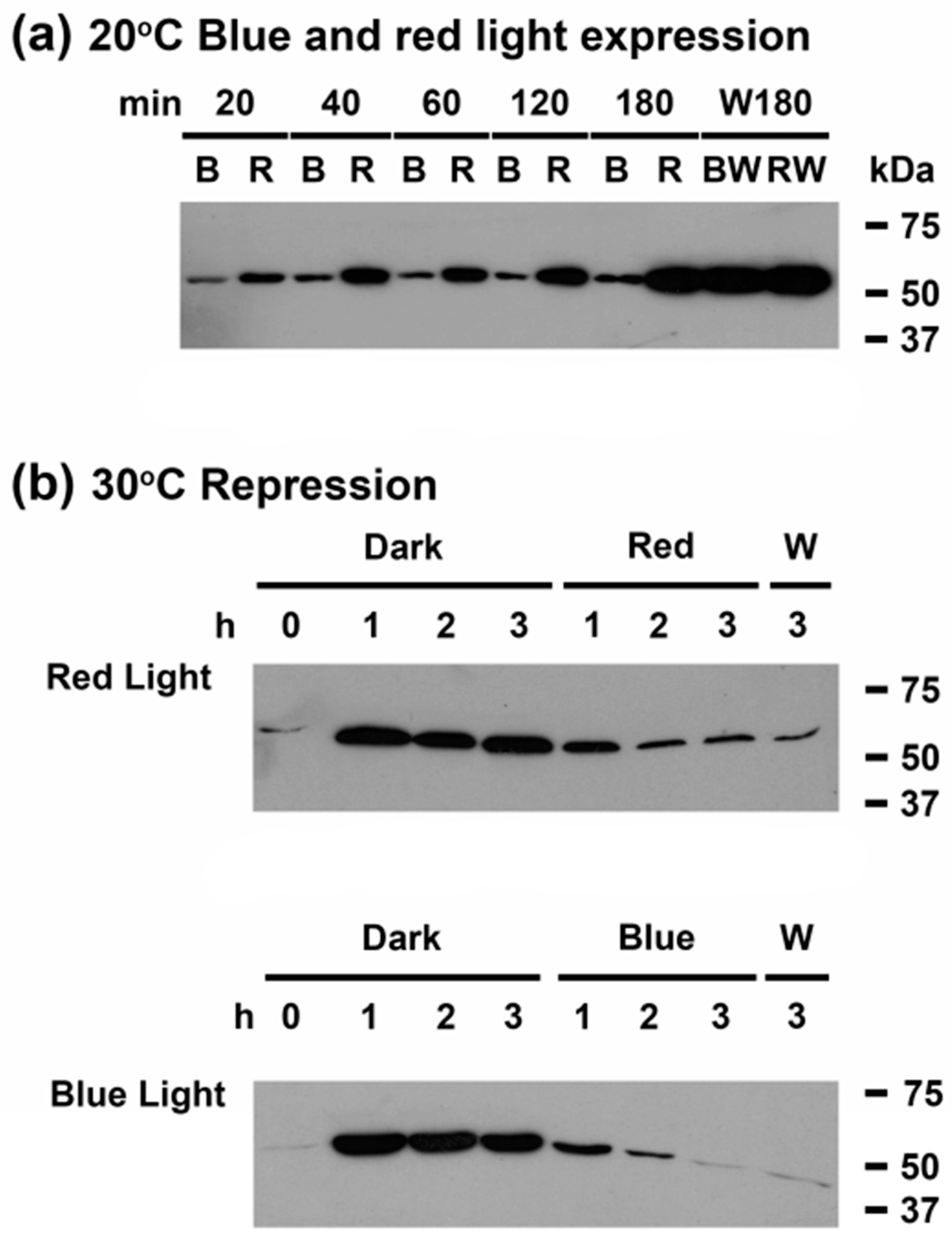
3.4. Light Quality Regulates CrhR Protein Expression at the Translational Level
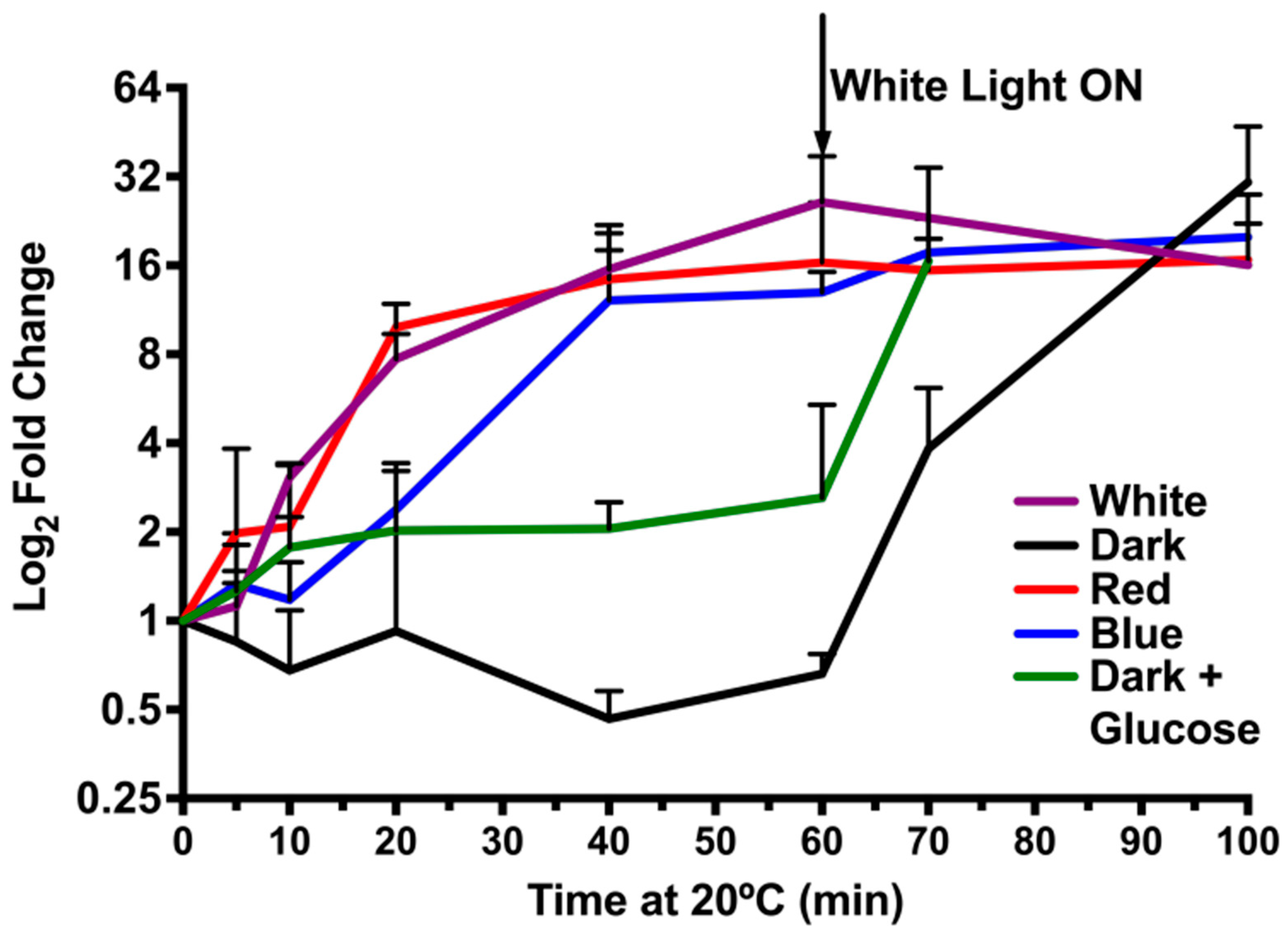
3.5. A light Signal Is Required to Repress CrhR Expression
3.6. Far-Red Light Suppresses Red Light Induction of CrhR Accumulation
3.7. Light Quality Regulation of crhR Transcript Accumulation
3.8. crhR Translation Is Pre-Initiated at 30 °C but Elongation Is Inhibited
4. Discussion
4.1. The Role of Light-Mediated Alteration of the PQ Redox Poise
4.2. Light Quality Regulates crhR Translation
4.3. A Light Signal Regulates CrhR Proteolysis
4.4. Dark Incubation Supersedes All Three Light Signal Responses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Montgomery, B.L. Sensing the light: Photoreceptive systems and signal transduction in cyanobacteria. Mol. Microbiol. 2007, 64, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Gutu, A.; Kehoe, D.M. Emerging perspectives on the mechanisms, regulation, and distribution of light color acclimation in cyanobacteria. Mol. Plant 2012, 5, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rockwell, N.C.; Lagarias, J.C. Phytochrome diversification in cyanobacteria and eukaryotic algae. Curr. Opin. Plant Biol. 2017, 37, 87–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujisawa, T.; Masuda, S. Light-induced chromophore and protein responses and mechanical signal transduction of BLUF proteins. Biophys. Rev. 2018, 10, 327–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiltbank, L.B.; Kehoe, D.M. Two cyanobacterial photoreceptors regulate photosynthetic light harvesting by sensing teal, green, yellow and red light. mBio 2016, 7, e02130-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiltbank, L.B.; Kehoe, D.M. Diverse light responses of cyanobacteria mediated by phytochrome superfamily photoreceptors. Nat. Rev. Microbiol. 2019, 17, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, I.M.; Puthiyaveetil, S.; Allen, J.F. A two-component regulatory system in transcriptional control of photosystem stoichiometry: Redox-dependent and sodium ion-dependent phosphoryl transfer from cyanobacterial histidine kinase Hik2 to response regulators Rre1 and RppA. Front. Plant Sci. 2016, 12, 196–212. [Google Scholar] [CrossRef] [Green Version]
- Locato, V.; Cimini, S.; De Gara, L. ROS and redox balance as multifaceted players of cross-tolerance: Epigenetic and retrograde control of gene expression. J. Exp. Bot. 2018, 69, 3373–3391. [Google Scholar] [CrossRef] [Green Version]
- Westermark, S.; Steuer, R. Toward multiscale models of cyanobacterial growth: A modular approach. Front. Bioeng. Biotechnol. 2016, 4, 95. [Google Scholar] [CrossRef] [Green Version]
- Kujat, S.L.; Owttrim, G.W. Redox-regulated RNA helicase expression. Plant Physiol. 2000, 124, 703–714. [Google Scholar] [CrossRef]
- Ritter, S.P.A.; Lewis, A.C.; Vincent, S.L.; Lo, L.L.; Cunha, A.P.A.; Chamot, D.; Ensminger, I.; Espie, G.S.; Owttrim, G.W. Evidence for convergent sensing of multiple abiotic stresses in cyanobacteria. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129462. [Google Scholar] [CrossRef] [PubMed]
- Barth, J.; Bergner, S.V.; Jaeger, D.; Niehues, A.; Schulze, S.; Scholz, M.; Fufezan, C. The interplay of light and oxygen in the reactive oxygen stress response of Chlamydomonas reinhardtii dissected by quantitative mass spectrometry. Mol. Cell. Proteom. 2014, 13, 969–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luimstra, V.M.; Schuurmans, J.M.; Verschoor, A.M.; Hellingwerf, K.J.; Huisman, J.; Matthijs, H.C.P. Blue light reduces photosynthetic efficiency of cyanobacteria through an imbalance between photosystems I and II. Photosynth. Res. 2018, 138, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossman, A.R.; Schaefer, M.R.; Chiang, G.G.; Collier, J.L. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol. Rev. 1993, 57, 725–749. [Google Scholar] [CrossRef] [PubMed]
- Hübschmann, T.; Yamamoto, H.; Gieler, T.; Murata, N.; Börner, T. Red and far-red light alter the transcript profile in the cyanobacterium Synechocystis sp. PCC 6803: Impact of cyanobacterial phytochromes. FEBS Lett. 2005, 579, 1613–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarcina, M.; Bouzovitis, N.; Mullineaux, C.W. Mobilization of photosystem II induced by intense red light in the Cyanobacterium Synechococcus sp. PCC7942. Plant Cell 2006, 18, 457–464. [Google Scholar] [CrossRef] [Green Version]
- Mironov, K.S.; Sidorov, R.A.; Kreslavski, V.D.; Bedbenov, V.S.; Tsydendambaev, V.D.; Los, D.A. Cold-induced gene expression and ω(3) fatty acid unsaturation is controlled by red light in Synechocystis. J. Photochem. Photobiol. B 2014, 137, 84–88. [Google Scholar] [CrossRef]
- Angerer, V.; Schwenk, P.; Wallner, T.; Kaever, V.; Hiltbrunner, A.; Wilde, A. The protein Slr1143 is an active diguanylate cyclase in Synechocystis sp. PCC 6803 and interacts with the photoreceptor Cph2. Microbiology 2017, 163, 920–930. [Google Scholar] [CrossRef]
- Fiedler, B.; Broc, D.; Schubert, H.; Rediger, A.; Börner, T.; Wilde, A. Involvement of cyanobacterial phytochromes in growth under different light qualities and quantities. Photochem. Photobiol. 2004, 79, 551–555. [Google Scholar] [CrossRef]
- Anderson, S.L.; McIntosh, L. Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803: A blue-light-requiring process. J. Bacteriol. 1991, 173, 2761–2767. [Google Scholar] [CrossRef]
- Luimstra, V.M.; Schuurmans, J.M.; Hellingwerf, K.J.; Matthijs, H.C.P.; Huisman, J. Blue light induces major changes in the gene expression profile of the cyanobacterium Synechocystis sp. PCC 6803. Physiol. Plant 2020, 170, 10–26. [Google Scholar] [CrossRef] [PubMed]
- Linder, P.; Jankowsky, E. From unwinding to clamping—The DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011, 12, 505–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linder, P.; Owttrim, G.W. Plant RNA helicases: Linking aberrant and silencing RNA. Trends Plant Sci. 2009, 14, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Owttrim, G.W. RNA helicases: Diverse roles in prokaryotic response to abiotic stress. RNA Biol. 2013, 10, 96–110. [Google Scholar] [CrossRef] [Green Version]
- Georg, J.; Rosana, A.R.R.; Chamot, D.; Migur, A.; Hess, W.R.; Owttrim, G.W. Inactivation of the RNA helicase CrhR impacts a specific subset of the transcriptome in the cyanobacterium Synechocystis sp. PCC 6803. RNA Biol. 2019, 16, 1205–1214. [Google Scholar] [CrossRef] [Green Version]
- Rosana, A.R.R.; Whitford, D.S.; Migur, A.; Steglich, C.; Kujat-Choy, S.L.; Hess, W.R.; Owttrim, G.W. RNA helicase-regulated processing of the Synechocystis rimO-crhR operon results in differential cistron expression and accumulation of two sRNAs. J. Biol. Chem. 2020, 295, 6372–6386. [Google Scholar] [CrossRef] [Green Version]
- Rosana, A.R.; Chamot, D.; Owttrim, G.W. Autoregulation of RNA helicase expression in response to temperature stress in Synechocystis sp. PCC 6803. PLoS ONE 2012, 7, e48683. [Google Scholar] [CrossRef] [Green Version]
- Tarassova, O.S.; Chamot, D.; Owttrim, G.W. Conditional, temperature-induced proteolytic regulation of cyanobacterial RNA helicase expression. J. Bacteriol. 2014, 196, 1560–1568. [Google Scholar] [CrossRef] [Green Version]
- Rosana, A.R.R.; Ventakesh, M.; Chamot, D.; Patterson-Fortin, L.M.; Tarassova, O.; Espie, G.S.; Owttrim, G.W. Inactivation of a low temperature-induced RNA helicase in Synechocystis sp. PCC 6803: Physiological and morphological consequences. Plant Cell Physiol. 2012, 53, 646–658. [Google Scholar] [CrossRef]
- Chen, W.; Fang, L.; Huang, X.; Ge, H.; Wang, J.; Wang, X.; Zhang, Y.; Sui, N.; Xu, W.; Wang, Y. Systematic identification of light-regulated cold-responsive proteome in a model cyanobacterium. J. Proteom. 2018, 179, 100–109. [Google Scholar] [CrossRef]
- Mironov, K.S.; Sidorov, R.A.; Trofimova, M.S.; Bedbenov, V.S.; Tsydendambaev, V.D.; Allakhverdiev, S.I.; Los, D.A. Light-dependent cold-induced fatty acid unsaturation, changes in membrane fluidity, and alterations in gene expression in Synechocystis. Biochim. Biophys. Acta 2012, 1817, 1352–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owttrim, G.W. RNA helicases in cyanobacteria: Biochemical and molecular approaches. Methods Enzymol. 2012, 511, 385–403. [Google Scholar] [CrossRef] [PubMed]
- Pinto, F.; Pacheco, C.C.; Ferreira, D.; Moradas-Ferreira, P.; Tamagnini, P. Selection of suitable reference genes for RT-qPCR analyses in cyanobacteria. PLoS ONE 2012, 4, e34983. [Google Scholar] [CrossRef]
- Rosana, A.R.; Whitford, D.S.; Fahlman, R.P.; Owttrim, G.W. Cyanobacterial RNA helicase CrhR localizes to the thylakoid membrane region and cosediments with degradosome and polysome complexes in Synechocystis sp. strain PCC 6803. J. Bacteriol. 2016, 198, 2089–2099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Migur, A.; Heyl, F.; Fuss, J.; Srikumar, A.; Huettel, B.; Steglich, C.; Prakash, J.S.S.; Reinhardt, R.; Backofen, R.; Owttrim, G.W.; et al. The temperature-regulated DEAD-box RNA helicase CrhR interactome: Autoregulation and photosynthesis-related transcripts. J. Exp. Bot. 2021, 72, 7564–7579. [Google Scholar] [CrossRef]
- Campbell, D.; Hurry, V.; Clarke, A.K.; Gustafsson, P.; Oquist, G. Chlorophyll fluorescence analysis of cyanobacterial photosynthesis and acclimation. Microbiol. Mol. Biol. Rev. 1998, 62, 667–683. [Google Scholar] [CrossRef] [Green Version]
- Misumi, M.; Katoh, H.; Tomo, T.; Sonoike, K. Relationship between photochemical quenching and non-photochemical quenching in six species of cyanobacteria reveals species difference in redox state and species commonality in energy dissipation. Plant Cell Physiol. 2016, 57, 1510–1517. [Google Scholar] [CrossRef]
- Schuurmans, R.M.; Matthijs, J.C.; Hellingwerf, K.J. Transition from exponential to linear photoautotrophic growth changes the physiology of Synechocystis sp. PCC 6803. Photosynth. Res. 2017, 132, 69–82. [Google Scholar] [CrossRef] [Green Version]
- El Bissati, K.; Kirilovsky, D. Regulation of psbA and psaE expression by light quality in Synechocystis species PCC 6803. A redox control mechanism. Plant Physiol. 2001, 125, 1988–2000. [Google Scholar] [CrossRef] [Green Version]
- Hughes, J.; Lamparter, T.; Mittmann, F.; Hartmann, E.; Gärtner, W.; Wilde, A.; Börner, T. A prokaryotic phytochrome. Nature 1997, 386, 663. [Google Scholar] [CrossRef]
- Park, C.M.; Kim, J.I.; Yang, S.S.; Kang, J.G.; Kang, J.H.; Shim, J.Y.; Chung, Y.H.; Park, Y.M.; Song, P.S. A second photochromic bacteriophytochrome from Synechocystis sp. PCC 6803: Spectral analysis and down-regulation by light. Biochemistry 2000, 39, 10840–10847. [Google Scholar] [CrossRef]
- Hirose, Y.; Shimada, T.; Narikawa, R.; Katayama, M.; Ikeuchi, M. Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc. Natl. Acad. Sci. USA 2008, 105, 9528–9533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.S.; Chung, Y.H.; Moon, Y.J.; Kim, C.; Watanabe, M.; Song, P.S.; Joe, C.O.; Bogorad, L.; Park, Y.M. Photomovement of the gliding cyanobacterium Synechocystis sp. PCC 6803. Photochem. Photobiol. 1999, 70, 95–102. [Google Scholar] [CrossRef]
- Wilde, A.; Fiedler, B.; Börner, T. The cyanobacterial phytochrome Cph2 inhibits phototaxis towards blue light. Mol. Microbiol. 2002, 44, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.L.; Hagen, K.D.; Chen, R.; Risser, D.D.; Ferreira, D.P.; Meeks, J.C. Genetic analysis reveals the identity of the photoreceptor for phototaxis in hormogonium filaments of Nostoc punctiforme. J. Bacteriol. 2015, 197, 782–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyystjärvi, T.; Herranen, M.; Aro, E.M. Regulation of translation elongation in cyanobacteria: Membrane targeting of the ribosome nascent-chain complexes controls the synthesis of D1 protein. Mol. Microbiol. 2001, 240, 476–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Jung, J.H.; Park, C.M. Light inhibits COP1-mediated degradation of ICE transcription factors to induce stomatal development in Arabidopsis. Plant Cell 2017, 29, 2817–2830. [Google Scholar] [CrossRef] [Green Version]
- Park, E.; Kim, Y.; Choi, G. Phytochrome B requires PIF degradation and sequestration to induce light responses across a wide range of light conditions. Plant Cell 2018, 30, 1277–1292. [Google Scholar] [CrossRef]
- Boehm, M.; Yu, J.; Krynicka, V.; Barker, M.; Tichy, M.; Komenda, J.; Nixon, P.J.; Nield, J. Subunit organization of a Synechocystis hetero-oligomeric thylakoid FtsH complex involved in photosystem II repair. Plant Cell 2012, 24, 3669–3683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baier, A.; Winkler, W.; Korte, T.; Lockau, W.; Karradt, A. Degradation of phycobilisomes in Synechocystis sp. PCC6803: Evidence for essential formation of an NblA1/NblA2 heterodimer and its codegradation by A Clp protease complex. J. Biol. Chem. 2014, 289, 11755–11766. [Google Scholar] [CrossRef] [PubMed]
- Köbler, C.; Schultz, S.J.; Kopp, D.; Voigt, K.; Wilde, A. The role of the Synechocystis sp. PCC 6803 homolog of the circadian clock output regulator RpaA in day-night transitions. Mol. Microbiol. 2018, 110, 847–861. [Google Scholar] [CrossRef] [PubMed]
- Welkie, D.G.; Rubin, B.E.; Chang, Y.G.; Diamond, S.; Rifkin, S.A.; LiWang, A.; Golden, S.S. Genome-wide fitness assessment during diurnal growth reveals an expanded role of the cyanobacterial circadian clock protein KaiA. Proc. Natl. Acad. Sci. USA 2018, 115, E7174–E7183. [Google Scholar] [CrossRef] [PubMed]
- Ritter, S.P.A.; Brand, L.A.; Owttrim, G.W. Redox poise of the electron transport chain is sensed through QB and transmitted by Hik8-RpaA-RpaB signaling to regulate CrhR RNA helicase accumulation in Synechocystis PCC 6803. Cells, 2022; submitted. [Google Scholar]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ritter, S.P.A.; Brand, L.A.; Vincent, S.L.; Rosana, A.R.R.; Lewis, A.C.; Whitford, D.S.; Owttrim, G.W. Multiple Light-Dark Signals Regulate Expression of the DEAD-Box RNA Helicase CrhR in Synechocystis PCC 6803. Cells 2022, 11, 3397. https://doi.org/10.3390/cells11213397
Ritter SPA, Brand LA, Vincent SL, Rosana ARR, Lewis AC, Whitford DS, Owttrim GW. Multiple Light-Dark Signals Regulate Expression of the DEAD-Box RNA Helicase CrhR in Synechocystis PCC 6803. Cells. 2022; 11(21):3397. https://doi.org/10.3390/cells11213397
Chicago/Turabian StyleRitter, Sean P. A., Logan A. Brand, Shelby L. Vincent, Albert Remus R. Rosana, Allison C. Lewis, Denise S. Whitford, and George W. Owttrim. 2022. "Multiple Light-Dark Signals Regulate Expression of the DEAD-Box RNA Helicase CrhR in Synechocystis PCC 6803" Cells 11, no. 21: 3397. https://doi.org/10.3390/cells11213397




