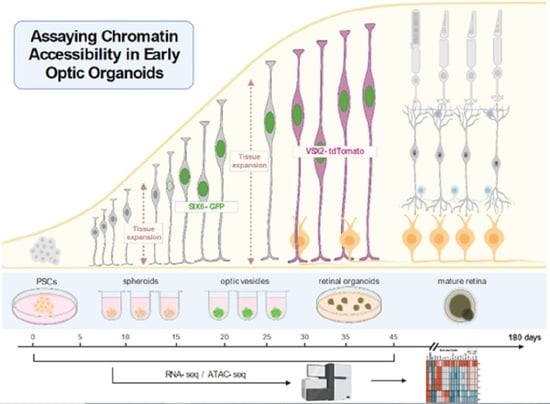
Chromatin Accessibility and Transcriptional Differences in Human Stem Cell-Derived Early-Stage Retinal Organoids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture of Pluripotent Stem Cells
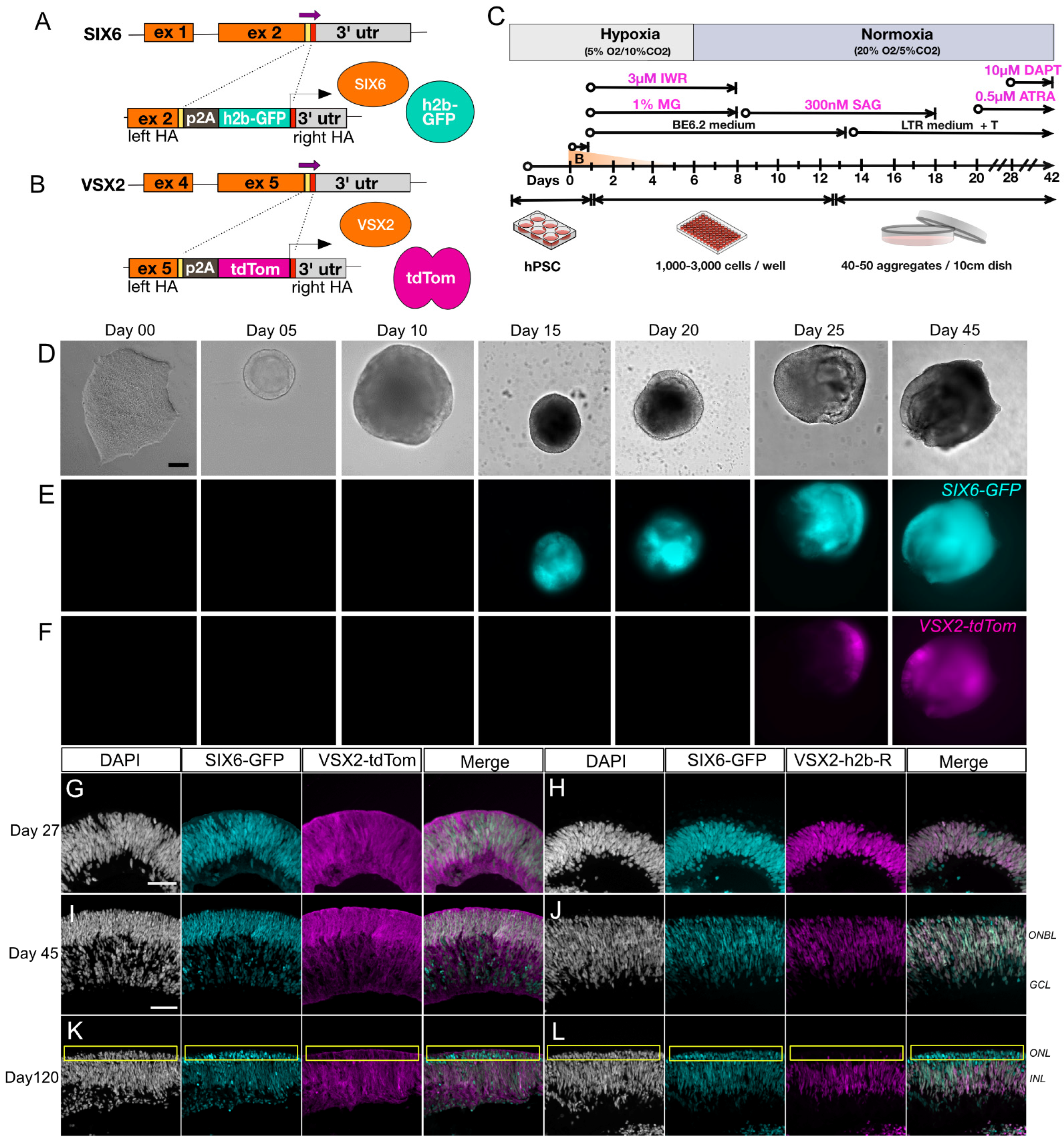
2.2. Engineering Cell Reporters
2.3. Generation of Retinal Organoids
2.4. Flow Cytometry
2.5. Sample Collection
2.6. Preparation and Imaging of Sectioned Organoids
2.7. qRT-PCR
2.8. Nuclei Isolation and ATAC-seq Libraries
2.9. RNA Isolation and RNA-seq Libraries
2.10. ATAC-seq and RNA-seq Data Analyses
3. Results
3.1. Generation of a Dual Fluorescent Reporter hPSC Line for Tracking Retinal Development
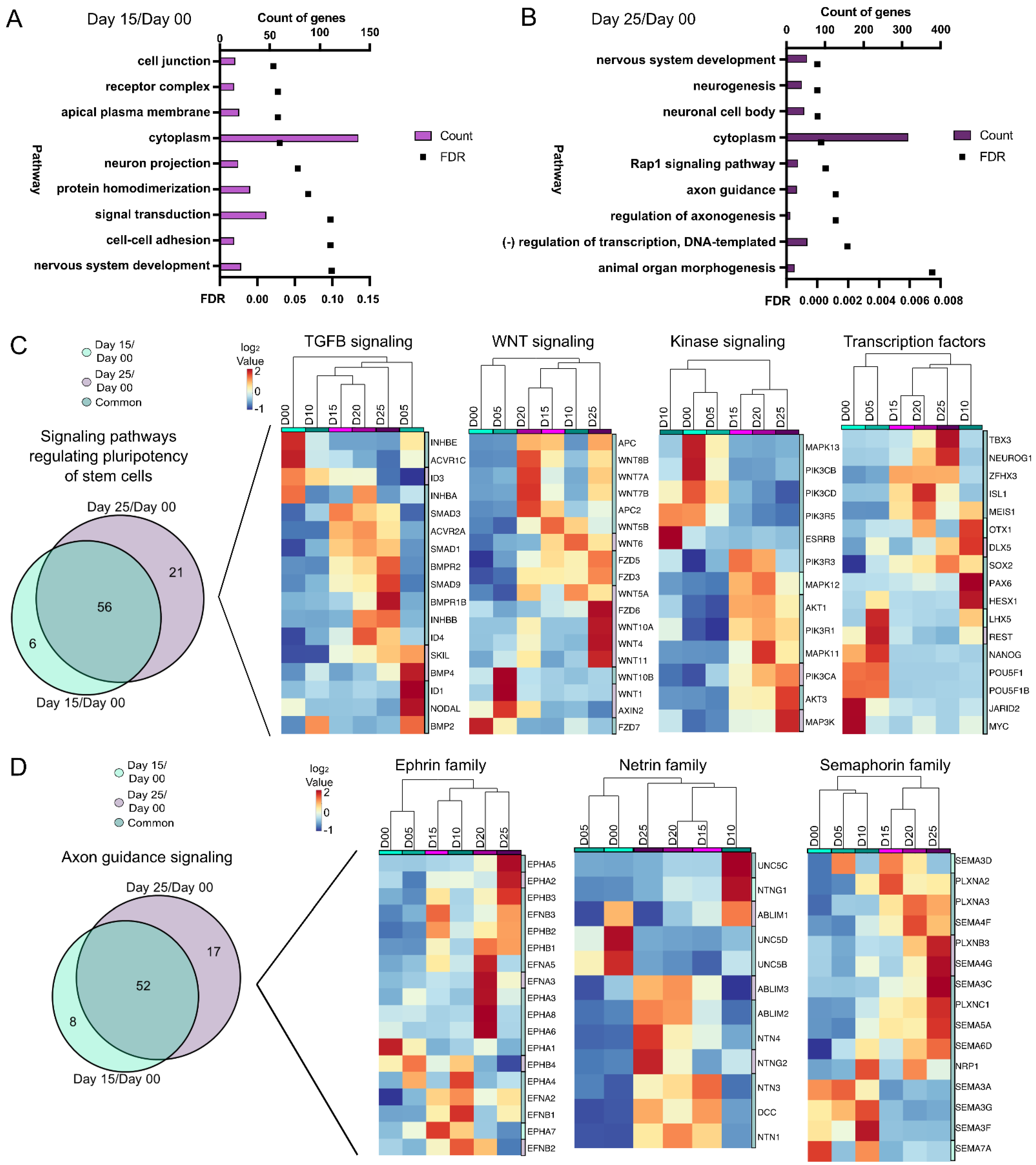
3.2. Chromatin Accessibility and Gene Expression Changes during the Stem Cell to Retinal Organoid Transition
3.3. Signaling Pathways during Optic Vesicle Differentiation
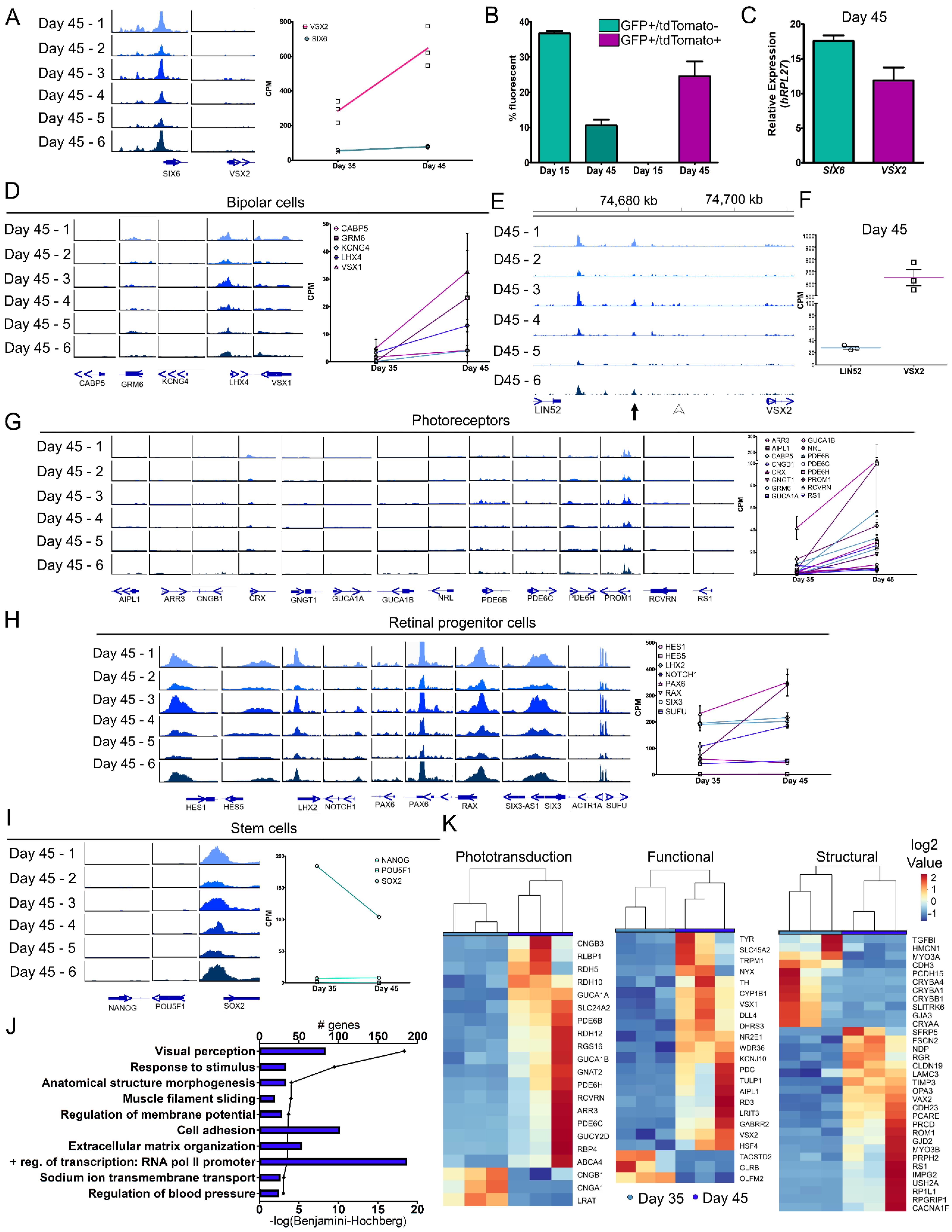
3.4. Molecular Differences during Early Retinal Cell Specification
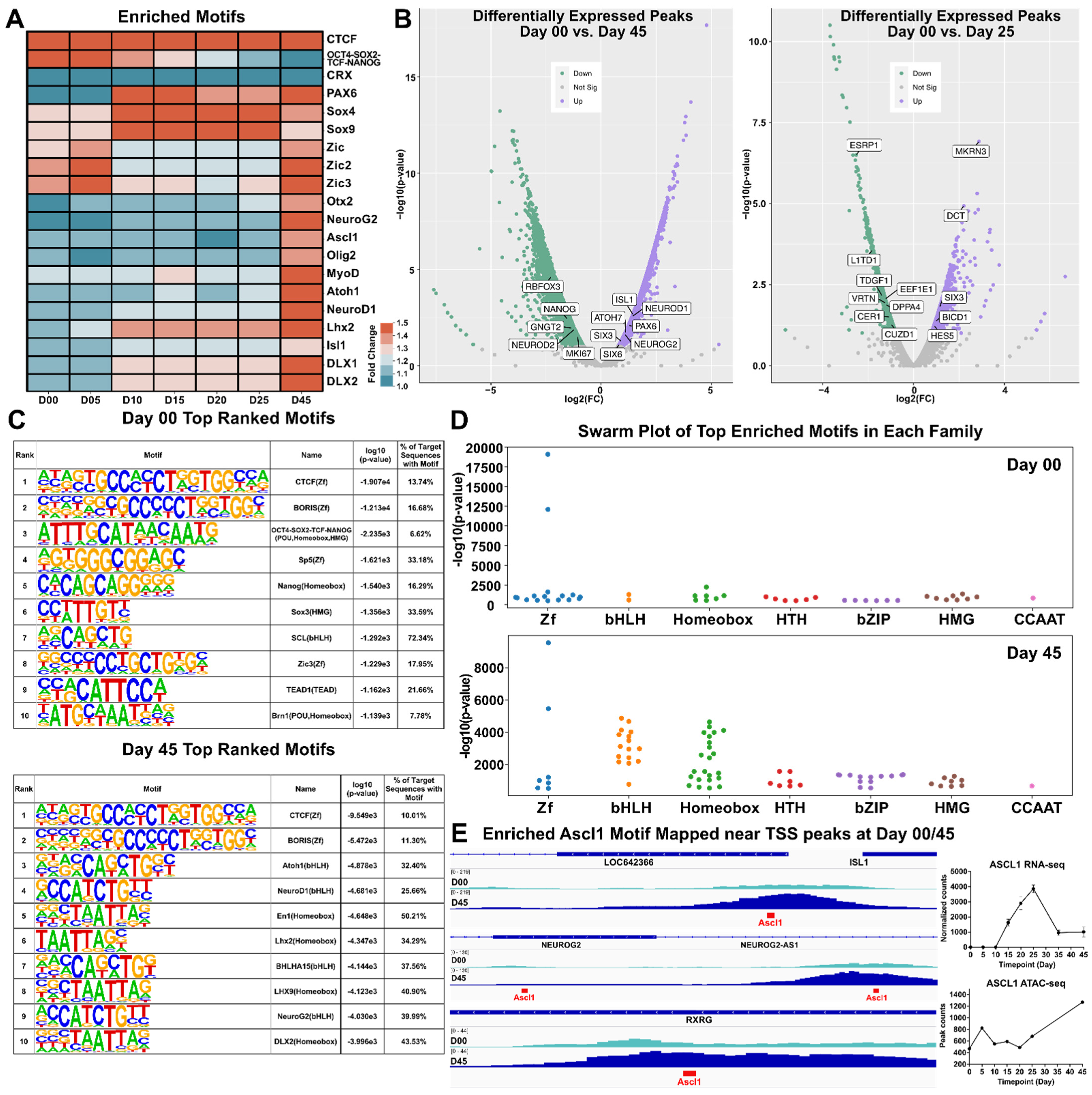
3.5. Motif Discovery and Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artero Castro, A.; Rodriguez Jimenez, F.J.; Jendelova, P.; Erceg, S. Deciphering retinal diseases through the generation of three dimensional stem cell-derived organoids: Concise Review. Stem Cells 2019, 37, 1496–1504. [Google Scholar] [CrossRef] [Green Version]
- Eiraku, M.; Sasai, Y. Mouse embryonic stem cell culture for generation of three-dimensional retinal and cortical tissues. Nat. Protoc. 2011, 7, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Mellough, C.B.; Collin, J.; Queen, R.; Hilgen, G.; Dorgau, B.; Zerti, D.; Felemban, M.; White, K.; Sernagor, E.; Lako, M. Systematic Comparison of Retinal Organoid Differentiation from Human Pluripotent Stem Cells Reveals Stage Specific, Cell Line, and Methodological Differences. Stem Cells Transl. Med. 2019, 8, 694–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahlin, K.J.; Maruotti, J.A.; Sripathi, S.R.; Ball, J.; Angueyra, J.M.; Kim, C.; Grebe, R.; Li, W.; Jones, B.W.; Zack, D.J. Photoreceptor Outer Segment-like Structures in Long-Term 3D Retinas from Human Pluripotent Stem Cells. Sci. Rep. 2017, 7, 766. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.Y.; Kaya, K.D.; Dong, L.; Swaroop, A. Three-dimensional retinal organoids from mouse pluripotent stem cells mimic in vivo development with enhanced stratification and rod photoreceptor differentiation. Mol. Vis. 2016, 22, 1077–1094. [Google Scholar]
- Kaewkhaw, R.; Kaya, K.D.; Brooks, M.; Homma, K.; Zou, J.; Chaitankar, V.; Rao, M.; Swaroop, A. Transcriptome Dynamics of Developing Photoreceptors in Three-Dimensional Retina Cultures Recapitulates Temporal Sequence of Human Cone and Rod Differentiation Revealing Cell Surface Markers and Gene Networks. Stem Cells 2015, 33, 3504–3518. [Google Scholar] [CrossRef]
- Volkner, M.; Zschatzsch, M.; Rostovskaya, M.; Overall, R.W.; Busskamp, V.; Anastassiadis, K.; Karl, M.O. Retinal Organoids from Pluripotent Stem Cells Efficiently Recapitulate Retinogenesis. Stem Cell Rep. 2016, 6, 525–538. [Google Scholar] [CrossRef] [Green Version]
- Lam, P.T.; Gutierrez, C.; Del Rio-Tsonis, K.; Robinson, M.L. Generation of a Retina Reporter hiPSC Line to Label Progenitor, Ganglion, and Photoreceptor Cell Types. Transl. Vis. Sci. Technol. 2020, 9, 21. [Google Scholar] [CrossRef] [Green Version]
- Wahlin, K.J.; Cheng, J.; Jurlina, S.L.; Jones, M.K.; Dash, N.R.; Ogata, A.; Kibria, N.; Ray, S.; Eldred, K.C.; Kim, C.; et al. CRISPR Generated SIX6 and POU4F2 Reporters Allow Identification of Brain and Optic Transcriptional Differences in Human PSC-Derived Organoids. Front. Cell Dev. Biol. 2021, 9, 764725. [Google Scholar] [CrossRef]
- Phillips, M.J.; Jiang, P.; Howden, S.; Barney, P.; Min, J.; York, N.W.; Chu, L.F.; Capowski, E.E.; Cash, A.; Jain, S.; et al. A Novel Approach to Single Cell RNA-Sequence Analysis Facilitates In Silico Gene Reporting of Human Pluripotent Stem Cell-Derived Retinal Cell Types. Stem Cells 2018, 36, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.J.; Capowski, E.E.; Petersen, A.; Jansen, A.D.; Barlow, K.; Edwards, K.L.; Gamm, D.M. Generation of a rod-specific NRL reporter line in human pluripotent stem cells. Sci. Rep. 2018, 8, 2370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langouet-Astrie, C.J.; Meinsen, A.L.; Grunwald, E.R.; Turner, S.D.; Enke, R.A. RNA sequencing analysis of the developing chicken retina. Sci. Data 2016, 3, 160117. [Google Scholar] [CrossRef] [PubMed]
- Laboissonniere, L.A.; Martin, G.M.; Goetz, J.J.; Bi, R.; Pope, B.; Weinand, K.; Ellson, L.; Fru, D.; Lee, M.; Wester, A.K.; et al. Single cell transcriptome profiling of developing chick retinal cells. J. Comp. Neurol. 2017, 525, 2735–2781. [Google Scholar] [CrossRef] [Green Version]
- Clark, B.S.; Stein-O’Brien, G.L.; Shiau, F.; Cannon, G.H.; Davis-Marcisak, E.; Sherman, T.; Santiago, C.P.; Hoang, T.V.; Rajaii, F.; James-Esposito, R.E.; et al. Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification. Neuron 2019, 102, 1111–1126.e5. [Google Scholar] [CrossRef]
- Hoshino, A.; Ratnapriya, R.; Brooks, M.J.; Chaitankar, V.; Wilken, M.S.; Zhang, C.; Starostik, M.R.; Gieser, L.; La Torre, A.; Nishio, M.; et al. Molecular Anatomy of the Developing Human Retina. Dev. Cell 2017, 43, 763–779.e4. [Google Scholar] [CrossRef] [Green Version]
- Schumacker, S.T.; Coppage, K.R.; Enke, R.A. RNA sequencing analysis of the human retina and associated ocular tissues. Sci. Data 2020, 7, 199. [Google Scholar] [CrossRef]
- Sridhar, A.; Hoshino, A.; Finkbeiner, C.R.; Chitsazan, A.; Dai, L.; Haugan, A.K.; Eschenbacher, K.M.; Jackson, D.L.; Trapnell, C.; Bermingham-McDonogh, O.; et al. Single-Cell Transcriptomic Comparison of Human Fetal Retina, hPSC-Derived Retinal Organoids, and Long-Term Retinal Cultures. Cell Rep. 2020, 30, 1644–1659.e4. [Google Scholar] [CrossRef] [PubMed]
- Welby, E.; Lakowski, J.; Di Foggia, V.; Budinger, D.; Gonzalez-Cordero, A.; Lun, A.T.L.; Epstein, M.; Patel, A.; Cuevas, E.; Kruczek, K.; et al. Isolation and Comparative Transcriptome Analysis of Human Fetal and iPSC-Derived Cone Photoreceptor Cells. Stem Cell Rep. 2017, 9, 1898–1915. [Google Scholar] [CrossRef] [Green Version]
- Hughes, A.E.; Enright, J.M.; Myers, C.A.; Shen, S.Q.; Corbo, J.C. Cell Type-Specific Epigenomic Analysis Reveals a Uniquely Closed Chromatin Architecture in Mouse Rod Photoreceptors. Sci. Rep. 2017, 7, 43184. [Google Scholar] [CrossRef] [Green Version]
- Mo, A.; Luo, C.; Davis, F.P.; Mukamel, E.A.; Henry, G.L.; Nery, J.R.; Urich, M.A.; Picard, S.; Lister, R.; Eddy, S.R.; et al. Epigenomic landscapes of retinal rods and cones. elife 2016, 5, e11613. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.P.; Hughes, A.E.; Lawrence, K.A.; Myers, C.A.; Corbo, J.C. Cis-regulatory basis of sister cell type divergence in the vertebrate retina. elife 2019, 8, e48216. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Zhang, W.; Zhang, M.; Akhtar, T.; Li, Y.; Yi, W.; Sun, X.; Zuo, Z.; Wei, M.; Fang, X.; et al. Chromatin accessibility analysis reveals regulatory dynamics of developing human retina and hiPSC-derived retinal organoids. Sci. Adv. 2020, 6, eaay5247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drexler, H.G.; Uphoff, C.C. Mycoplasma contamination of cell cultures: Incidence, sources, effects, detection, elimination, prevention. Cytotechnology 2002, 39, 75–90. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Frankish, A.; Diekhans, M.; Ferreira, A.M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010, 11, R25. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Duttke, S.H.; Chang, M.W.; Heinz, S.; Benner, C. Identification and dynamic quantification of regulatory elements using total RNA. Genome Res. 2019, 29, 1836–1846. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdottir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Ortiz, J.A.; Taing, L.; Meyer, C.A.; Lee, B.; Zhang, Y.; Shin, H.; Wong, S.S.; Ma, J.; Lei, Y.; et al. Cistrome: An integrative platform for transcriptional regulation studies. Genome Biol. 2011, 12, R83. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Rios, J.; Tessmar, K.; Loosli, F.; Wittbrodt, J.; Bovolenta, P. Six3 and Six6 activity is modulated by members of the groucho family. Development 2003, 130, 185–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toy, J.; Yang, J.M.; Leppert, G.S.; Sundin, O.H. The optx2 homeobox gene is expressed in early precursors of the eye and activates retina-specific genes. Proc. Natl. Acad. Sci. USA 1998, 95, 10643–10648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burmeister, M.; Novak, J.; Liang, M.Y.; Basu, S.; Ploder, L.; Hawes, N.L.; Vidgen, D.; Hoover, F.; Goldman, D.; Kalnins, V.I.; et al. Ocular retardation mouse caused by Chx10 homeobox null allele: Impaired retinal progenitor proliferation and bipolar cell differentiation. Nat. Genet. 1996, 12, 376–384. [Google Scholar] [CrossRef]
- Thomas, K.C.; Zheng, X.F.; Garces Suarez, F.; Raftery, J.M.; Quinlan, K.G.; Yang, N.; North, K.N.; Houweling, P.J. Evidence based selection of commonly used RT-qPCR reference genes for the analysis of mouse skeletal muscle. PLoS ONE 2014, 9, e88653. [Google Scholar] [CrossRef] [Green Version]
- Alldredge, A.; Fuhrmann, S. Loss of Axin2 Causes Ocular Defects During Mouse Eye Development. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5253–5262. [Google Scholar] [CrossRef] [Green Version]
- Bowman, A.N.; van Amerongen, R.; Palmer, T.D.; Nusse, R. Lineage tracing with Axin2 reveals distinct developmental and adult populations of Wnt/beta-catenin-responsive neural stem cells. Proc. Natl. Acad. Sci. USA 2013, 110, 7324–7329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhou, J.; Fang, Z.; Jiang, M.; Chen, X. Noggin versus basic fibroblast growth factor on the differentiation of human embryonic stem cells. Neural Regen Res. 2013, 8, 2171–2177. [Google Scholar] [CrossRef] [PubMed]
- Messina, A.; Incitti, T.; Bozza, A.; Bozzi, Y.; Casarosa, S. Noggin Expression in the Adult Retina Suggests a Conserved Role during Vertebrate Evolution. J. Histochem. Cytochem. 2014, 62, 532–540. [Google Scholar] [CrossRef] [Green Version]
- Fagoonee, S.; Bearzi, C.; Di Cunto, F.; Clohessy, J.G.; Rizzi, R.; Reschke, M.; Tolosano, E.; Provero, P.; Pandolfi, P.P.; Silengo, L.; et al. The RNA binding protein ESRP1 fine-tunes the expression of pluripotency-related factors in mouse embryonic stem cells. PLoS ONE 2013, 8, e72300. [Google Scholar] [CrossRef]
- Cepko, C.L.; Austin, C.P.; Yang, X.; Alexiades, M.; Ezzeddine, D. Cell fate determination in the vertebrate retina. Proc. Natl. Acad. Sci. USA 1996, 93, 589–595. [Google Scholar] [CrossRef] [Green Version]
- Livesey, R.; Cepko, C. Neurobiology. Developing order. Nature 2001, 413, 471–473. [Google Scholar] [CrossRef]
- Diri, I.A.; Valentine, M.; Xu, B.; Putnam, D.; Griffiths, L.; Lupo, M.; Norrie, J.; Zhang, J.; Johnson, D.; Easton, J.; et al. The Nucleome of Developing Murine Rod Photoreceptors. bioRxiv 2018, 369702. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.S.; Matsuda, T.; Cepko, C.L. A core paired-type and POU homeodomain-containing transcription factor program drives retinal bipolar cell gene expression. J. Neurosci. 2008, 28, 7748–7764. [Google Scholar] [CrossRef] [PubMed]
- Norrie, J.L.; Lupo, M.S.; Xu, B.; Al Diri, I.; Valentine, M.; Putnam, D.; Griffiths, L.; Zhang, J.; Johnson, D.; Easton, J.; et al. Nucleome Dynamics during Retinal Development. Neuron 2019, 104, 512–528. [Google Scholar] [CrossRef] [PubMed]
- Livne-Bar, I.; Pacal, M.; Cheung, M.C.; Hankin, M.; Trogadis, J.; Chen, D.; Dorval, K.M.; Bremner, R. Chx10 is required to block photoreceptor differentiation but is dispensable for progenitor proliferation in the postnatal retina. Proc. Natl. Acad. Sci. USA 2006, 103, 4988–4993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, M.; Chen, Y.; Yang, M.; Guo, A.; Xu, Y.; Xu, L.; Koeffler, H.P. dbCoRC: A database of core transcriptional regulatory circuitries modeled by H3K27ac ChIP-seq signals. Nucleic Acids Res. 2018, 46, D71–D77. [Google Scholar] [CrossRef] [Green Version]
- Capowski, E.E.; Wright, L.S.; Liang, K.; Phillips, M.J.; Wallace, K.; Petelinsek, A.; Hagstrom, A.; Pinilla, I.; Borys, K.; Lien, J.; et al. Regulation of WNT Signaling by VSX2 During Optic Vesicle Patterning in Human Induced Pluripotent Stem Cells. Stem Cells 2016, 34, 2625–2634. [Google Scholar] [CrossRef]
- Sadasivam, S.; DeCaprio, J.A. The DREAM complex: Master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer 2013, 13, 585–595. [Google Scholar] [CrossRef] [Green Version]
- Lewis, P.W.; Sahoo, D.; Geng, C.; Bell, M.; Lipsick, J.S.; Botchan, M.R. Drosophila lin-52 acts in opposition to repressive components of the Myb-MuvB/dREAM complex. Mol. Cell Biol. 2012, 32, 3218–3227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaya, K.D.; Chen, H.Y.; Brooks, M.J.; Kelley, R.A.; Shimada, H.; Nagashima, K.; de Val, N.; Drinnan, C.T.; Gieser, L.; Kruczek, K.; et al. Transcriptome-based molecular staging of human stem cell-derived retinal organoids uncovers accelerated photoreceptor differentiation by 9-cis retinal. bioRxiv 2019, 733071. [Google Scholar] [CrossRef] [Green Version]
- Ortmann, D.; Vallier, L. Variability of human pluripotent stem cell lines. Curr. Opin. Genet. Dev. 2017, 46, 179–185. [Google Scholar] [CrossRef]
- Chichagova, V.; Dorgau, B.; Felemban, M.; Georgiou, M.; Armstrong, L.; Lako, M. Differentiation of Retinal Organoids from Human Pluripotent Stem Cells. Curr. Protoc. Stem Cell Biol. 2019, 50, e95. [Google Scholar] [CrossRef] [PubMed]
- Hallam, D.; Hilgen, G.; Dorgau, B.; Zhu, L.; Yu, M.; Bojic, S.; Hewitt, P.; Schmitt, M.; Uteng, M.; Kustermann, S.; et al. Human-Induced Pluripotent Stem Cells Generate Light Responsive Retinal Organoids with Variable and Nutrient-Dependent Efficiency. Stem Cells 2018, 36, 1535–1551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaner, N.C.; Steinbach, P.A.; Tsien, R.Y. A guide to choosing fluorescent proteins. Nat. Methods 2005, 2, 905–909. [Google Scholar] [CrossRef]
- Hunt, N.C.; Hallam, D.; Karimi, A.; Mellough, C.B.; Chen, J.; Steel, D.H.W.; Lako, M. 3D culture of human pluripotent stem cells in RGD-alginate hydrogel improves retinal tissue development. Acta Biomater. 2017, 49, 329–343. [Google Scholar] [CrossRef] [Green Version]
- Yan, W.; Peng, Y.R.; van Zyl, T.; Regev, A.; Shekhar, K.; Juric, D.; Sanes, J.R. Cell Atlas of The Human Fovea and Peripheral Retina. Sci. Rep. 2020, 10, 9802. [Google Scholar] [CrossRef]
- Taylor, S.M.; Alvarez-Delfin, K.; Saade, C.J.; Thomas, J.L.; Thummel, R.; Fadool, J.M.; Hitchcock, P.F. The bHLH Transcription Factor NeuroD Governs Photoreceptor Genesis and Regeneration Through Delta-Notch Signaling. Investig. Ophthalmol. Vis. Sci. 2015, 56, 7496–7515. [Google Scholar] [CrossRef]
- Graham, V.; Khudyakov, J.; Ellis, P.; Pevny, L. SOX2 functions to maintain neural progenitor identity. Neuron 2003, 39, 749–765. [Google Scholar] [CrossRef] [Green Version]
- Brafman, D.A.; Moya, N.; Allen-Soltero, S.; Fellner, T.; Robinson, M.; McMillen, Z.L.; Gaasterland, T.; Willert, K. Analysis of SOX2-expressing cell populations derived from human pluripotent stem cells. Stem Cell Rep. 2013, 1, 464–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodato, M.A.; Ng, C.W.; Wamstad, J.A.; Cheng, A.W.; Thai, K.K.; Fraenkel, E.; Jaenisch, R.; Boyer, L.A. SOX2 co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specify cell state. PLoS Genet. 2013, 9, e1003288. [Google Scholar] [CrossRef] [Green Version]
- Heavner, W.E.; Andoniadou, C.L.; Pevny, L.H. Establishment of the neurogenic boundary of the mouse retina requires cooperation of SOX2 and WNT signaling. Neural Dev. 2014, 9, 27. [Google Scholar] [CrossRef] [Green Version]
- Hagey, D.W.; Klum, S.; Kurtsdotter, I.; Zaouter, C.; Topcic, D.; Andersson, O.; Bergsland, M.; Muhr, J. SOX2 regulates common and specific stem cell features in the CNS and endoderm derived organs. PLoS Genet. 2018, 14, e1007224. [Google Scholar] [CrossRef] [PubMed]
- Amador-Arjona, A.; Cimadamore, F.; Huang, C.T.; Wright, R.; Lewis, S.; Gage, F.H.; Terskikh, A.V. SOX2 primes the epigenetic landscape in neural precursors enabling proper gene activation during hippocampal neurogenesis. Proc. Natl. Acad. Sci. USA 2015, 112, E1936–E1945. [Google Scholar] [CrossRef] [Green Version]
- Fuhrmann, S. Wnt signaling in eye organogenesis. Organogenesis 2008, 4, 60–67. [Google Scholar] [CrossRef] [Green Version]
- Lad, E.M.; Cheshier, S.H.; Kalani, M.Y. Wnt-signaling in retinal development and disease. Stem Cells Dev. 2009, 18, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Udeh, A.; Dvoriantchikova, G.; Carmy, T.; Ivanov, D.; Hackam, A.S. Wnt signaling induces neurite outgrowth in mouse retinal ganglion cells. Exp. Eye Res. 2019, 182, 39–43. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jones, M.K.; Agarwal, D.; Mazo, K.W.; Chopra, M.; Jurlina, S.L.; Dash, N.; Xu, Q.; Ogata, A.R.; Chow, M.; Hill, A.D.; et al. Chromatin Accessibility and Transcriptional Differences in Human Stem Cell-Derived Early-Stage Retinal Organoids. Cells 2022, 11, 3412. https://doi.org/10.3390/cells11213412
Jones MK, Agarwal D, Mazo KW, Chopra M, Jurlina SL, Dash N, Xu Q, Ogata AR, Chow M, Hill AD, et al. Chromatin Accessibility and Transcriptional Differences in Human Stem Cell-Derived Early-Stage Retinal Organoids. Cells. 2022; 11(21):3412. https://doi.org/10.3390/cells11213412
Chicago/Turabian StyleJones, Melissa K., Devansh Agarwal, Kevin W. Mazo, Manan Chopra, Shawna L. Jurlina, Nicholas Dash, Qianlan Xu, Anna R. Ogata, Melissa Chow, Alex D. Hill, and et al. 2022. "Chromatin Accessibility and Transcriptional Differences in Human Stem Cell-Derived Early-Stage Retinal Organoids" Cells 11, no. 21: 3412. https://doi.org/10.3390/cells11213412
APA StyleJones, M. K., Agarwal, D., Mazo, K. W., Chopra, M., Jurlina, S. L., Dash, N., Xu, Q., Ogata, A. R., Chow, M., Hill, A. D., Kambli, N. K., Xu, G., Sasik, R., Birmingham, A., Fisch, K. M., Weinreb, R. N., Enke, R. A., Skowronska-Krawczyk, D., & Wahlin, K. J. (2022). Chromatin Accessibility and Transcriptional Differences in Human Stem Cell-Derived Early-Stage Retinal Organoids. Cells, 11(21), 3412. https://doi.org/10.3390/cells11213412