Further Extension of Lifespan by Unc-43/CaMKII and Egl-8/PLCβ Mutations in Germline-Deficient Caenorhabditis elegans
Abstract
:1. Introduction
2. Materials and Methods
2.1. C. elegans Strains and Culture
2.2. Lifespan Analysis
2.3. Stress Resistance Assays
2.4. Growing Worms for RNA-Extraction
2.5. RNA-Extraction
2.6. RNA-Sequencing
2.7. RNA-Seq Data Analysis
2.8. Overlap of DEG-Lists
2.9. GO Term Enrichment Analysis and Collagen/Matrisome Classification of DEG-Lists
2.10. Dauer Analysis
2.11. qPCR
2.12. Statistical Analysis
3. Results
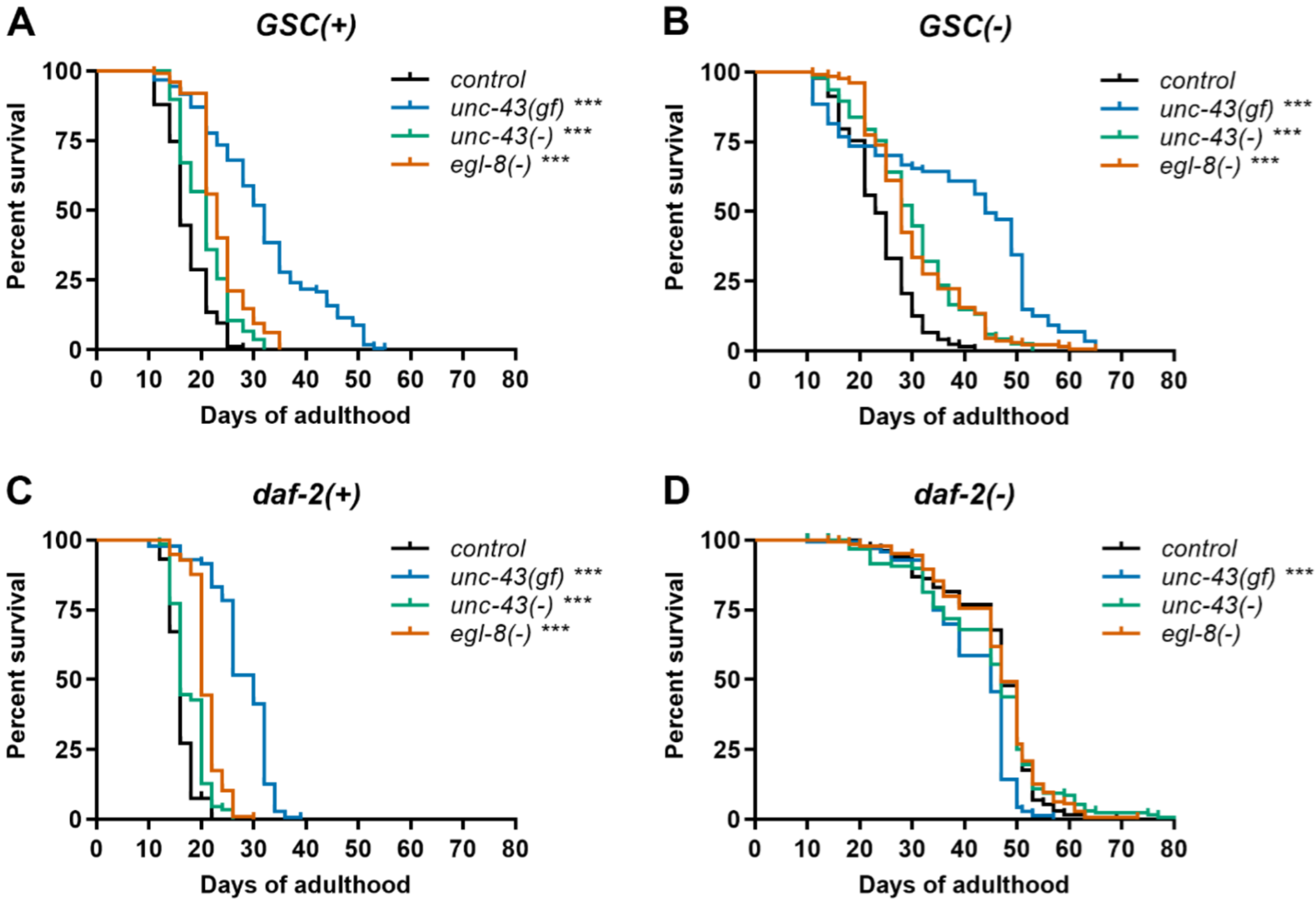
3.1. Unc-43(gf) and Unc-43(−) Further Extend Lifespan of Germline-Deficient C. elegans
3.2. Loss of Egl-8 Also Extends Lifespan of Germline-Deficient C. elegans
3.3. Unc-43(gf), Unc-43(−) and Egl-8(−) Do Not Further Extend Lifespan of Daf-2(−) Worms
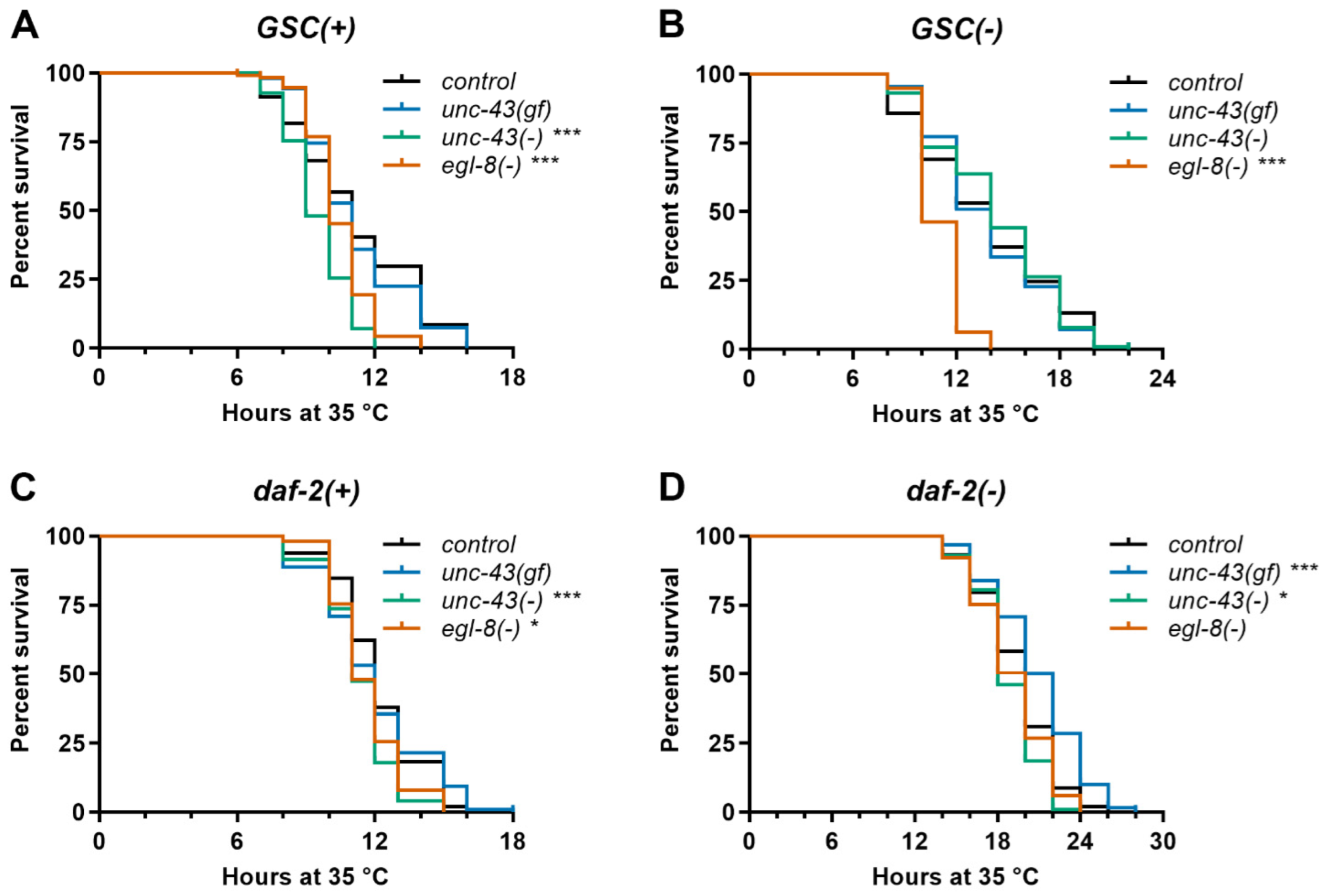
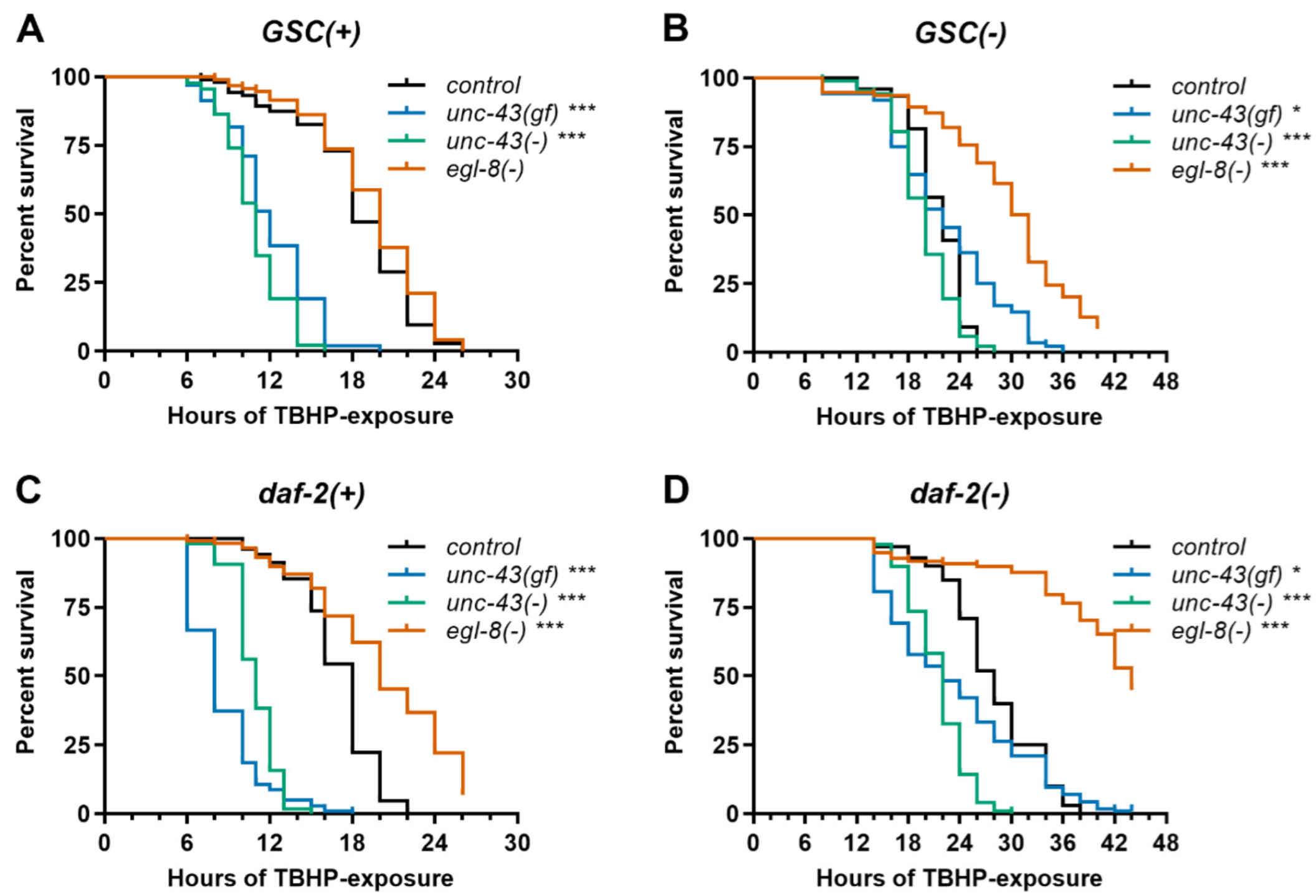
3.4. Unc-43(gf), Unc-43(−) and Egl-8(−) Differentially Affect Stress Resistance of Wildtype, GSC(−) and Daf-2(−) Worms
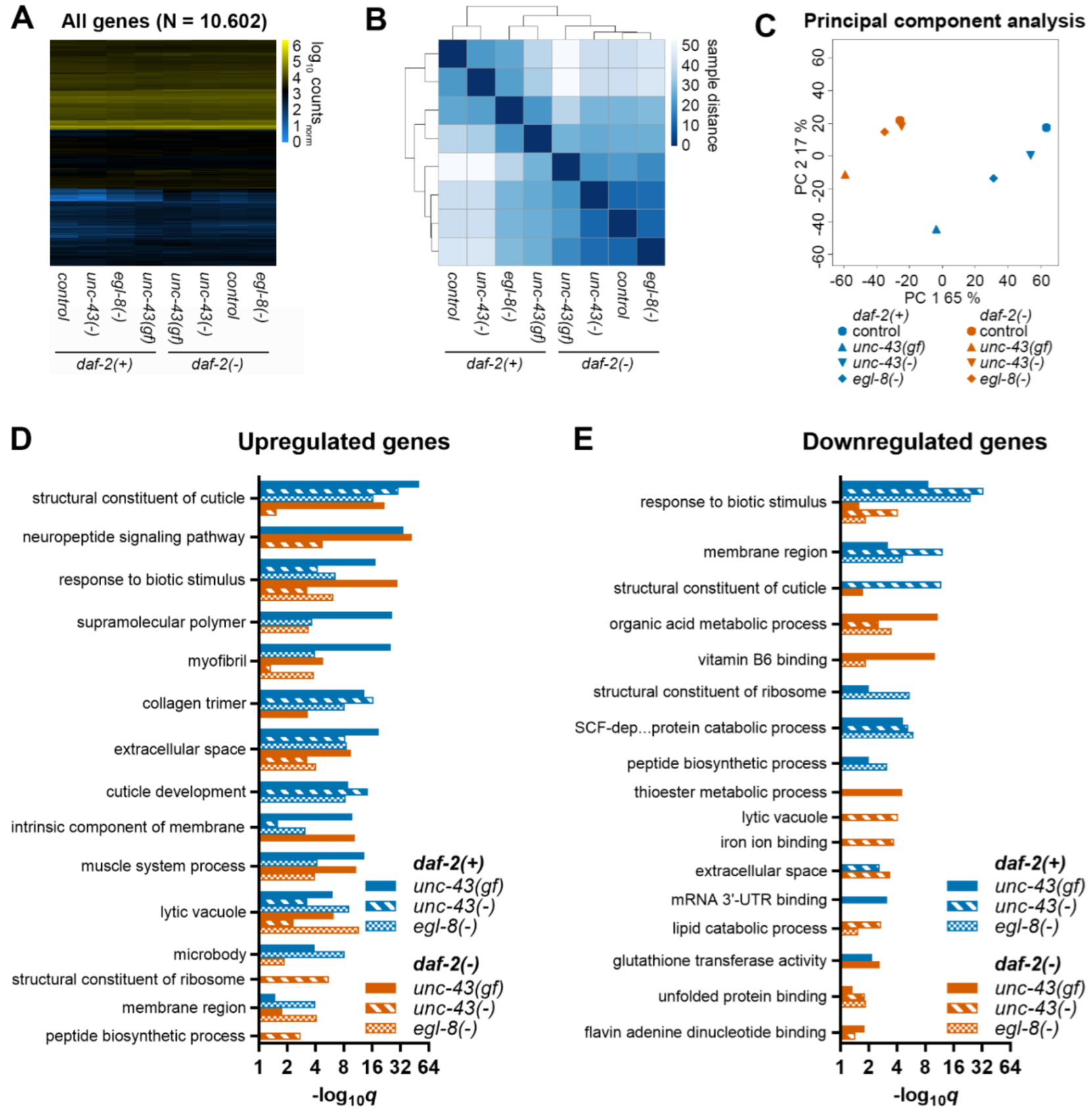
3.5. Unc-43(gf) and Egl-8(−) Globally Shift Gene Expression towards Daf-2(−)
3.6. Unc-43 and Egl-8 Regulated Genes Are Enriched for Collagen-Related Processes
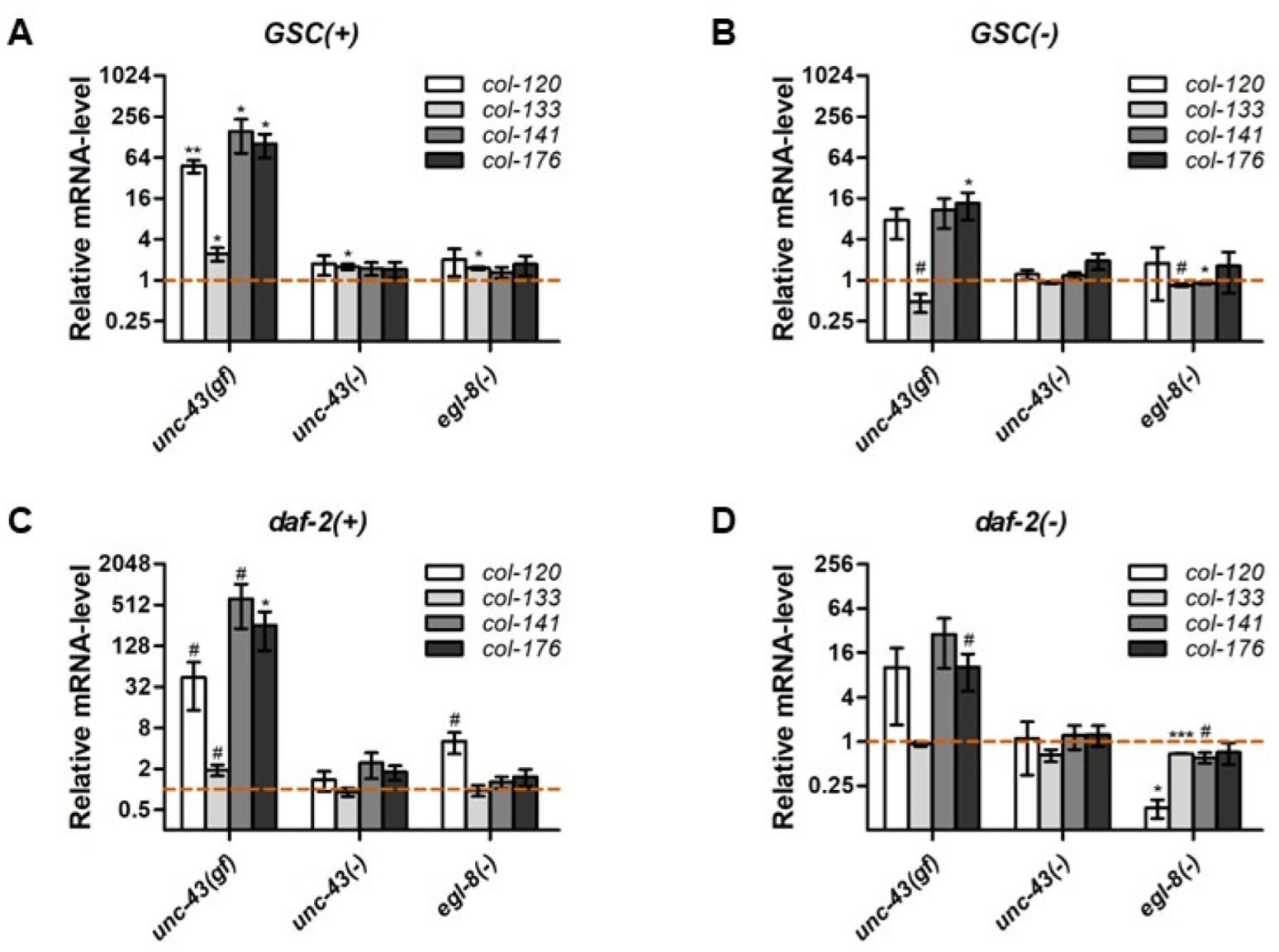
3.7. Unc-43 and Egl-8 Modulate the Expression of Non-Dauer Longevity-Associated Collagen Genes
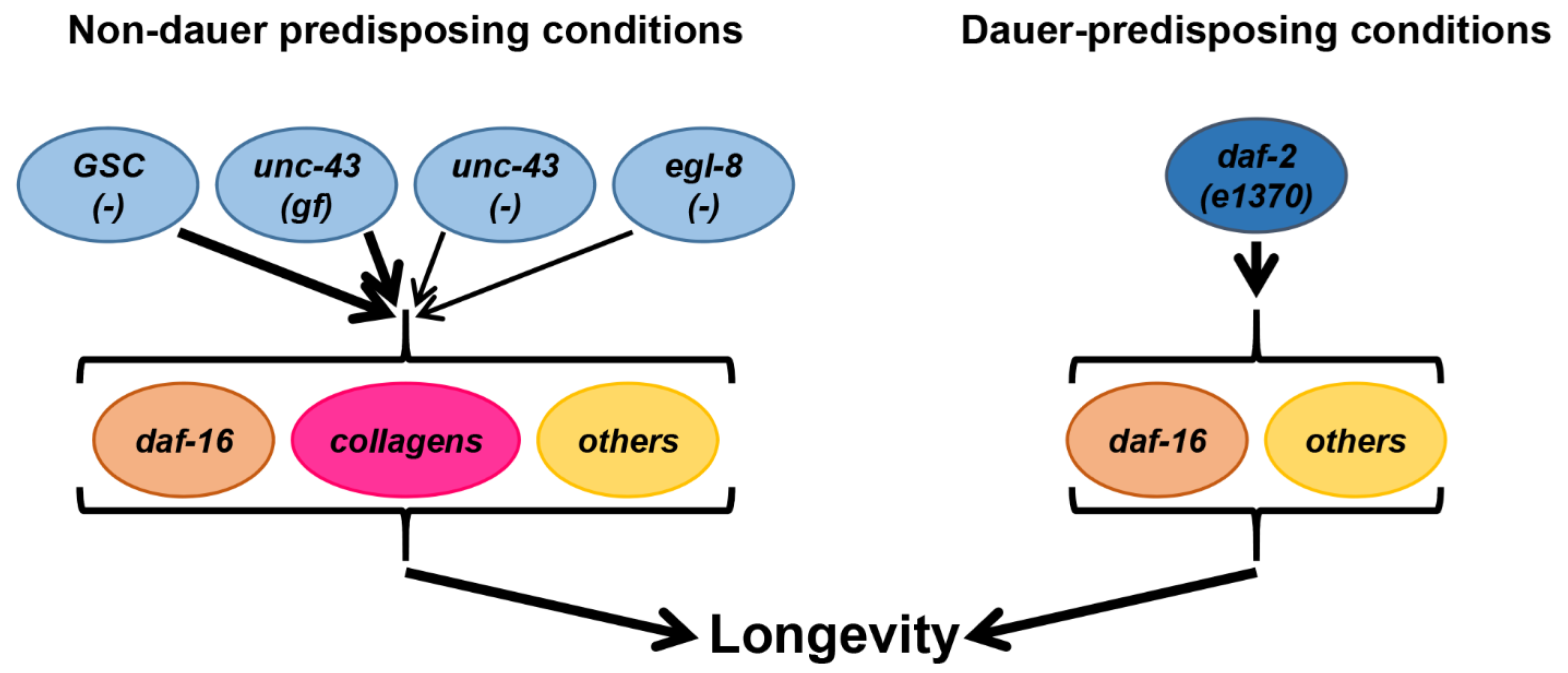
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kenyon, C.J. The genetics of ageing. Nature 2010, 464, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Morris, B.J.; Willcox, D.C.; Donlon, T.A.; Willcox, B.J. FOXO3: A Major Gene for Human Longevity—A Mini-Review. Gerontology 2015, 61, 515–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsin, H.; Kenyon, C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature 1999, 399, 362–366. [Google Scholar] [CrossRef]
- Arantes-Oliveira, N.; Apfeld, J.; Dillin, A.; Kenyon, C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science 2002, 295, 502–505. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Xie, Q.; Ding, Y.H.; Li, S.T.; Peng, S.; Zhang, Y.P.; Tan, D.; Yuan, Z.; Dong, M.Q. CAMKII and calcineurin regulate the lifespan of Caenorhabditis elegans through the FOXO transcription factor DAF-16. Elife 2013, 2, e00518. [Google Scholar] [CrossRef]
- Hansen, M.; Hsu, A.L.; Dillin, A.; Kenyon, C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005, 1, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Lapierre, L.R.; De Magalhaes Filho, C.D.; McQuary, P.R.; Chu, C.C.; Visvikis, O.; Chang, J.T.; Gelino, S.; Ong, B.; Davis, A.E.; Irazoqui, J.E.; et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 2013, 4, 2267. [Google Scholar] [CrossRef] [Green Version]
- Steinbaugh, M.J.; Narasimhan, S.D.; Robida-Stubbs, S.; Moronetti Mazzeo, L.E.; Dreyfuss, J.M.; Hourihan, J.M.; Raghavan, P.; Operana, T.N.; Esmaillie, R.; Blackwell, T.K. Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. Elife 2015, 4, e07836. [Google Scholar] [CrossRef]
- Wei, Y.; Kenyon, C. Roles for ROS and hydrogen sulfide in the longevity response to germline loss in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2016, 113, E2832–E2841. [Google Scholar] [CrossRef] [Green Version]
- Berman, J.R.; Kenyon, C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell 2006, 124, 1055–1068. [Google Scholar] [CrossRef]
- Ghazi, A.; Henis-Korenblit, S.; Kenyon, C. A transcription elongation factor that links signals from the reproductive system to lifespan extension in Caenorhabditis elegans. PLoS Genet. 2009, 5, e1000639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Libina, N.; Berman, J.R.; Kenyon, C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 2003, 115, 489–502. [Google Scholar] [CrossRef] [Green Version]
- Denzel, M.S.; Lapierre, L.R.; Mack, H.I.D. Emerging topics in C. elegans aging research: Transcriptional regulation, stress response and epigenetics. Mech. Ageing Dev. 2019, 177, 4–21. [Google Scholar] [CrossRef]
- Bayer, K.U.; Schulman, H. CaM Kinase: Still Inspiring at 40. Neuron 2019, 103, 380–394. [Google Scholar] [CrossRef]
- Baylis, H.A.; Vazquez-Manrique, R.P. Genetic analysis of IP3 and calcium signalling pathways in C. elegans. Biochim. Biophys. Acta 2012, 1820, 1253–1268. [Google Scholar] [CrossRef] [PubMed]
- Kawli, T.; Wu, C.; Tan, M.W. Systemic and cell intrinsic roles of Gqalpha signaling in the regulation of innate immunity, oxidative stress, and longevity in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2010, 107, 13788–13793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chun, L.; Gong, J.; Yuan, F.; Zhang, B.; Liu, H.; Zheng, T.; Yu, T.; Xu, X.Z.; Liu, J. Metabotropic GABA signalling modulates longevity in C. elegans. Nat. Commun. 2015, 6, 8828. [Google Scholar] [CrossRef] [Green Version]
- Ch’ng, Q.; Sieburth, D.; Kaplan, J.M. Profiling synaptic proteins identifies regulators of insulin secretion and lifespan. PLoS Genet. 2008, 4, e1000283. [Google Scholar] [CrossRef] [Green Version]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 2015, 12, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S.; Furio-Tari, P.; Turra, D.; Pietro, A.D.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res 2015, 43, e140. [Google Scholar] [CrossRef] [Green Version]
- Tarazona, S.; Garcia-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res 2011, 21, 2213–2223. [Google Scholar] [CrossRef] [Green Version]
- Dillies, M.A.; Rau, A.; Aubert, J.; Hennequet-Antier, C.; Jeanmougin, M.; Servant, N.; Keime, C.; Marot, G.; Castel, D.; Estelle, J.; et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform 2013, 14, 671–683. [Google Scholar] [CrossRef] [Green Version]
- Angeles-Albores, D.; RY, N.L.; Chan, J.; Sternberg, P.W. Tissue enrichment analysis for C. elegans genomics. BMC Bioinform. 2016, 17, 366. [Google Scholar] [CrossRef] [Green Version]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Holdorf, A.D.; Higgins, D.P.; Hart, A.C.; Boag, P.R.; Pazour, G.J.; Walhout, A.J.M.; Walker, A.K. WormCat: An Online Tool for Annotation and Visualization of Caenorhabditis elegans Genome-Scale Data. Genetics 2020, 214, 279–294. [Google Scholar] [CrossRef]
- Teuscher, A.C.; Jongsma, E.; Davis, M.N.; Statzer, C.; Gebauer, J.M.; Naba, A.; Ewald, C.Y. The in-silico characterization of the Caenorhabditis elegans matrisome and proposal of a novel collagen classification. Matrix Biol Plus 2019, 1, 100001. [Google Scholar] [CrossRef]
- Hoogewijs, D.; Houthoofd, K.; Matthijssens, F.; Vandesompele, J.; Vanfleteren, J.R. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol 2008, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, D.; Smith, M.A.; Zhang, B.; Pan, X. Selection of reliable reference genes in Caenorhabditis elegans for analysis of nanotoxicity. PLoS ONE 2012, 7, e31849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ewald, C.Y.; Landis, J.N.; Porter Abate, J.; Murphy, C.T.; Blackwell, T.K. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature 2015, 519, 97–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gems, D.; Sutton, A.J.; Sundermeyer, M.L.; Albert, P.S.; King, K.V.; Edgley, M.L.; Larsen, P.L.; Riddle, D.L. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 1998, 150, 129–155. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Seo, K.; Hwang, W.; Koo, H.J.; Hahm, J.H.; Yang, J.S.; Han, S.K.; Hwang, D.; Kim, S.; Jang, S.K.; et al. RNA helicase HEL-1 promotes longevity by specifically activating DAF-16/FOXO transcription factor signaling in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2015, 112, E4246–E4255. [Google Scholar] [CrossRef] [Green Version]
- Senchuk, M.M.; Dues, D.J.; Schaar, C.E.; Johnson, B.K.; Madaj, Z.B.; Bowman, M.J.; Winn, M.E.; Van Raamsdonk, J.M. Activation of DAF-16/FOXO by reactive oxygen species contributes to longevity in long-lived mitochondrial mutants in Caenorhabditis elegans. PLoS Genet. 2018, 14, e1007268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, A.T.; Guo, C.; Itani, O.A.; Budaitis, B.G.; Williams, T.W.; Hopkins, C.E.; McEachin, R.C.; Pande, M.; Grant, A.R.; Yoshina, S.; et al. Longevity Genes Revealed by Integrative Analysis of Isoform-Specific daf-16/FoxO Mutants of Caenorhabditis elegans. Genetics 2015, 201, 613–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaletsky, R.; Lakhina, V.; Arey, R.; Williams, A.; Landis, J.; Ashraf, J.; Murphy, C.T. The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature 2016, 529, 92–96. [Google Scholar] [CrossRef] [Green Version]
- Murphy, C.T.; McCarroll, S.A.; Bargmann, C.I.; Fraser, A.; Kamath, R.S.; Ahringer, J.; Li, H.; Kenyon, C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 2003, 424, 277–283. [Google Scholar] [CrossRef]
- Tepper, R.G.; Ashraf, J.; Kaletsky, R.; Kleemann, G.; Murphy, C.T.; Bussemaker, H.J. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell 2013, 154, 676–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiner, D.J.; Newton, E.M.; Tian, H.; Thomas, J.H. Diverse behavioural defects caused by mutations in Caenorhabditis elegans unc-43 CaM kinase II. Nature 1999, 402, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.N.; Corkins, M.E.; Li, J.C.; Singh, K.; Parsons, S.; Tucey, T.M.; Sorkac, A.; Huang, H.; Dimitriadi, M.; Sinclair, D.A.; et al. C. elegans lifespan extension by osmotic stress requires FUdR, base excision repair, FOXO, and sirtuins. Mech. Ageing Dev. 2016, 154, 30–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, H.; Fletcher, M.; Primitivo, M.; Leonard, A.; Sutphin, G.L.; Rintala, N.; Kaeberlein, M.; Leiser, S.F. Genetic interaction with temperature is an important determinant of nematode longevity. Aging Cell 2017, 16, 1425–1429. [Google Scholar] [CrossRef] [Green Version]
- Yamawaki, T.M.; Arantes-Oliveira, N.; Berman, J.R.; Zhang, P.; Kenyon, C. Distinct activities of the germline and somatic reproductive tissues in the regulation of Caenorhabditis elegans’ longevity. Genetics 2008, 178, 513–526. [Google Scholar] [CrossRef] [Green Version]
- Fraser, A.G.; Kamath, R.S.; Zipperlen, P.; Martinez-Campos, M.; Sohrmann, M.; Ahringer, J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 2000, 408, 325–330. [Google Scholar] [CrossRef]
- Kamath, R.S.; Martinez-Campos, M.; Zipperlen, P.; Fraser, A.G.; Ahringer, J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001, 2, RESEARCH0002. [Google Scholar] [CrossRef] [Green Version]
- Kenyon, C.; Chang, J.; Gensch, E.; Rudner, A.; Tabtiang, R. A C. elegans mutant that lives twice as long as wild type. Nature 1993, 366, 461–464. [Google Scholar] [CrossRef]
- Oliveira, R.P.; Porter Abate, J.; Dilks, K.; Landis, J.; Ashraf, J.; Murphy, C.T.; Blackwell, T.K. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 2009, 8, 524–541. [Google Scholar] [CrossRef] [Green Version]
- Feng, H.; Ren, M.; Chen, L.; Rubin, C.S. Properties, regulation, and in vivo functions of a novel protein kinase D: Caenorhabditis elegans DKF-2 links diacylglycerol second messenger to the regulation of stress responses and life span. J. Biol. Chem. 2007, 282, 31273–31288. [Google Scholar] [CrossRef]
- Garigan, D.; Hsu, A.L.; Fraser, A.G.; Kamath, R.S.; Ahringer, J.; Kenyon, C. Genetic analysis of tissue aging in Caenorhabditis elegans: A role for heat-shock factor and bacterial proliferation. Genetics 2002, 161, 1101–1112. [Google Scholar] [CrossRef] [PubMed]
- Mack, H.I.D.; Zhang, P.; Fonslow, B.R.; Yates, J.R. The protein kinase MBK-1 contributes to lifespan extension in daf-2 mutant and germline-deficient Caenorhabditis elegans. Aging 2017, 9, 1414–1432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, E.C.; Horvitz, H.R. Mutations with dominant effects on the behavior and morphology of the nematode Caenorhabditis elegans. Genetics 1986, 113, 821–852. [Google Scholar] [CrossRef] [PubMed]
- Yook, K.; Hodgkin, J. Mos1 mutagenesis reveals a diversity of mechanisms affecting response of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics 2007, 175, 681–697. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mack, H.I.D.; Buck, L.G.; Skalet, S.; Kremer, J.; Li, H.; Mack, E.K.M. Further Extension of Lifespan by Unc-43/CaMKII and Egl-8/PLCβ Mutations in Germline-Deficient Caenorhabditis elegans. Cells 2022, 11, 3527. https://doi.org/10.3390/cells11223527
Mack HID, Buck LG, Skalet S, Kremer J, Li H, Mack EKM. Further Extension of Lifespan by Unc-43/CaMKII and Egl-8/PLCβ Mutations in Germline-Deficient Caenorhabditis elegans. Cells. 2022; 11(22):3527. https://doi.org/10.3390/cells11223527
Chicago/Turabian StyleMack, Hildegard I. D., Laura G. Buck, Sonja Skalet, Jennifer Kremer, Hao Li, and Elisabeth K. M. Mack. 2022. "Further Extension of Lifespan by Unc-43/CaMKII and Egl-8/PLCβ Mutations in Germline-Deficient Caenorhabditis elegans" Cells 11, no. 22: 3527. https://doi.org/10.3390/cells11223527




