Genomic Landscape Highlights Molecular Mechanisms Involved in Silicate Solubilization, Stress Tolerance, and Potential Growth-Promoting Activity of Bacterium Enterobacter sp. LR6
Abstract
1. Introduction
2. Materials and Methods
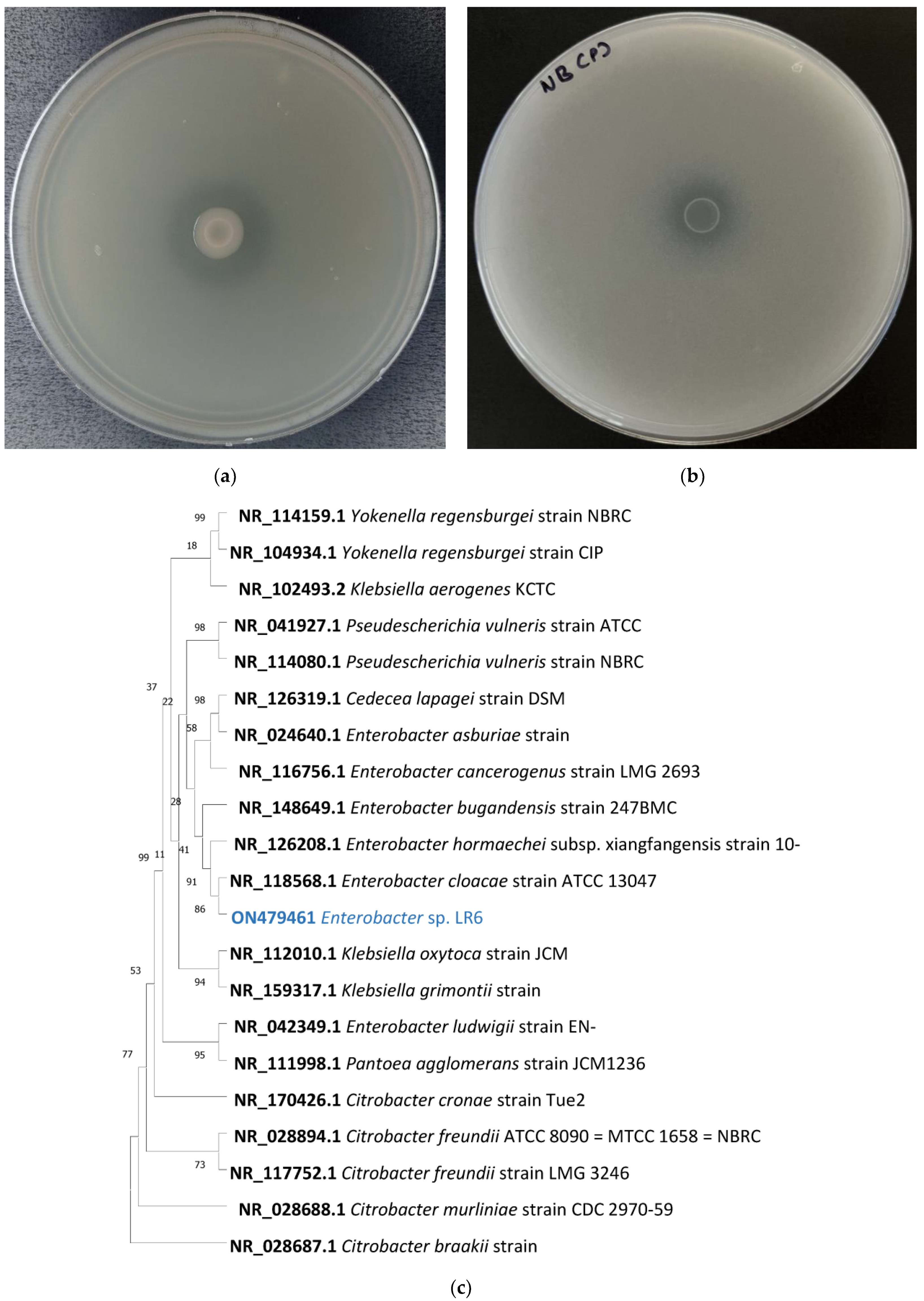
2.1. Identification and Phylogenetic Analysis of Bacterial Strain
2.2. Genome Sequencing and De Novo Assembly
2.3. Whole-Genome-Based Taxonomic Analysis
2.4. Gene Prediction and Functional Annotation
2.5. Comparative Genomic Analysis
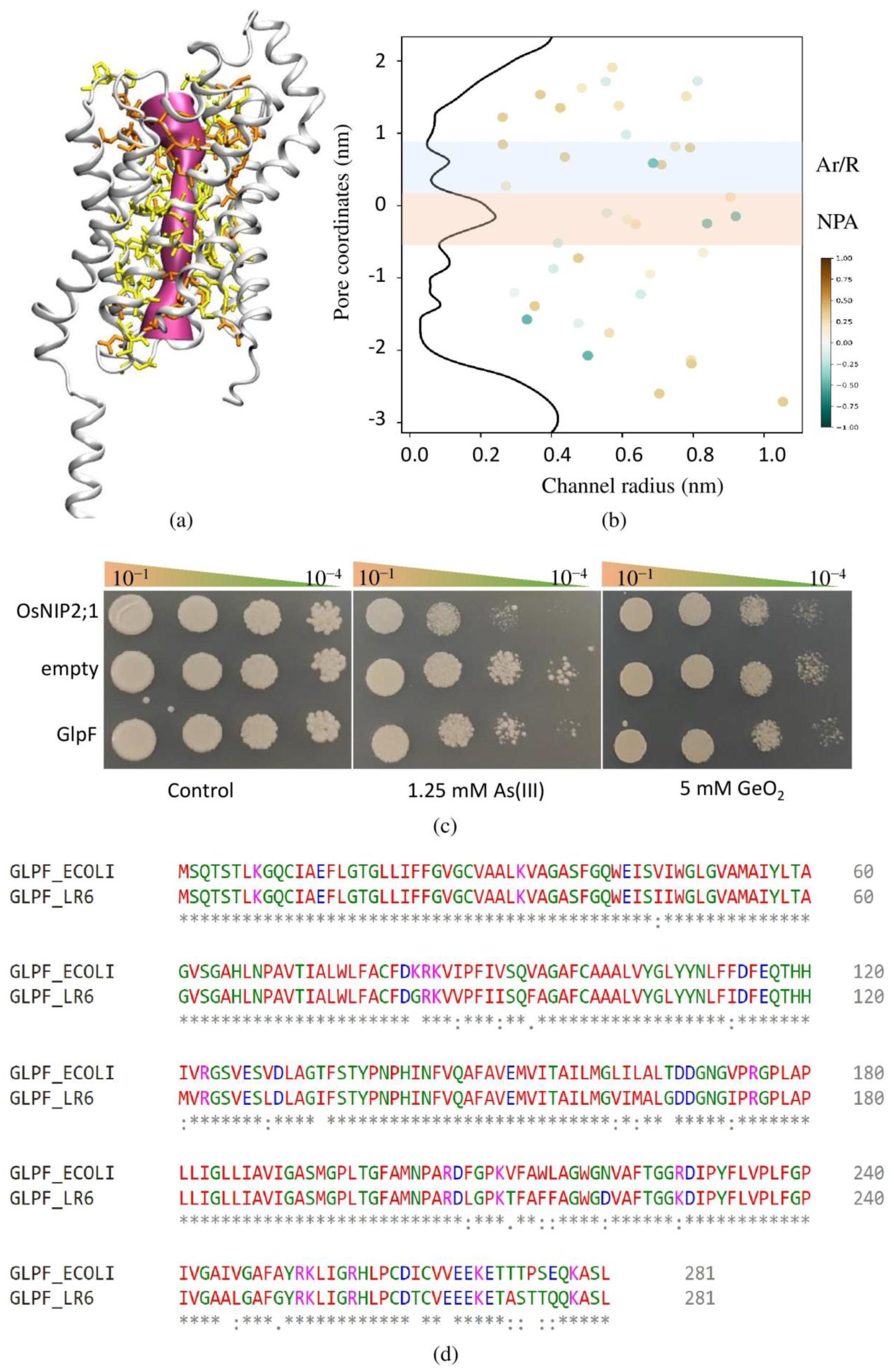
2.6. Identification, Characterization, and 3D Structure of Aquaporin
2.7. Functional Validation of GlpF for Metalloid Transportation
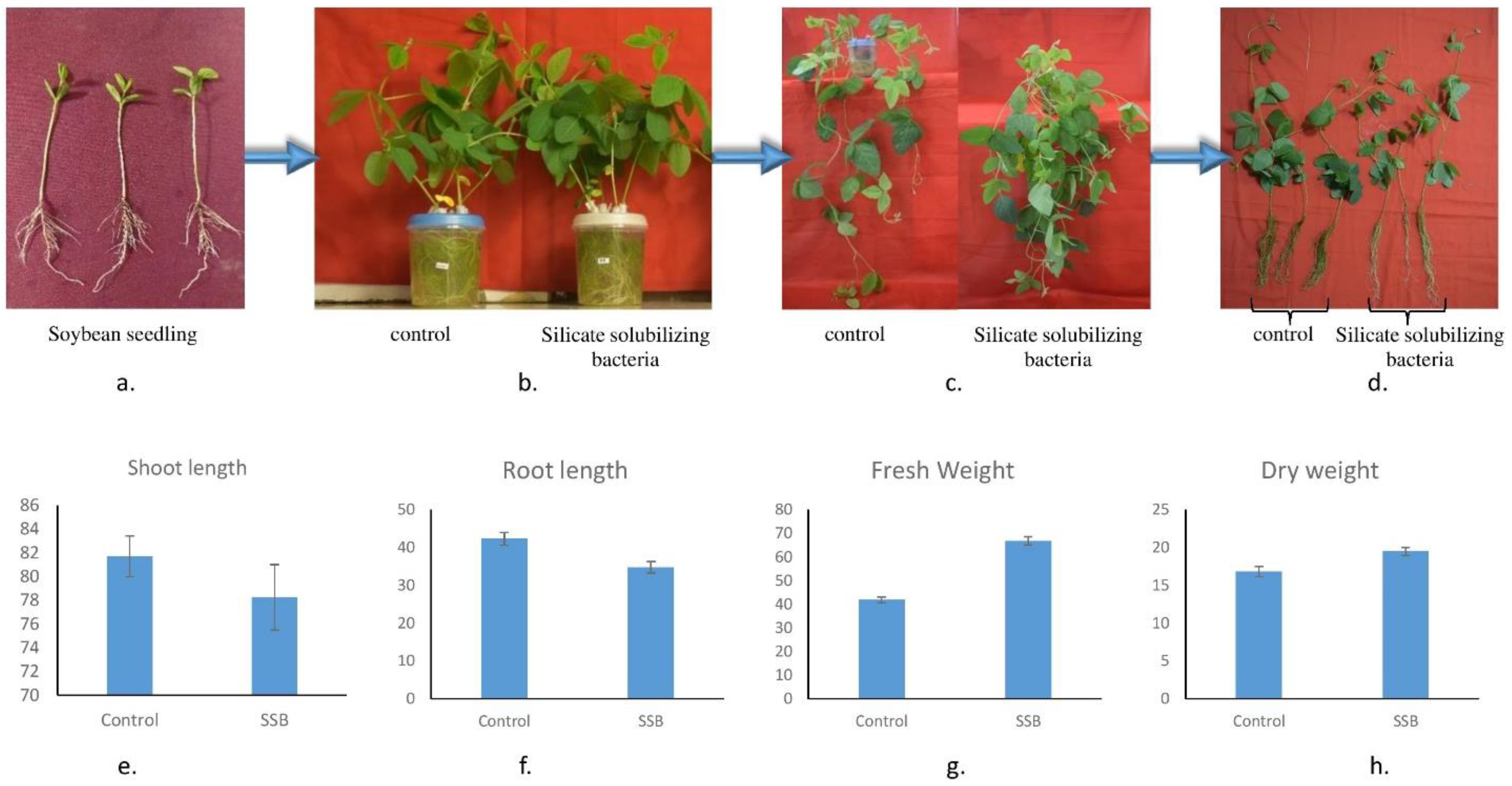
2.8. Effect of LR6 on Plant Growth
2.9. Data Availability
3. Results and Discussion
3.1. Silicate-Solubilizing Activity of Enterobacter sp. LR6
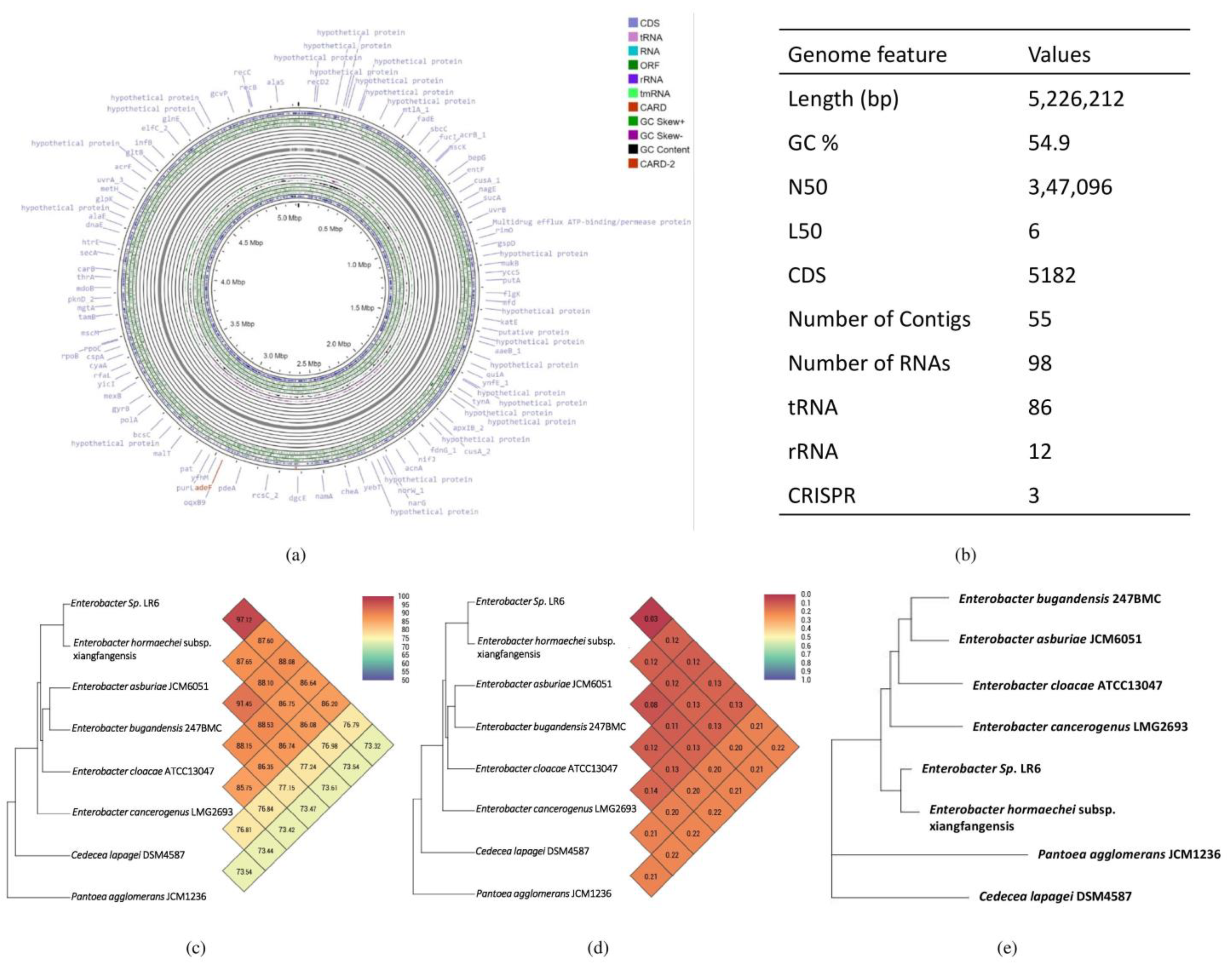
3.2. Genome Structural Feature
3.3. Phylogenetic and Taxogenomic Analysis
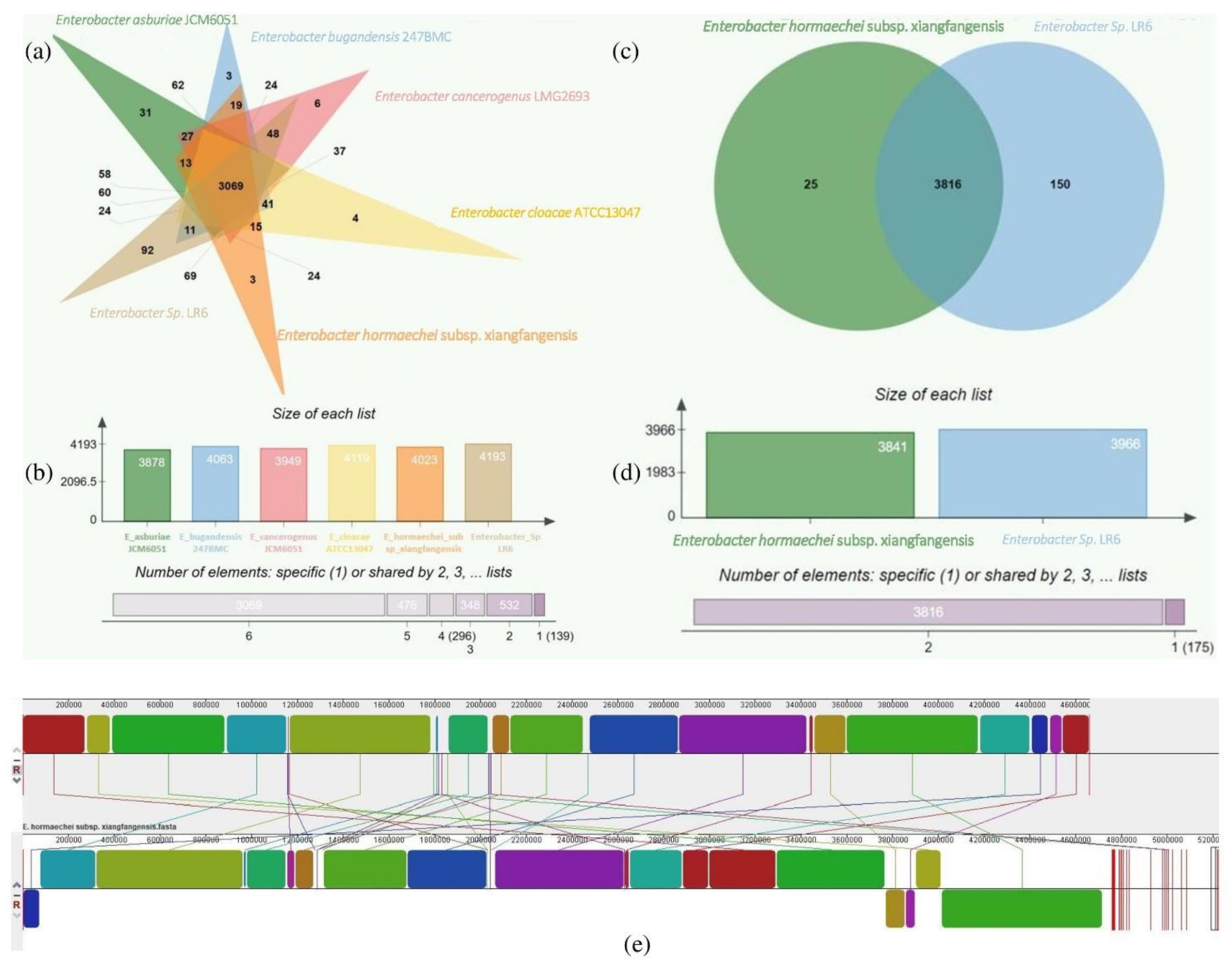
3.4. Comparative Genome Analysis
3.5. Functional Annotation of the Genome
3.6. Prediction of Gene Clusters Encoding Secondary Metabolite Stress Tolerance and Colonization-Related Protein
3.7. Genetic Potential of Enterobacter sp. LR6 for Survival in Plant Rhizosphere
3.7.1. Movements towards Rhizosphere: Motility/Chemotaxis
3.7.2. Hemagglutinin
3.7.3. Counteracting the Plant’s Defense Mechanism
3.7.4. Stress Management
3.8. Plant-Growth-Promoting Activity
3.9. Identification, Characterization, and Functional Validation of GlpF
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kang, S.-M.; Waqas, M.; Shahzad, R.; You, Y.-H.; Asaf, S.; Khan, M.A.; Lee, K.-E.; Joo, G.-J.; Kim, S.-J.; Lee, I.-J. Isolation and characterization of a novel silicate-solubilizing bacterial strain Burkholderia eburnea CS4-2 that promotes growth of japonica rice (Oryza sativa L. cv. Dongjin). Soil Sci. Plant Nutr. 2017, 63, 233–241. [Google Scholar]
- Sahebi, M.; Hanafi, M.M.; Siti Nor Akmar, A.; Rafii, M.Y.; Azizi, P.; Tengoua, F.; Nurul Mayzaitul Azwa, J.; Shabanimofrad, M. Importance of silicon and mechanisms of biosilica formation in plants. BioMed Res. Int. 2015, 2015, 396010. [Google Scholar] [CrossRef] [PubMed]
- Raturi, G.; Sharma, Y.; Rana, V.; Thakral, V.; Myaka, B.; Salvi, P.; Singh, M.; Dhar, H.; Deshmukh, R. Exploration of silicate solubilizing bacteria for sustainable agriculture and silicon biogeochemical cycle. Plant Physiol. Biochem. 2021, 166, 827–838. [Google Scholar] [CrossRef]
- Chandrakala, C.; Voleti, S.; Bandeppa, S.; Sunil Kumar, N.; Latha, P. Silicate solubilization and plant growth promoting potential of Rhizobium sp. isolated from rice rhizosphere. Silicon 2019, 11, 2895–2906. [Google Scholar] [CrossRef]
- Vasanthi, N.; Saleena, L.M.; Raj, S.A. Silicon in day today life. World Appl. Sci. J. 2012, 17, 1425–1440. [Google Scholar]
- Fauteux, F.; Rémus-Borel, W.; Menzies, J.G.; Bélanger, R.R. Silicon and plant disease resistance against pathogenic fungi. FEMS Microbiol. Lett. 2005, 249, 1–6. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Yang, Y. Mechanisms of silicon-induced fungal disease resistance in plants. Plant Physiol. Biochem. 2021, 165, 200–206. [Google Scholar] [CrossRef]
- Deshmukh, R.K.; Sonah, H.; Bélanger, R.R. Plant Aquaporins: Genome-wide identification, transcriptomics, proteomics, and advanced analytical tools. Front. Plant Sci. 2016, 7, 1896. [Google Scholar] [CrossRef]
- Raturi, G.; Kumawat, S.; Mandlik, R.; Duhan, D.; Thakral, V.; Sudhakaran, S.; Ram, C.; Sonah, H.; Deshmukh, R. Deciphering the Role of Aquaporins Under Different Abiotic Stress Conditions in Watermelon (Citrullus lanatus). J. Plant Growth Regul. 2022, 1–13. [Google Scholar] [CrossRef]
- Meli, R.; Pirozzi, C.; Pelagalli, A. New perspectives on the potential role of aquaporins (AQPs) in the physiology of inflammation. Front. Physiol. 2018, 9, 101. [Google Scholar] [CrossRef]
- Pettersson, N.; Filipsson, C.; Becit, E.; Brive, L.; Hohmann, S. Aquaporins in yeasts and filamentous fungi. Biol. Cell 2005, 97, 487–500. [Google Scholar] [CrossRef]
- Kayingo, G.; Bill, R.M.; Calamita, G.; Hohmann, S.; Prior, B.A. Microbial water channels and glycerol facilitators. Curr. Top. Membr. 2001, 51, 335–370. [Google Scholar]
- Tong, H.; Hu, Q.; Zhu, L.; Dong, X. Prokaryotic aquaporins. Cells 2019, 8, 1316. [Google Scholar] [CrossRef] [PubMed]
- Elhaissoufi, W.; Ghoulam, C.; Barakat, A.; Zeroual, Y.; Bargaz, A. Phosphate bacterial solubilization: A key rhizosphere driving force enabling higher P use efficiency and crop productivity. J. Adv. Res. 2021, 38, 13–28. [Google Scholar] [CrossRef]
- Kwak, Y.; Park, G.-S.; Shin, J.-H. High quality draft genome sequence of the type strain of Pseudomonas lutea OK2T, a phosphate-solubilizing rhizospheric bacterium. Stand. Genom. Sci. 2016, 11, 1–10. [Google Scholar] [CrossRef]
- Uroz, S.; Picard, L.; Turpault, M.-P. Recent progress in understanding the ecology and molecular genetics of soil mineral weathering bacteria. Trends Microbiol. 2022, 30, 882–897. [Google Scholar] [CrossRef] [PubMed]
- Bist, V.; Niranjan, A.; Ranjan, M.; Lehri, A.; Seem, K.; Srivastava, S. Silicon-solubilizing media and its implication for characterization of bacteria to mitigate biotic stress. Front. Plant Sci. 2020, 11, 28. [Google Scholar] [CrossRef]
- Premono, M.E.; Moawad, A.; Vlek, P. Effect of phosphate-solubilizing Pseudomonas putida on the growth of maize and its survival in the rhizosphere. Indones. J. Crop Sci. 1996, 11, 13. [Google Scholar]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 5 May 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grazziotin, A.L.; Vidal, N.M.; Hoepers, P.G.; Reis, T.F.; Mesa, D.; Caron, L.F.; Ingberman, M.; Beirão, B.C.; Zuffo, J.P.; Fonseca, B.B. Comparative genomics of a novel clade shed light on the evolution of the genus Erysipelothrix and characterise an emerging species. Sci. Rep. 2021, 11, 3383. [Google Scholar]
- Yoon, S.-H.; Ha, S.-M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Kim, Y.O.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; Van Nimwegen, E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 2014, 31, 1077–1088. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Klesse, G.; Rao, S.; Sansom, M.S.; Tucker, S.J. CHAP: A versatile tool for the structural and functional annotation of ion channel pores. J. Mol. Biol. 2019, 431, 3353–3365. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H.; Willems, A.R.; Woods, R.A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 1995, 11, 355–360. [Google Scholar] [CrossRef]
- Pommerrenig, B.; Diehn, T.A.; Bernhardt, N.; Bienert, M.D.; Mitani-Ueno, N.; Fuge, J.; Bieber, A.; Spitzer, C.; Bräutigam, A.; Ma, J.F. Functional evolution of nodulin 26-like intrinsic proteins: From bacterial arsenic detoxification to plant nutrient transport. New Phytol. 2020, 225, 1383–1396. [Google Scholar] [CrossRef]
- Santi, L.; Goenadi, D. Solubilization of silicate from quartz mineral by potential silicate solubilizing bacteria (Pelarutan silika asal mineral kuarsa oleh bakteri pelarut silika potensial). E-J. Menara Perkeb. 2017, 85, 2. [Google Scholar] [CrossRef]
- Pande, A.; Kaushik, S.; Pandey, P.; Negi, A. Isolation, characterization, and identification of phosphate-solubilizing Burkholderia cepacia from the sweet corn cv. Golden Bantam rhizosphere soil and effect on growth-promoting activities. Int. J. Veg. Sci. 2020, 26, 591–607. [Google Scholar] [CrossRef]
- Trujillo, M.E.; Bacigalupe, R.; Pujic, P.; Igarashi, Y.; Benito, P.; Riesco, R.; Medigue, C.; Normand, P. Genome features of the endophytic actinobacterium Micromonospora lupini strain Lupac 08: On the process of adaptation to an endophytic life style? PLoS ONE 2014, 9, e108522. [Google Scholar] [CrossRef]
- Lee, Z.M.-P.; Bussema III, C.; Schmidt, T.M. rrn DB: Documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 2009, 37, D489–D493. [Google Scholar] [CrossRef]
- Shrestha, P.M.; Noll, M.; Liesack, W. Phylogenetic identity, growth-response time and rRNA operon copy number of soil bacteria indicate different stages of community succession. Environ. Microbiol. 2007, 9, 2464–2474. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Annavajhala, M.K.; Gomez-Simmonds, A.; Uhlemann, A.-C. Multidrug-resistant Enterobacter cloacae complex emerging as a global, diversifying threat. Front. Microbiol. 2019, 10, 44. [Google Scholar] [CrossRef]
- Sum, R.; Swaminathan, M.; Rastogi, S.K.; Piloto, O.; Cheong, I. Beta-hemolytic bacteria selectively trigger liposome lysis, enabling rapid and accurate pathogen detection. ACS Sens. 2017, 2, 1441–1451. [Google Scholar] [CrossRef]
- Scavino, A.F.; Pedraza, R.O. The role of siderophores in plant growth-promoting bacteria. In Bacteria in Agrobiology: Crop Productivity; Springer: Berlin/Heidelberg, Germany, 2013; pp. 265–285. [Google Scholar]
- Schöner, T.A.; Gassel, S.; Osawa, A.; Tobias, N.J.; Okuno, Y.; Sakakibara, Y.; Shindo, K.; Sandmann, G.; Bode, H.B. Aryl polyenes, a highly abundant class of bacterial natural products, are functionally related to antioxidative carotenoids. ChemBioChem 2016, 17, 247–253. [Google Scholar] [CrossRef]
- Ney, B.; Ahmed, F.H.; Carere, C.R.; Biswas, A.; Warden, A.C.; Morales, S.E.; Pandey, G.; Watt, S.J.; Oakeshott, J.G.; Taylor, M.C. The methanogenic redox cofactor F420 is widely synthesized by aerobic soil bacteria. ISME J. 2017, 11, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, E.; Chauhan, S.; Sareen, D. Thiopeptides encoding biosynthetic gene clusters mined from bacterial genomes. J. Biosci. 2021, 46, 1–12. [Google Scholar] [CrossRef]
- van der Lelie, D.; Taghavi, S.; Monchy, S.; Schwender, J.; Miller, L.; Ferrieri, R.; Rogers, A.; Wu, X.; Zhu, W.; Weyens, N. Poplar and its bacterial endophytes: Coexistence and harmony. Crit. Rev. Plant Sci. 2009, 28, 346–358. [Google Scholar] [CrossRef]
- Gottig, N.; Garavaglia, B.S.; Garofalo, C.G.; Orellano, E.G.; Ottado, J. A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS ONE 2009, 4, e4358. [Google Scholar] [CrossRef] [PubMed]
- Najmuldeen, H.; Alghamdi, R.; Alghofaili, F.; Yesilkaya, H. Functional assessment of microbial superoxide dismutase isozymes suggests a differential role for each isozyme. Free. Radic. Biol. Med. 2019, 134, 215–228. [Google Scholar] [CrossRef]
- Katsuwon, J.; Anderson, A. Response of plant-colonizing pseudomonads to hydrogen peroxide. Appl. Environ. Microbiol. 1989, 55, 2985–2989. [Google Scholar] [CrossRef]
- Shea, R.J.; Mulks, M.H. ohr, Encoding an organic hydroperoxide reductase, is an in vivo-induced gene in Actinobacillus pleuropneumoniae. Infect. Immun. 2002, 70, 794–802. [Google Scholar] [CrossRef]
- Rocha, E.R.; Smith, C.J. Role of the alkyl hydroperoxide reductase (ahpCF) gene in oxidative stress defense of the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 1999, 181, 5701–5710. [Google Scholar] [CrossRef]
- Rho, B.-S.; Hung, L.-W.; Holton, J.M.; Vigil, D.; Kim, S.-I.; Park, M.S.; Terwilliger, T.C.; Pédelacq, J.-D. Functional and structural characterization of a thiol peroxidase from Mycobacterium tuberculosis. J. Mol. Biol. 2006, 361, 850–863. [Google Scholar] [CrossRef]
- Poole, R.K. Flavohaemoglobin: The pre-eminent nitric oxide–detoxifying machine of microorganisms. F1000Research 2020, 9, 7. [Google Scholar] [CrossRef]
- Gardner, A.M.; Gessner, C.R.; Gardner, P.R. Regulation of the Nitric Oxide Reduction Operon (norRVW) in Escherichia coli: ROLE OF NorR AND ς54 IN THE NITRIC OXIDE STRESS RESPONSE. J. Biol. Chem. 2003, 278, 10081–10086. [Google Scholar] [CrossRef]
- Burse, A.; Weingart, H.; Ullrich, M.S. The phytoalexin-inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora. Mol. Plant-Microbe Interact. 2004, 17, 43–54. [Google Scholar] [CrossRef]
- Ben Fekih, I.; Zhang, C.; Li, Y.P.; Zhao, Y.; Alwathnani, H.A.; Saquib, Q.; Rensing, C.; Cervantes, C. Distribution of arsenic resistance genes in prokaryotes. Front. Microbiol. 2018, 9, 2473. [Google Scholar] [CrossRef]
- Williams, C.L.; Neu, H.M.; Alamneh, Y.A.; Reddinger, R.M.; Jacobs, A.C.; Singh, S.; Abu-Taleb, R.; Michel, S.L.; Zurawski, D.V.; Merrell, D.S. Characterization of Acinetobacter baumannii copper resistance reveals a role in virulence. Front. Microbiol. 2020, 11, 16. [Google Scholar] [CrossRef]
- Thatoi, H.; Das, S.; Mishra, J.; Rath, B.P.; Das, N. Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: A review. J. Environ. Manag. 2014, 146, 383–399. [Google Scholar] [CrossRef]
- Lee, S.M.; Grass, G.; Haney, C.J.; Fan, B.; Rosen, B.P.; Anton, A.; Nies, D.H.; Rensing, C. Functional analysis of the Escherichia coli zinc transporter ZitB. FEMS Microbiol. Lett. 2002, 215, 273–278. [Google Scholar] [CrossRef]
- Osman, D.; Cooke, A.; Young, T.R.; Deery, E.; Robinson, N.J.; Warren, M.J. The requirement for cobalt in vitamin B12: A paradigm for protein metalation. Biochim. et Biophys. Acta (BBA)-Mol. Cell Res. 2021, 1868, 118896. [Google Scholar] [CrossRef]
- Dai, J.; Wei, H.; Tian, C.; Damron, F.H.; Zhou, J.; Qiu, D. An extracytoplasmic function sigma factor-dependent periplasmic glutathione peroxidase is involved in oxidative stress response of Shewanella oneidensis. BMC Microbiol. 2015, 15, 34. [Google Scholar] [CrossRef]
- Keto-Timonen, R.; Hietala, N.; Palonen, E.; Hakakorpi, A.; Lindström, M.; Korkeala, H. Cold shock proteins: A minireview with special emphasis on Csp-family of enteropathogenic Yersinia. Front. Microbiol. 2016, 7, 1151. [Google Scholar] [CrossRef]
- Maleki, F.; Khosravi, A.; Nasser, A.; Taghinejad, H.; Azizian, M. Bacterial heat shock protein activity. J. Clin. Diagn. Res. JCDR 2016, 10, BE01. [Google Scholar] [CrossRef]
- Guinote, I.B.; Moreira, R.N.; Freire, P.; Arraiano, C.M. Characterization of the BolA homolog IbaG: A new gene involved in acid resistance. J. Microbiol. Biotechnol. 2012, 22, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Shabayek, S.; Spellerberg, B. Acid stress response mechanisms of group B streptococci. Front. Cell. Infect. Microbiol. 2017, 7, 395. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, A.; Ketkar, O.A.; Irfan, S.; Rana, V.; Rahi, P.; Deshmukh, R.; Kaur, J.; Dhar, H. Genomic Insights into Omega-3 Polyunsaturated Fatty Acid Producing Shewanella sp. N2AIL from Fish Gut. Biology 2022, 11, 632. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Wei, B.; Shi, M.; Gao, H. Functional assessment of EnvZ/OmpR two-component system in Shewanella oneidensis. PLoS ONE 2011, 6, e23701. [Google Scholar] [CrossRef]
- Toender, J.; Borchert, M. Use of Enzymes Having Silicase Activity. Novozymes AS US8822188B2, 2 September 2009. [Google Scholar]
- Kaur, P.; Sharma, A.; Bhardwaj, N.K.; Singh, A.; Dalal, S.; Sharma, J. A novel, simple, and quick plate assay to screen silicolytic bacteria and silicase production using different substrates. Bioresour. Technol. Rep. 2022, 17, 100971. [Google Scholar] [CrossRef]
- Schütz, A.; Golbik, R.; Tittmann, K.; Svergun, D.I.; Koch, M.H.; Hübner, G.; König, S. Studies on structure–function relationships of indolepyruvate decarboxylase from Enterobacter cloacae, a key enzyme of the indole acetic acid pathway. Eur. J. Biochem. 2003, 270, 2322–2331. [Google Scholar] [CrossRef]
- Ramesh, A.; Sharma, S.K.; Sharma, M.P.; Yadav, N.; Joshi, O.P. Plant growth-promoting traits in Enterobacter cloacae subsp. dissolvens MDSR9 isolated from soybean rhizosphere and its impact on growth and nutrition of soybean and wheat upon inoculation. Agric. Res. 2014, 3, 53–66. [Google Scholar] [CrossRef]
- Calamita, G. The Escherichia coli aquaporin-Z water channel: MicroReview. Mol. Microbiol. 2000, 37, 254–262. [Google Scholar] [CrossRef]
- Jensen, M.Ø.; Park, S.; Tajkhorshid, E.; Schulten, K. Energetics of glycerol conduction through aquaglyceroporin GlpF. Proc. Natl. Acad. Sci. USA 2002, 99, 6731–6736. [Google Scholar] [CrossRef]
- Meng, Y.-L.; Liu, Z.; Rosen, B.P. As (III) and Sb (III) uptake by GlpF and efflux by ArsB in Escherichia coli. J. Biol. Chem. 2004, 279, 18334–18341. [Google Scholar] [CrossRef]
| Stress Type | Protein Type | Genes/Operon/Protein | Location | Function | References | |
|---|---|---|---|---|---|---|
| Oxidative stress | Superoxide dismutase | SodA (Mn superoxide dismutase); SodB (Fe superoxide dismutase); and SodC (Cu/Zn superoxide dismutase) | sequence01_2888396_2887776, sequence01_1115542_1114961, sequence01_1123281_1123799 | Provides superoxide resistance | [47] | |
| Catalase | KatE, manganese catalase, and KatG | sequence01_4550870_4549998, sequence01_3881760_3883940 | Protection against the H2O2 | [48] | ||
| Hydroperoxide reductases | Organic hydroperoxide resistance protein, organic hydroperoxide resistance transcriptional regulator | sequence01_3509831_3509403, sequence01_3510368_3509916 | Detoxification of organic hydroperoxide | [49] | ||
| Alkyl hydroperoxide reductase | Alkyl hydroperoxide reductase protein F (ahpF), alkyl hydroperoxide reductase protein C (ahpC), alkyl hydroperoxide reductase subunit C-like protein | sequence01_419756_421321, sequence01_419004_419567, sequence01_178141_177539 | Oxidative stress defense | [50] | ||
| Thiol peroxidases | Thiol peroxidase, Tpx-type, thiol peroxidase, Bcp-type | sequence01_1694758_1695264, sequence01_2514125_2514595 | Reduces t-butyl hydroperoxide, H2O2, and cumene hydroperoxide | [51] | ||
| Nitric oxide dioxygenase | Flavohemoglobin/nitric oxide dioxygenase, nitric oxide reductase FlRd-NAD(+) reductase, anaerobic nitric oxide reductase flavorubredoxin | sequence01_2597555_2598745, sequence01_4699018_4697885, sequence01_4700463_4699015 | Detoxifies free radical nitric oxide | [52] | ||
| Nitrate reductase | Anaerobic nitric oxide reductase transcription regulator NorR | sequence01_4700651_4702165 | Required for the expression of anaerobic nitric oxide (NO) reductase | [53] | ||
| RND transporters | AcrAB | sequence01_266663_267316 | Export of pytoalexins | [54] | ||
| Heavy metal resistance | Arsenic | Arsenate reductase, arsenic resistance protein, arsenite/antimonite:H+ antiporter, arsenical pump | ArsB, ArsA | sequence01_673660_672371, sequence01_4740871_4739582, sequence31_15724_17007, sequence01_4734229_4735986, sequence01_4742670_4740919 | Provides resistance to arsenic | [55] |
| copper | Copper resistance protein, copper resistance protein | CopD, Cop O, CopC, CopB, CueR | sequence01_1510825_1509896, sequence01_1931658_1930789, sequence01_1511210_1510830, sequence01_1932031_1931660, sequence01_1512146_1511250, sequence01_290196_290606 | Provides resistance to copper | [56] | |
| chromate | Chromate reductase, transport and resistance protein | ChrA, ChrB | sequence01_4722045_4720666, sequence01_4722976_4721999 | Provides resistance to chromate | [57] | |
| Cobalt, zinc and cadmium | Zinc transporter | ZitB, RcnR-like protein, RcnA, RcnB | sequence01_543130_542192, sequence01_1425411_1425686, sequence01_254854_254039, sequence01_4727409_4728539, sequence01_2243421_2243753 | Cobalt–zinc–cadmium resistance | [58,59] | |
| Temperature stress | RNA polymerase sigma factor | rpoE | sequence01_1576112_1575555, sequence01_2635482_2634907 | Regulates the degQ and support growth at low and high temperature | [60] | |
| Stress sensor | degQ | sequence01_4075345_4073978 | Enables bacteria to grow at high temperature | [60] | ||
| Csp family | CspB, CspC, CspD, CspE | sequence01_1202383_1202165, sequence01_1668932_1668744, sequence01_1913740_1913531, sequence01_730174_729953, sequence01_441915_442124 | Protects the bacteria during rapid downshift of temperature | [61] | ||
| Hsp family | HspQ, HslJ, GrpE | sequence01_824127_823810, sequence01_1671988_1672425, sequence01_3834576_3835169 | Involved in protein folding and refolding, expressed in high temperature | [62] | ||
| Acid stress | Acid stress protein | IbaG | sequence01_4123024_4123278 | Changes mRNA expression pattern to provide acid resistance | [63] | |
| F0F1-ATPase transporter | F0F1-ATPase | sequence01_2972737_2973117, sequence01_2973126_2973941, sequence01_2974764_2975297, sequence01_2975310_2976851, sequence01_2976903_2977766, sequence01_2977798_2979180, sequence01_2979201_2979620 | Induces acid tolerance response | [64] | ||
| Arginine deiminase, ornithine carbamoyltransferase | arcA, arcB | sequence01_3548646_3547930, sequence01_4107667_4110000 | Leads to production of alkaline molecules, such as ammonia to maintain pH. | [64] | ||
| Alkaline stress | Membrane-bound Na+/H+ antiporter | nhaA, nhaB, nhaP2 | sequence01_3565391_3566566, sequence01_1886487_1888025, sequence01_1879396_1881129 | A membrane-bound Na+/H+ antiporter system adapts the bacteria to alkaline stress | [65] | |
| Osmotic stress | Osmolarity sensory histidine kinase, transcriptional response regulator, OmpR family | EnvZ/OmpR two-component system | sequence01_2738070_2736724, sequence01_2738786_2738067, sequence01_685891_686598, sequence01_1485204_1485944 | Mediates osmotic stress response in a number of Gram-negative bacteria | [66] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raturi, G.; Sharma, Y.; Mandlik, R.; Kumawat, S.; Rana, N.; Dhar, H.; Tripathi, D.K.; Sonah, H.; Sharma, T.R.; Deshmukh, R. Genomic Landscape Highlights Molecular Mechanisms Involved in Silicate Solubilization, Stress Tolerance, and Potential Growth-Promoting Activity of Bacterium Enterobacter sp. LR6. Cells 2022, 11, 3622. https://doi.org/10.3390/cells11223622
Raturi G, Sharma Y, Mandlik R, Kumawat S, Rana N, Dhar H, Tripathi DK, Sonah H, Sharma TR, Deshmukh R. Genomic Landscape Highlights Molecular Mechanisms Involved in Silicate Solubilization, Stress Tolerance, and Potential Growth-Promoting Activity of Bacterium Enterobacter sp. LR6. Cells. 2022; 11(22):3622. https://doi.org/10.3390/cells11223622
Chicago/Turabian StyleRaturi, Gaurav, Yogesh Sharma, Rushil Mandlik, Surbhi Kumawat, Nitika Rana, Hena Dhar, Durgesh Kumar Tripathi, Humira Sonah, Tilak Raj Sharma, and Rupesh Deshmukh. 2022. "Genomic Landscape Highlights Molecular Mechanisms Involved in Silicate Solubilization, Stress Tolerance, and Potential Growth-Promoting Activity of Bacterium Enterobacter sp. LR6" Cells 11, no. 22: 3622. https://doi.org/10.3390/cells11223622
APA StyleRaturi, G., Sharma, Y., Mandlik, R., Kumawat, S., Rana, N., Dhar, H., Tripathi, D. K., Sonah, H., Sharma, T. R., & Deshmukh, R. (2022). Genomic Landscape Highlights Molecular Mechanisms Involved in Silicate Solubilization, Stress Tolerance, and Potential Growth-Promoting Activity of Bacterium Enterobacter sp. LR6. Cells, 11(22), 3622. https://doi.org/10.3390/cells11223622








