Label-Free Digital Holographic Microscopy for In Vitro Cytotoxic Effect Quantification of Organic Nanoparticles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Nanomaterials
2.3. Cell Preparation for Time-Lapse DHM QPI Proliferation Experiments
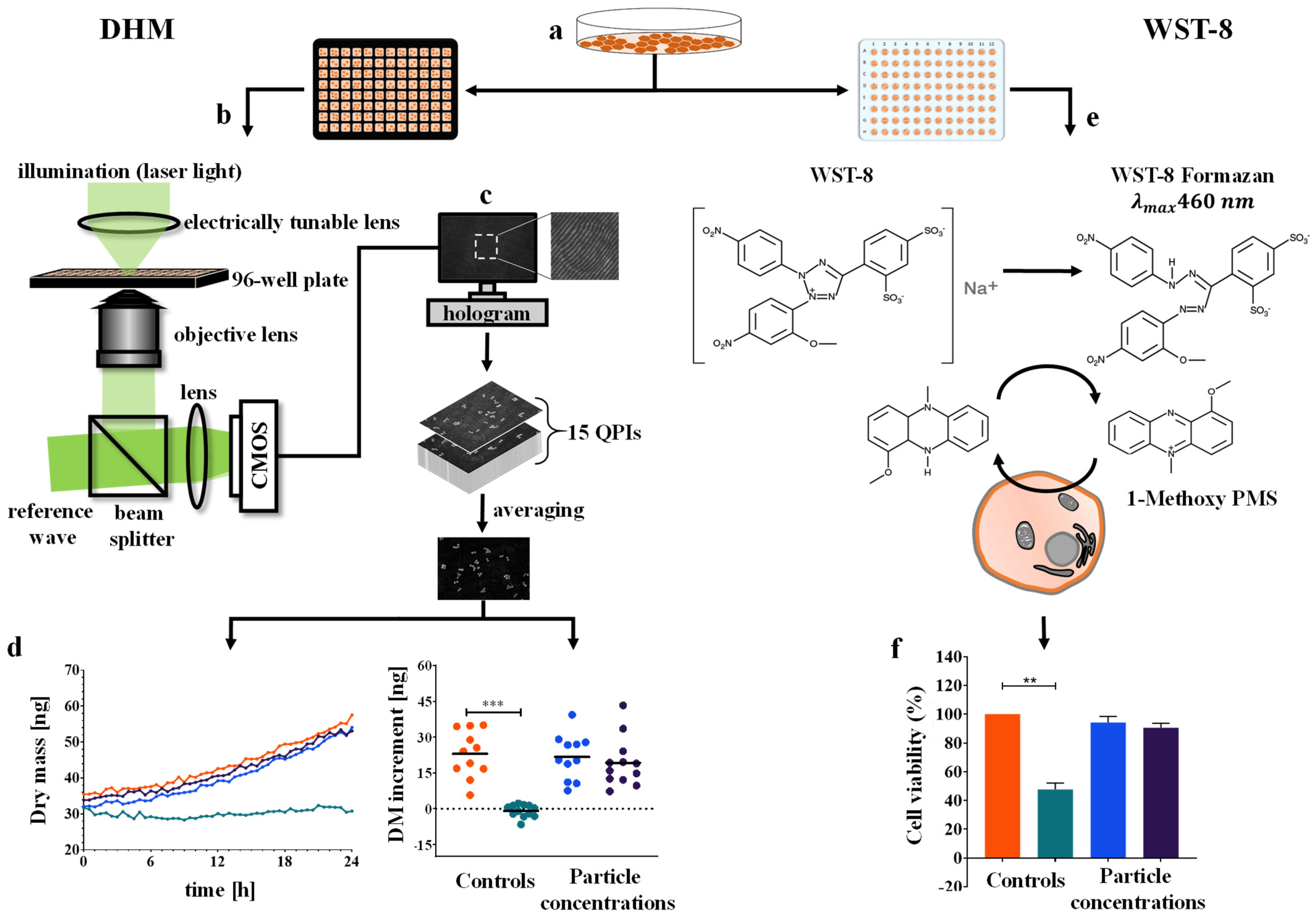
2.4. Time-Lapse QPI by DHM
2.5. Determination of the Cellular Dry Mass from Quantitative Phase Images
2.6. WST-8 Cell Viability Assay
2.7. Statistical Analysis
3. Results
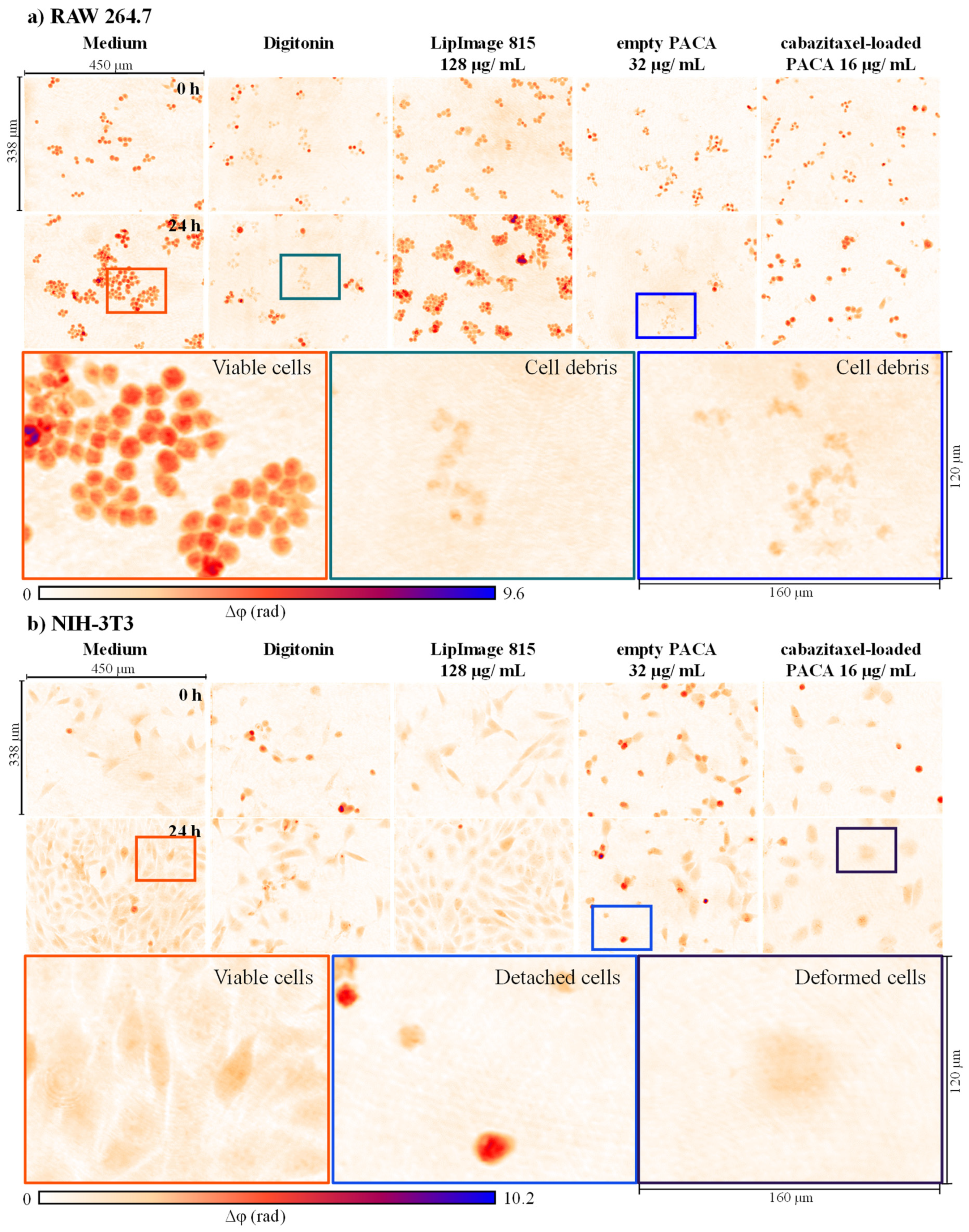
3.1. Qualitative Analysis of Cell Morphology and Proliferation Alterations in DHM QPI Images after Incubation with Nanoparticles
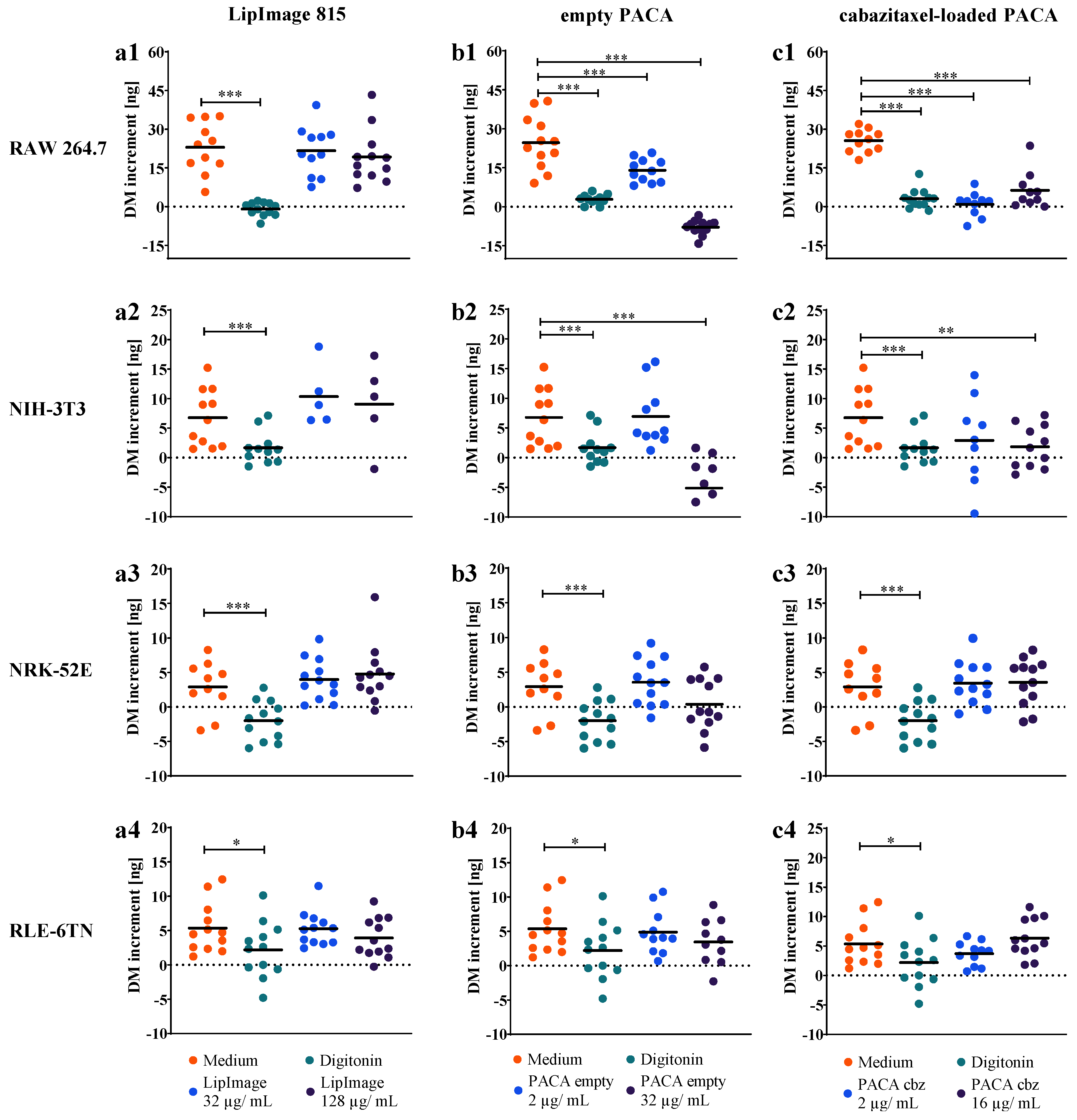
3.2. Time-Resolved Dry Mass Dynamics of Nanoparticle Incubated Cell Populations
3.3. Dry Mass Increment as Endpoint for Cell Proliferation and Nanoparticles Cytotoxicity
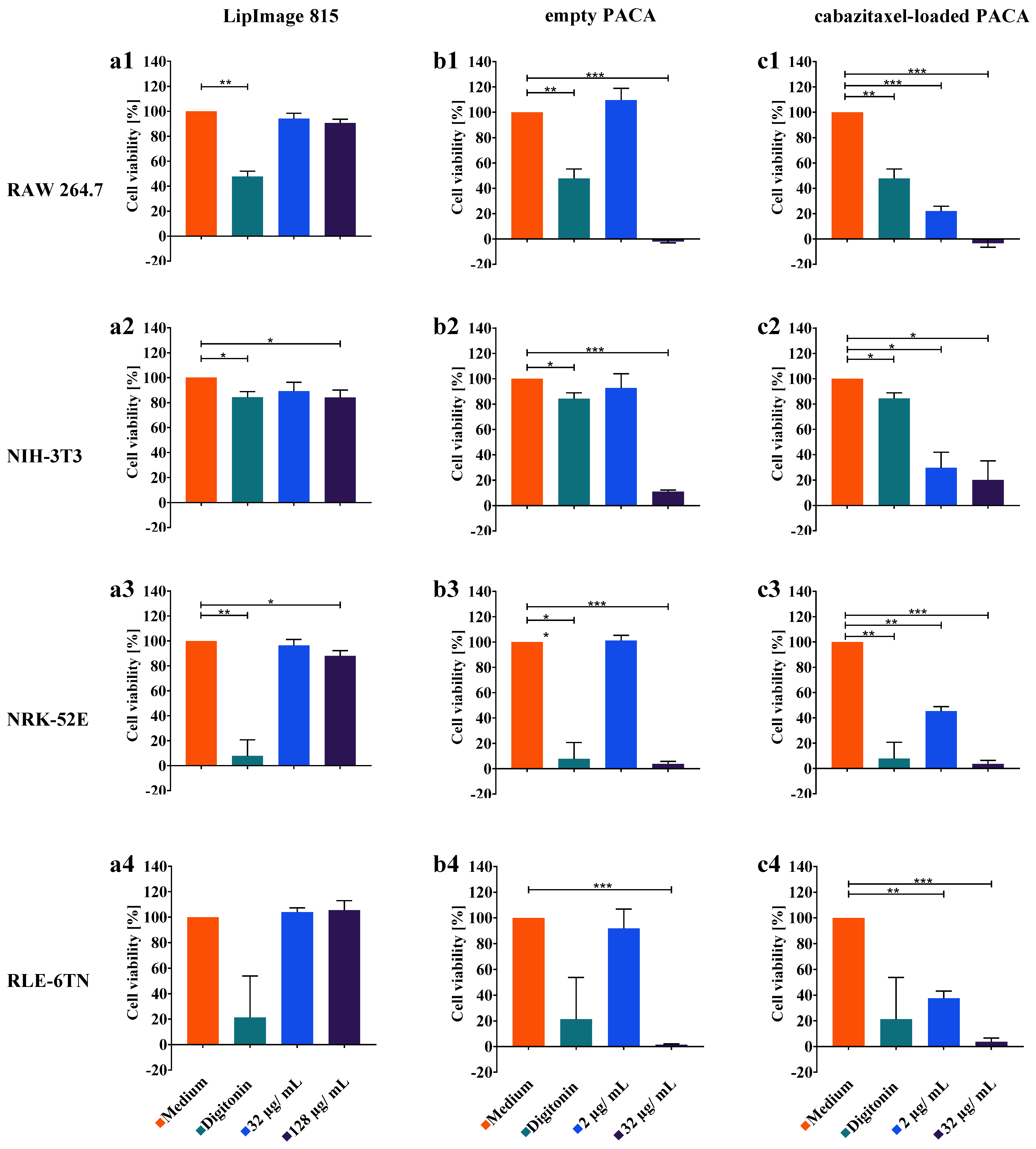
3.4. WST-8 Cell Viability Assay
4. Discussion
5. Conclusions and Future Prospects
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sadrieh, N.; Tyner, K.M. Nanotechnology and therapeutic delivery: A drug regulation perspective. Ther. Deliv. 2010, 1, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.H.; Harford, J.B.; Eaton, M.A.; Boisseau, P.M.; Dube, A.; Hayeshi, R.; Swai, H.; Lee, D.S. Nanomedicine: Past, present and future—A global perspective. Biochem. Biophys. Res. Commun. 2015, 468, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Etheridge, M.L.; Campbell, S.A.; Erdman, A.G.; Haynes, C.L.; Wolf, S.M.; McCullough, J. The big picture on nanomedicine: The state of investigational and approved nanomedicine products. Nanomedicine 2013, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheheltani, R.; Ezzibdeh, R.M.; Chhour, P.; Pulaparthi, K.; Kim, J.; Jurcova, M.; Hsu, J.C.; Blundell, C.; Litt, H.I.; Ferrari, V.A.; et al. Tunable, biodegradable gold nanoparticles as contrast agents for computed tomography and photoacoustic imaging. Biomaterials 2016, 102, 87–97. [Google Scholar] [CrossRef] [Green Version]
- Jeon, M.; Halbert, M.V.; Stephen, Z.R.; Zhang, M. Iron Oxide Nanoparticles as T1 Contrast Agents for Magnetic Resonance Imaging: Fundamentals, Challenges, Applications, and Prospectives. Adv. Mater. 2021, 33, 1906539. [Google Scholar] [CrossRef]
- Poon, W.; Kingston, B.R.; Ouyang, B.; Ngo, W.; Chan, W.C.W. A framework for designing delivery systems. Nat. Nanotechnol. 2020, 15, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Malam, Y.; Loizidou, M.; Seifalian, A.M. Liposomes and nanoparticles: Nanosized vehicles for drug delivery in cancer. Trends Pharmacol. Sci. 2009, 30, 592–599. [Google Scholar] [CrossRef]
- Fries, C.N.; Curvino, E.J.; Chen, J.-L.; Permar, S.R.; Fouda, G.G.; Collier, J.H. Advances in nanomaterial vaccine strategies to address infectious diseases impacting global health. Nat. Nanotechnol. 2020. [Google Scholar] [CrossRef]
- Shin, M.D.; Shukla, S.; Chung, Y.H.; Beiss, V.; Chan, S.K.; Ortega-Rivera, O.A.; Wirth, D.M.; Chen, A.; Sack, M.; Pokorski, J.K.; et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020, 15, 646–655. [Google Scholar] [CrossRef]
- Lewinski, N.; Colvin, V.; Drezek, R. Cytotoxicity of nanoparticles. Small 2008, 4, 26–49. [Google Scholar] [CrossRef]
- Arora, S.; Rajwade, J.M.; Paknikar, K.M. Nanotoxicology and in vitro studies: The need of the hour. Toxicol. Appl. Pharmacol. 2012, 258, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Andraos, C.; Yu, I.J.; Gulumian, M. Interference: A Much-Neglected Aspect in High-Throughput Screening of Nanoparticles. Int. J. Toxicol. 2020, 39, 397–421. [Google Scholar] [CrossRef] [PubMed]
- Piret, J.-P.; Bondarenko, O.M.; Boyles, M.S.P.; Himly, M.; Ribeiro, A.R.; Benetti, F.; Smal, C.; Lima, B.; Potthoff, A.; Simion, M.; et al. Pan-European inter-laboratory studies on a panel of in vitro cytotoxicity and pro-inflammation assays for nanoparticles. Arch. Toxicol. 2017, 91, 2315–2330. [Google Scholar] [CrossRef] [PubMed]
- Mues, S.; Ketelhut, S.; Kemper, B.; Schnekenburger, J. Digital Holographic Microscopy as Multimodal Read Out for In Vitro Nanomaterial Cytotoxicity Testing; SPIE: Bellingham, WA, USA, 2017; Volume 10414. [Google Scholar]
- Kroll, A.; Dierker, C.; Rommel, C.; Hahn, D.; Wohlleben, W.; Schulze-Isfort, C.; Göbbert, C.; Voetz, M.; Hardinghaus, F.; Schnekenburger, J. Cytotoxicity screening of 23 engineered nanomaterials using a test matrix of ten cell lines and three different assays. Part. Fibre Toxicol. 2011, 8, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohmer, N.; Rippl, A.; May, S.; Walter, A.; Heo, M.B.; Kwak, M.; Roesslein, M.; Song, N.W.; Wick, P.; Hirsch, C. Interference of engineered nanomaterials in flow cytometry: A case study. Colloids Surf. B Biointerfaces 2018, 172, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Sohaebuddin, S.K.; Thevenot, P.T.; Baker, D.; Eaton, J.W.; Tang, L. Nanomaterial cytotoxicity is composition, size, and cell type dependent. Part. Fibre Toxicol. 2010, 7, 22. [Google Scholar] [CrossRef] [Green Version]
- Kroll, A.; Pillukat, M.H.; Hahn, D.; Schnekenburger, J. Current in vitro methods in nanoparticle risk assessment: Limitations and challenges. Eur. J. Pharm. Biopharm. 2009, 72, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Bremer-Hoffmann, S.; Halamoda-Kenzaoui, B.; Borgos, S.E. Identification of regulatory needs for nanomedicines. J. Interdiscip. Nanomed. 2018, 3, 4–15. [Google Scholar] [CrossRef]
- Halamoda-Kenzaoui, B.; Holzwarth, U.; Roebben, G.; Bogni, A.; Bremer-Hoffmann, S. Mapping of the available standards against the regulatory needs for nanomedicines. WIREs Nanomed. Nanobiotechnol. 2019, 11, e1531. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.; Depeursinge, C.; Popescu, G. Quantitative phase imaging in biomedicine. Nat. Photonics 2018, 12, 578–589. [Google Scholar] [CrossRef]
- Lee, K.; Kim, K.; Jung, J.; Heo, J.; Cho, S.; Lee, S.; Chang, G.; Jo, Y.; Park, H.; Park, Y. Quantitative phase imaging techniques for the study of cell pathophysiology: From principles to applications. Sensors 2013, 13, 4170–4191. [Google Scholar] [CrossRef]
- Lenz, P.; Bruckner, M.; Ketelhut, S.; Heidemann, J.; Kemper, B.; Bettenworth, D. Multimodal Quantitative Phase Imaging with Digital Holographic Microscopy Accurately Assesses Intestinal Inflammation and Epithelial Wound Healing. J. Vis. Exp. 2016. [Google Scholar] [CrossRef] [PubMed]
- Kemper, B.; Carl, D.; Schnekenburger, J.; Bredebusch, I.; Schäfer, M.; Domschke, W.; von Bally, G. Investigation of living pancreas tumor cells by digital holographic microscopy. J. Biomed. Opt. 2006, 11, 34005. [Google Scholar] [CrossRef] [PubMed]
- Kamlund, S.; Strand, D.; Janicke, B.; Alm, K.; Oredsson, S. Influence of salinomycin treatment on division and movement of individual cancer cells cultured in normoxia or hypoxia evaluated with time-lapse digital holographic microscopy. Cell Cycle 2017, 16, 2128–2138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Leslie, K.A.; Guest, D.; Yeshcheulova, O.; Roy, I.J.; Piva, M.; Moriceau, G.; Zangle, T.A.; Lo, R.S.; Teitell, M.A.; et al. High-Speed Live-Cell Interferometry: A New Method for Quantifying Tumor Drug Resistance and Heterogeneity. Anal. Chem. 2018, 90, 3299–3306. [Google Scholar] [CrossRef]
- Bokemeyer, A.; Tepasse, P.R.; Quill, L.; Lenz, P.; Rijcken, E.; Vieth, M.; Ding, N.; Ketelhut, S.; Rieder, F.; Kemper, B.; et al. Quantitative Phase Imaging Using Digital Holographic Microscopy Reliably Assesses Morphology and Reflects Elastic Properties of Fibrotic Intestinal Tissue. Sci. Rep. 2019, 9, 19388. [Google Scholar] [CrossRef] [Green Version]
- Mugnano, M.; Memmolo, P.; Miccio, L.; Merola, F.; Bianco, V.; Bramanti, A.; Gambale, A.; Russo, R.; Andolfo, I.; Iolascon, A.; et al. Label-Free Optical Marker for Red-Blood-Cell Phenotyping of Inherited Anemias. Anal. Chem. 2018, 90, 7495–7501. [Google Scholar] [CrossRef]
- Kühn, J.; Shaffer, E.; Mena, J.; Breton, B.; Parent, J.; Rappaz, B.; Chambon, M.; Emery, Y.; Magistretti, P.; Depeursinge, C.; et al. Label-free cytotoxicity screening assay by digital holographic microscopy. Assay Drug Dev. Technol. 2013, 11, 101–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makdasi, E.; Laskar, O.; Milrot, E.; Schuster, O.; Shmaya, S.; Yitzhaki, S. Whole-Cell Multiparameter Assay for Ricin and Abrin Activity-Based Digital Holographic Microscopy. Toxins 2019, 11, 174. [Google Scholar] [CrossRef] [Green Version]
- Mugnano, M.; Memmolo, P.; Miccio, L.; Grilli, S.; Merola, F.; Calabuig, A.; Bramanti, A.; Mazzon, E.; Ferraro, P. In vitro cytotoxicity evaluation of cadmium by label-free holographic microscopy. J. Biophotonics 2018, 11, e201800099. [Google Scholar] [CrossRef]
- Mues, S.; Antunovic, J.; Ketelhut, S.; Kemper, B.; Schnekenburger, J. Novel Optical Approaches for Label.-Free Quantification of Nano-cytotoxic Effects; SPIE: Bellingham, WA, USA, 2016; Volume 9719. [Google Scholar]
- Rappaz, B.; Cano, E.; Colomb, T.; Kühn, J.; Depeursinge, C.; Simanis, V.; Magistretti, P.J.; Marquet, P. Noninvasive characterization of the fission yeast cell cycle by monitoring dry mass with digital holographic microscopy. J. Biomed. Opt. 2009, 14, 034049. [Google Scholar] [CrossRef]
- Popescu, G.; Park, Y.; Lue, N.; Best-Popescu, C.; Deflores, L.; Dasari, R.R.; Feld, M.S.; Badizadegan, K. Optical imaging of cell mass and growth dynamics. Am. J. Physiol. Cell Physiol. 2008, 295, C538–C544. [Google Scholar] [CrossRef] [Green Version]
- Kemper, B.; von Bally, G. Digital holographic microscopy for live cell applications and technical inspection. Appl. Opt. 2008, 47, A52–A61. [Google Scholar] [CrossRef]
- Gravier, J.; Garcia, F.P.N.Y.; Delmas, T.; Mittler, F.; Couffin, A.-C.; Vinet, F.; Texier-Nogues, I. Lipidots: Competitive organic alternative to quantum dots for in vivo fluorescence imaging. J. Biomed. Opt. 2011, 16, 096013. [Google Scholar] [CrossRef]
- Jacquart, A.; Keramidas, M.; Vollaire, J.; Boisgard, R.; Pottier, G.; Rustique, E.; Mittler, F.; Navarro, F.P.; Boutet, J.; Coll, J.L.; et al. LipImage 815: Novel dye-loaded lipid nanoparticles for long-term and sensitive in vivo near-infrared fluorescence imaging. J. Biomed. Opt. 2013, 18, 101311. [Google Scholar] [CrossRef]
- Sayag, D.; Cabon, Q.; Texier, I.; Navarro, F.P.; Boisgard, R.; Virieux-Watrelot, D.; Carozzo, C.; Ponce, F. Phase-0/phase-I study of dye-loaded lipid nanoparticles for near-infrared fluorescence imaging in healthy dogs. Eur. J. Pharm. Biopharm. 2016, 100, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Vauthier, C.; Dubernet, C.; Fattal, E.; Pinto-Alphandary, H.; Couvreur, P. Poly(alkylcyanoacrylates) as biodegradable materials for biomedical applications. Adv. Drug Deliv. Rev. 2003, 55, 519–548. [Google Scholar] [CrossRef]
- Sulheim, E.; Iversen, T.G.; To Nakstad, V.; Klinkenberg, G.; Sletta, H.; Schmid, R.; Hatletveit, A.R.; Wagbo, A.M.; Sundan, A.; Skotland, T.; et al. Cytotoxicity of Poly(Alkyl Cyanoacrylate) Nanoparticles. Int. J. Mol. Sci. 2017, 18, 2454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Øverbye, A.; Torgersen, M.L.; Sønstevold, T.; Iversen, T.G.; Mørch, Ý.; Skotland, T.; Sandvig, K. Cabazitaxel-loaded poly(alkyl cyanoacrylate) nanoparticles: Toxicity and changes in the proteome of breast, colon and prostate cancer cells. Nanotoxicology 2021, 15, 865–884. [Google Scholar] [CrossRef]
- Kastl, L.; Isbach, M.; Dirksen, D.; Schnekenburger, J.; Kemper, B. Quantitative phase imaging for cell culture quality control. Cytom. Part. A 2017, 91, 470–481. [Google Scholar] [CrossRef] [Green Version]
- Kemper, B.; Barroso, Á.; Eder, K.; Marzi, A.; Ritz, S.; Schnekenburger, J.; Ketelhut, S. Reduction of the Influence of Parasitic Interferences and Coherence Induced Disturbances in Digital Holography-Based Quantitative Phase Microscopy by Modulation of the Object Illumination with an Electrically Focus Tunable Lens; SPIE: Bellingham, WA, USA, 2021; Volume 11653. [Google Scholar]
- Min, J.; Yao, B.; Trendafilova, V.; Ketelhut, S.; Kastl, L.; Greve, B.; Kemper, B. Quantitative phase imaging of cells in a flow cytometry arrangement utilizing Michelson interferometer-based off-axis digital holographic microscopy. J. Biophotonics 2019, 12, e201900085. [Google Scholar] [CrossRef]
- Sternberg, S.R. Biomedical Image Processing. Computer 1983, 16, 22–34. [Google Scholar] [CrossRef]
- Barer, R. Refractometry and Interferometry of Living Cells. J. Opt. Soc. Am. 1957, 47, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Jedrzejczak-Silicka, M.; Mijowska, E. General Cytotoxicity and Its Application in Nanomaterial Analysis. In Cytotoxicity; IntechOpen Limited: London, UK, 2017. [Google Scholar] [CrossRef] [Green Version]
- Barer, R. Determination of dry mass, thickness, solid and water concentration in living cells. Nature 1953, 172, 1097–1098. [Google Scholar] [CrossRef] [PubMed]
- Barer, R.; Tkaczyk, S. Refractive Index of Concentrated Protein Solutions. Nature 1954, 173, 821–822. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Brown, P.H.; Schuck, P. On the distribution of protein refractive index increments. Biophys. J. 2011, 100, 2309–2317. [Google Scholar] [CrossRef] [Green Version]
- Dunigan, D.D.; Waters, S.B.; Owen, T.C. Aqueous soluble tetrazolium/formazan MTS as an indicator of NADH- and NADPH-dependent dehydrogenase activity. Biotechniques 1995, 19, 640–649. [Google Scholar]
- Eder, K.; Marzi, A.; Barroso, Á.; Kemper, B.; Schnekenburger, J. Medical Nanoparticle Impact on Macrophage Temporal Dry Mass Development Quantified In Vitro by Digital Holographic Microscopy; SPIE: Bellingham, WA, USA, 2021; Volume 11653. [Google Scholar]
- Pavillon, N.; Kühn, J.; Moratal, C.; Jourdain, P.; Depeursinge, C.; Magistretti, P.J.; Marquet, P. Early cell death detection with digital holographic microscopy. PLoS ONE 2012, 7, e30912. [Google Scholar] [CrossRef]
- Bardyn, M.; Rappaz, B.; Jaferzadeh, K.; Crettaz, D.; Tissot, J.D.; Moon, I.; Turcatti, G.; Lion, N.; Prudent, M. Red blood cells ageing markers: A multi-parametric analysis. Blood Transfus. 2017, 15, 239–248. [Google Scholar] [CrossRef]
- Mölder, A.; Sebesta, M.; Gustafsson, M.; Gisselson, L.; Wingren, A.G.; Alm, K. Non-invasive, label-free cell counting and quantitative analysis of adherent cells using digital holography. J. Microsc. 2008, 232, 240–247. [Google Scholar] [CrossRef]
- Rezaei, M.; Cao, J.; Friedrich, K.; Kemper, B.; Brendel, O.; Grosser, M.; Adrian, M.; Baretton, G.; Breier, G.; Schnittler, H.J. The expression of VE-cadherin in breast cancer cells modulates cell dynamics as a function of tumor differentiation and promotes tumor-endothelial cell interactions. Histochem. Cell Biol. 2018, 149, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Eder, K.; Kutscher, T.; Marzi, A.; Barroso, Á.; Schnekenburger, J.; Kemper, B. Automated Detection of Macrophages in Quantitative Phase Images by Deep Learning Using a Mask Region.-Based Convolutional Neural Network; SPIE: Bellingham, WA, USA, 2021; Volume 116551. [Google Scholar]
- Girshovitz, P.; Shaked, N.T. Generalized cell morphological parameters based on interferometric phase microscopy and their application to cell life cycle characterization. Biomed. Opt. Express 2012, 3, 1757–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belashov, A.V.; Zhikhoreva, A.A.; Belyaeva, T.N.; Salova, A.V.; Kornilova, E.S.; Semenova, I.V.; Vasyutinskii, O.S. Machine Learning Assisted Classification of Cell Lines and Cell States on Quantitative Phase Images. Cells 2021, 10, 2587. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eder, K.M.; Marzi, A.; Barroso, Á.; Ketelhut, S.; Kemper, B.; Schnekenburger, J. Label-Free Digital Holographic Microscopy for In Vitro Cytotoxic Effect Quantification of Organic Nanoparticles. Cells 2022, 11, 644. https://doi.org/10.3390/cells11040644
Eder KM, Marzi A, Barroso Á, Ketelhut S, Kemper B, Schnekenburger J. Label-Free Digital Holographic Microscopy for In Vitro Cytotoxic Effect Quantification of Organic Nanoparticles. Cells. 2022; 11(4):644. https://doi.org/10.3390/cells11040644
Chicago/Turabian StyleEder, Kai Moritz, Anne Marzi, Álvaro Barroso, Steffi Ketelhut, Björn Kemper, and Jürgen Schnekenburger. 2022. "Label-Free Digital Holographic Microscopy for In Vitro Cytotoxic Effect Quantification of Organic Nanoparticles" Cells 11, no. 4: 644. https://doi.org/10.3390/cells11040644
APA StyleEder, K. M., Marzi, A., Barroso, Á., Ketelhut, S., Kemper, B., & Schnekenburger, J. (2022). Label-Free Digital Holographic Microscopy for In Vitro Cytotoxic Effect Quantification of Organic Nanoparticles. Cells, 11(4), 644. https://doi.org/10.3390/cells11040644





