Static Magnetic Field Inhibits Growth of Escherichia coli Colonies via Restriction of Carbon Source Utilization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Stains and Mutant Strain Construction
2.2. SMF Treatments, Cell Culture and Measurement of Colonies
2.3. Analysis of Transcriptome Sequencing and Quantitative PCR
2.4. Metabolite Extraction and Profiling Analysis
3. Results
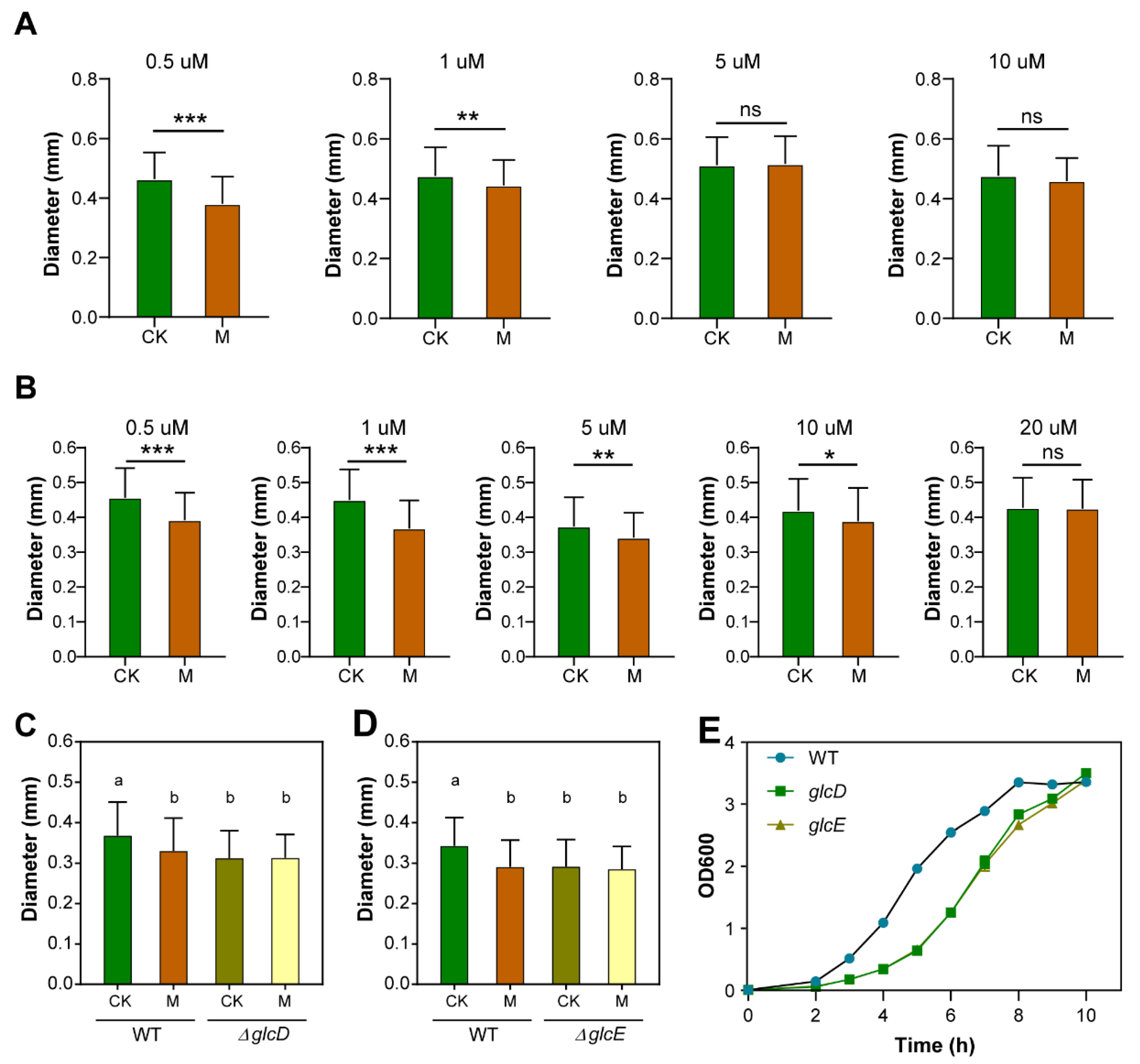
3.1. Moderate-Intensity SMF Inhibits Bacterial Growth
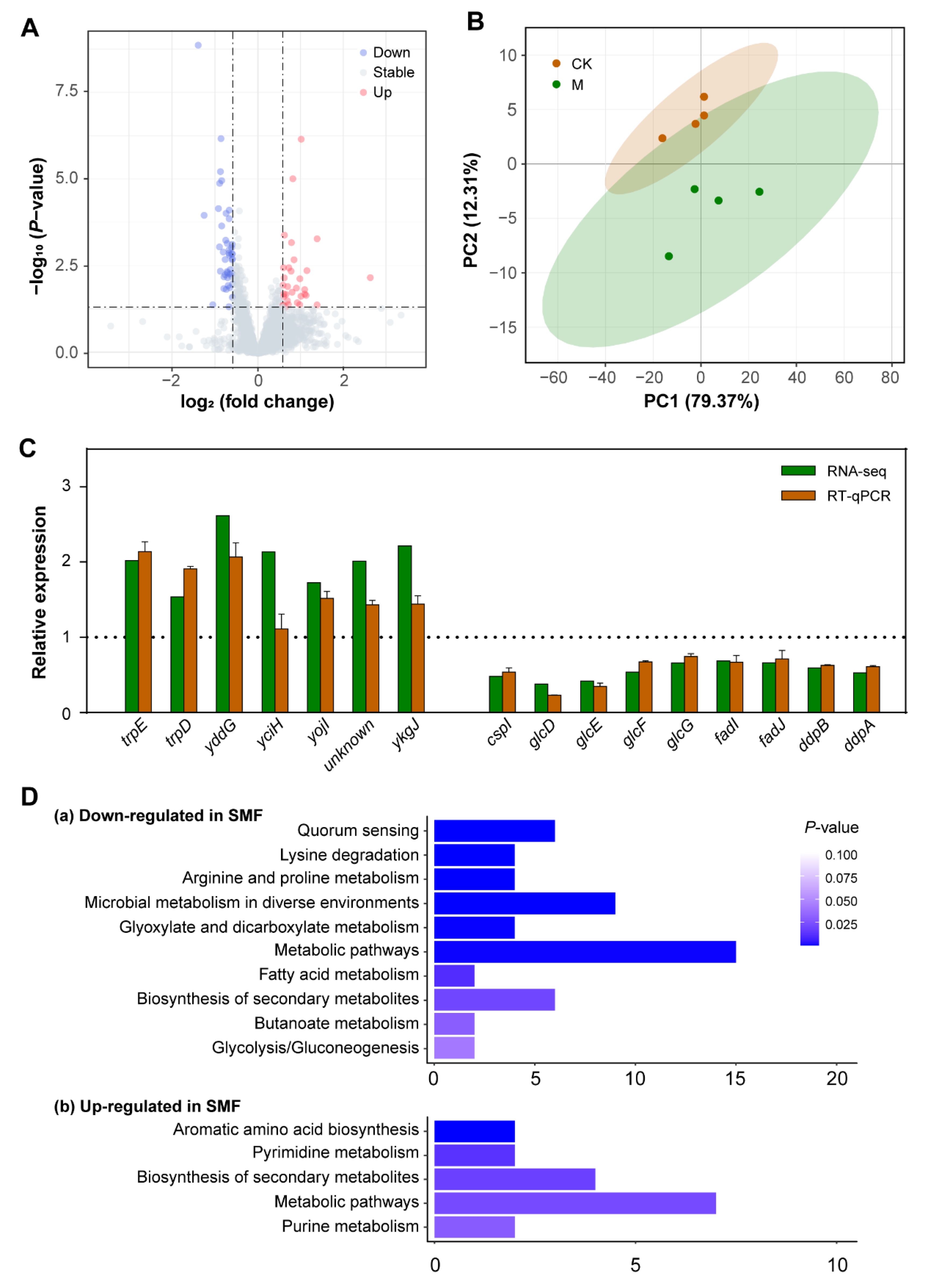
3.2. Transcriptomic Profiling Reveals That SMF-Regulated DEGs Are Enriched Mainly in Metabolism of Carbon Source Utilization and Stress Response
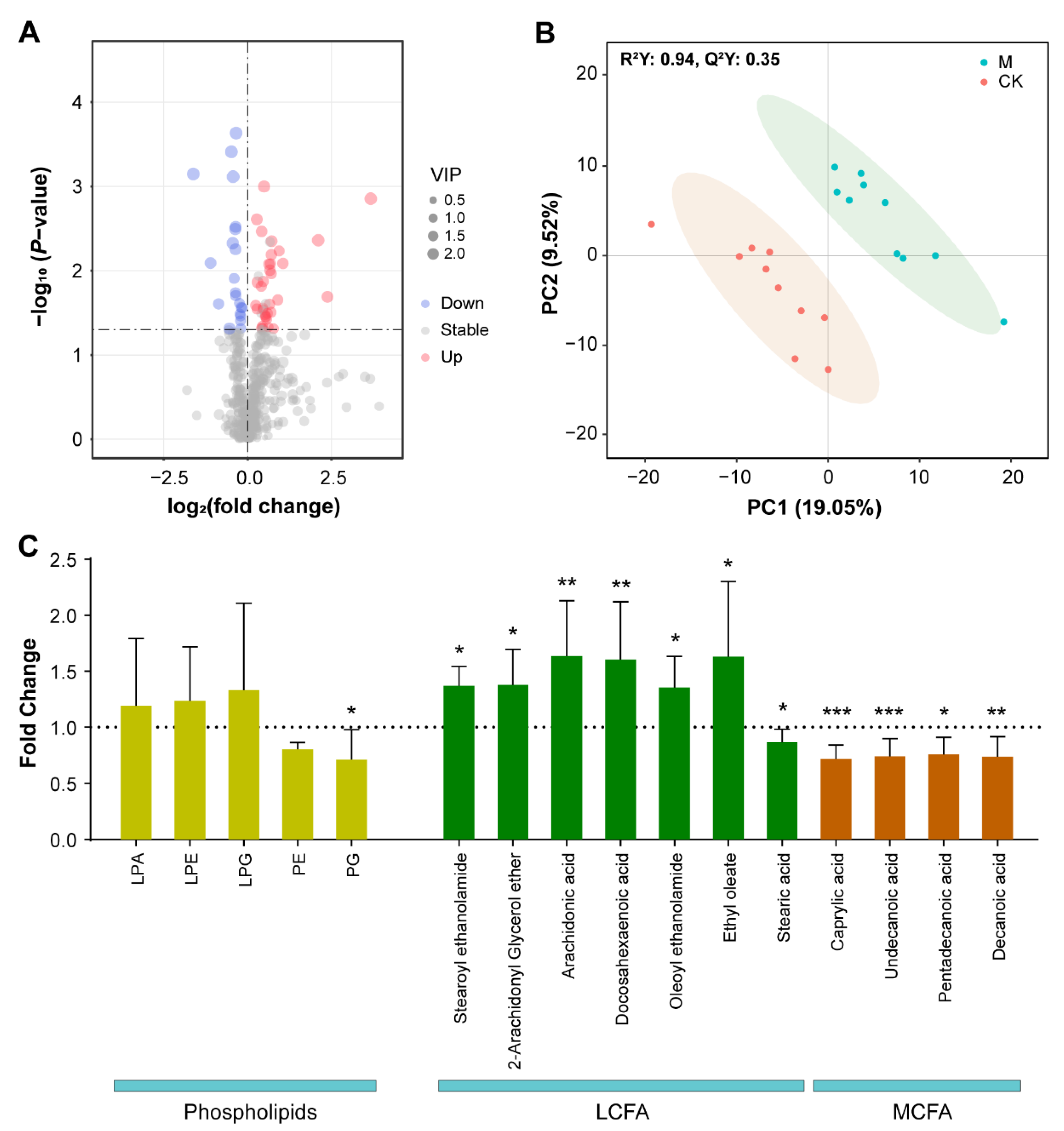
3.3. Metabolomic Assays Show That Metabolites Significantly Altered in Content by SMF Are Enriched Mainly in Phospholipids, Fatty Acids and Aromatic Amino Acids
3.4. Both Glycolate and Glyoxylate Rescue the SMF-Inhibited Growth Phenotype
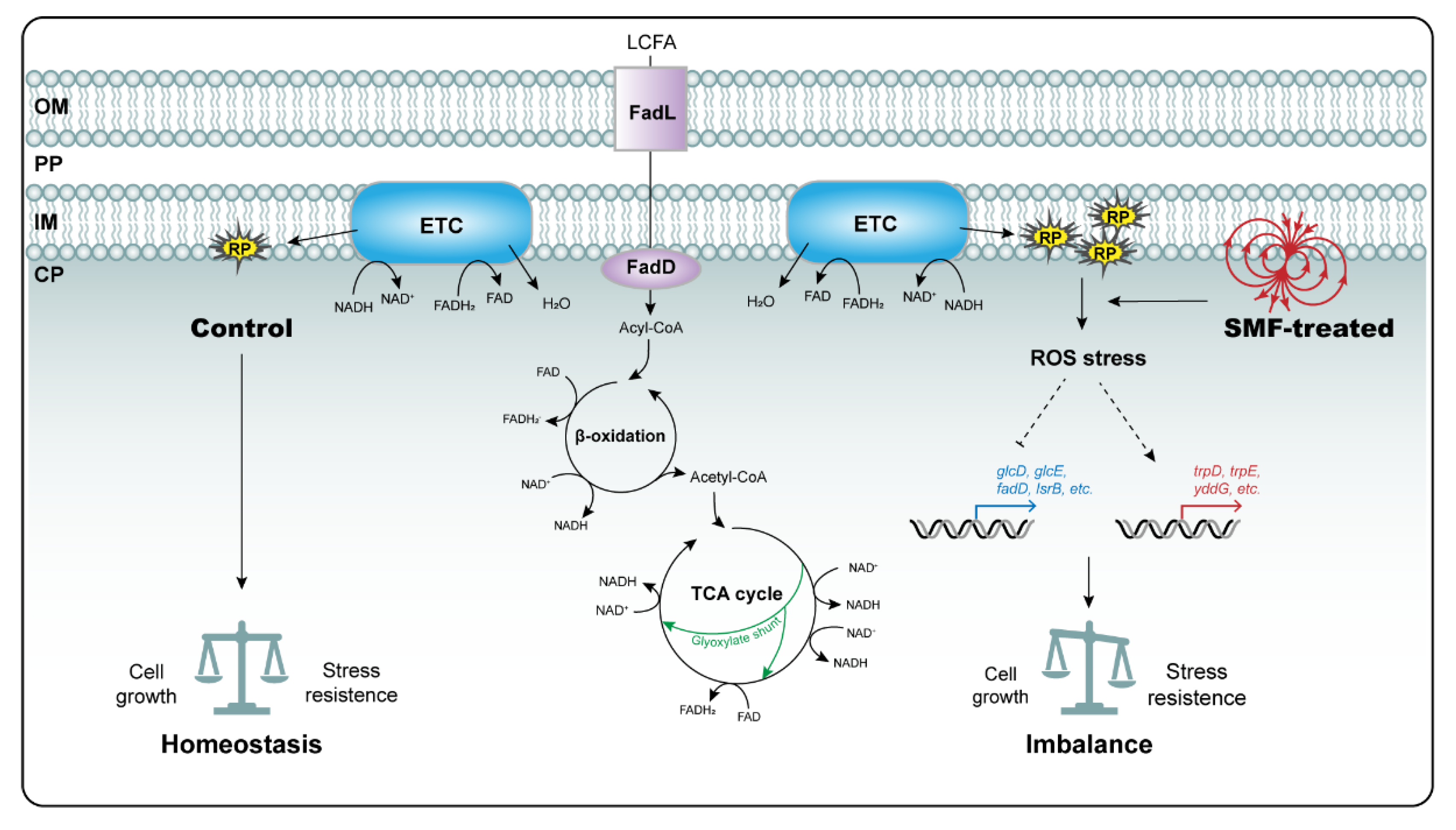
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Alken, P.; Thébault, E.; Beggan, C.D.; Amit, H.; Aubert, J.; Baerenzung, J.; Bondar, T.N.; Brown, W.J.; Califf, S.; Chambodut, A.; et al. International Geomagnetic Reference Field: The thirteenth generation. Earth Planets Space 2021, 73, 1–25. [Google Scholar] [CrossRef]
- Tarduno, J.A.; Cottrell, R.D.; Bono, R.K.; Oda, H.; Davis, W.J.; Fayek, M.; Erve, O.V.; Nimmo, F.; Huang, W.; Thern, E.R.; et al. Paleomagnetism indicates that primary magnetite in zircon records a strong Hadean geodynamo. Proc. Natl. Acad. Sci. USA 2020, 117, 2309–2318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouritsen, H. Long-distance navigation and magnetoreception in migratory animals. Nature 2018, 558, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Blakemore, R. Magnetotactic bacteria. Science 1975, 190, 377–379. [Google Scholar] [CrossRef] [PubMed]
- Binhi, V. Theoretical concepts in magnetobiology. Electro. Magn. 2001, 20, 43–58. [Google Scholar] [CrossRef]
- Clites, B.L.; Pierce, J.T. Identifying Cellular and Molecular Mechanisms for Magnetosensation. Annu. Rev. Neurosci. 2017, 40, 231–250. [Google Scholar] [CrossRef]
- Markov, M.S. Magnetic field therapy: A review. Electromagn. Biol. Med. 2007, 26, 1–23. [Google Scholar] [CrossRef]
- Lin, W.; Kirschvink, J.L.; Paterson, G.A.; Bazylinski, D.A.; Pan, Y. On the origin of microbial magnetoreception. Natl. Sci. Rev. 2020, 7, 472–479. [Google Scholar] [CrossRef] [Green Version]
- Binhi, V.N.; Rubin, A.B. Theoretical Concepts in Magnetobiology after 40 Years of Research. Cells 2022, 11, 247. [Google Scholar] [CrossRef]
- Hunt, R.W.; Zavalin, A.; Bhatnagar, A.; Chinnasamy, S.; Das, K.C. Electromagnetic biostimulation of living cultures for biotechnology, biofuel and bioenergy applications. Int. J. Mol. Sci. 2009, 10, 4515–4558. [Google Scholar] [CrossRef]
- Křiklavová, L.; Truhlář, M.; Škodová, P.; Lederer, T.; Jirků, V. Effects of a static magnetic field on phenol degradation effectiveness and Rhodococcus erythropolis growth and respiration in a fed-batch reactor. Bioresour. Technol. 2014, 167, 510–513. [Google Scholar] [CrossRef] [PubMed]
- Rosen, A.D. Mechanism of action of moderate-intensity static magnetic fields on biological systems. Cell Biochem. Biophys. 2003, 39, 163–173. [Google Scholar] [CrossRef]
- Ayrapetyan, S. The role of cell hydration in realization of biological effects of non-ionizing radiation (NIR). Electromagn. Biol. Med. 2015, 34, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.; Deamici, K.; Menestrino, B.; Garda-Buffon, J.; Costa, J.A. Magnetic treatment of microalgae for enhanced product formation. World J. Microbiol. Biotechnol. 2017, 33, 169. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Huang, H.; Deng, A.; Pan, C. Effects of static magnetic fields on Escherichia coli. Micron 2009, 40, 894–898. [Google Scholar] [CrossRef]
- Bajpai, I.; Saha, N.; Basu, B. Moderate intensity static magnetic field has bactericidal effect on E. coli and S. epidermidis on sintered hydroxyapatite. J. Biomed. Mater. Res. B Appl. Biomater. 2012, 100, 1206–1217. [Google Scholar] [CrossRef]
- Łebkowska, M.; Rutkowska-Narożniak, A.; Pajor, E.; Tabernacka, A.; Załęska-Radziwiłł, M. Impact of a static magnetic field on biodegradation of wastewater compounds and bacteria recombination. Environ. Sci. Pollut. Res. 2018, 25, 22571–22583. [Google Scholar] [CrossRef] [PubMed]
- Potenza, L.; Ubaldi, L.; De Sanctis, R.; De Bellis, R.; Cucchiarini, L.; Dachà, M. Effects of a static magnetic field on cell growth and gene expression in Escherichia coli. Mutat. Res. Genet. Toxicol. Environ. Mutagenesis 2004, 561, 53–62. [Google Scholar] [CrossRef]
- Letuta, U.G.; Berdinskiy, V.L.; Udagawa, C.; Tanimoto, Y. Enzymatic mechanisms of biological magnetic sensitivity. Bioelectromagnetics 2017, 38, 511–521. [Google Scholar] [CrossRef]
- Łebkowska, M.; Rutkowska-Narożniak, A.; Pajor, E.; Pochanke, Z. Effect of a static magnetic field on formaldehyde biodegradation in wastewater by activated sludge. Bioresour. Technol. 2011, 102, 8777–8782. [Google Scholar] [CrossRef]
- Filipic, J.; Kraigher, B.; Tepus, B.; Kokol, V.; Mandic-Mulec, I. Effects of low-density static magnetic fields on the growth and activities of wastewater bacteria Escherichia coli and Pseudomonas putida. Bioresour. Technol. 2012, 120, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Wang, X.; Hu, S.; Han, T.; He, S.; Zhang, G.; He, M.; Lin, X. Bioeffect of static magnetic field on photosynthetic bacteria: Evaluation of bioresources production and wastewater treatment efficiency. Water Environ. Res. 2020, 92, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Huang, Z.; Fan, B. Effects of prolonged exposure to moderate static magnetic field and its synergistic effects with alkaline pH on Enterococcus faecalis. Microb. Pathog. 2018, 115, 117–122. [Google Scholar] [CrossRef] [PubMed]
- El May, A.; Zouaoui, J.; Snoussi, S.; Ben Mouhoub, R.; Landoulsi, A. relA and spoT Gene Expression is Modulated in Salmonella Grown Under Static Magnetic Field. Curr. Microbiol. 2021, 78, 887–893. [Google Scholar] [CrossRef]
- Okano, H. Effects of static magnetic fields in biology: Role of free radicals. Front. Biosci. 2008, 13, 6106–6125. [Google Scholar] [CrossRef] [Green Version]
- Scaiano, J.C.; Cozens, F.L.; McLean, J. Model for the rationalization of magnetic field effects in vivo. Application of the radical-pair mechanism to biological systems. Photochem. Photobiol. 1994, 59, 585–589. [Google Scholar] [CrossRef]
- Serrano, G.; Miranda-Ostojic, C.; Ferrada, P.; Wulff-Zotelle, C.; Maureira, A.; Fuentealba, E.; Gallardo, K.; Zapata, M.; Rivas, M. Response to Static Magnetic Field-Induced Stress in Scenedesmus obliquus and Nannochloropsis gaditana. Mar. Drugs 2021, 19, 527. [Google Scholar] [CrossRef]
- Hanini, R.; Chatti, A.; Ghorbel, S.B.; Landoulsi, A. Role of Sod Gene in Response to Static Magnetic Fields in Pseudomonas aeruginosa. Curr. Microbiol. 2017, 74, 930–937. [Google Scholar] [CrossRef]
- Buchachenko, A.L.; Kouznetsov, D.A.; Orlova, M.A.; Markarian, A.A. Magnetic isotope effect of magnesium in phosphoglycerate kinase phosphorylation. Proc. Natl. Acad. Sci. USA 2005, 102, 10793–10796. [Google Scholar] [CrossRef] [Green Version]
- Shevchenko, U.G.; Avdeeva, E.I.; Berdinskii, V.L. Biological effects of the 25Mg magnetic isotope in E. coli cells. Russ. J. Phys. Chem. B 2012, 6, 531–537. [Google Scholar] [CrossRef]
- Letuta, U.G.; Berdinskiy, V.L. Biological effects of static magnetic fields and zinc isotopes on E. coli bacteria. Bioelectromagnetics 2019, 40, 62–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Zhou, H.; Dai, W.; Xiong, J.; Chen, F. Effect of Static Magnetic Field on Monascus ruber M7 Based on Transcriptome Analysis. J. Fungi 2021, 7, 256. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Ning, S.; Shi, S.; Tan, L. Improving Azo Dye Decolorization Performance and Halotolerance of Pichia occidentalis A2 by Static Magnetic Field and Possible Mechanisms Through Comparative Transcriptome Analysis. Front. Microbiol. 2020, 11, 712. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, B.; Duan, C.; Sun, B.; Yang, J.; Yang, S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015, 81, 2506–2514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.Y.; Yomo, T.; Ying, B.W. Clustering of Bacterial Growth Dynamics in Response to Growth Media by Dynamic Time Warping. Microorganisms 2020, 8, 331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, N.; Nakahigashi, K.; Nakamichi, T.; Yoshino, M.; Takai, Y.; Touda, Y.; Furubayashi, A.; Kinjyo, S.; Dose, H.; Hasegawa, M.; et al. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 2009, 5, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, B.; Liu, J.; Cheng, J.; Zhang, L.; Song, C.; Tian, X.; Fan, Y.; Lv, Y.; Zhang, X. A Static Magnetic Field Improves Iron Metabolism and Prevents High-Fat-Diet/Streptozocin-Induced Diabetes. Innovation 2021, 2, 100077. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic. Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [Green Version]
- Wen, B.; Mei, Z.; Zeng, C.; Liu, S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 183. [Google Scholar] [CrossRef] [Green Version]
- Vimala, A.; Harinarayanan, R. Transketolase activity modulates glycerol-3-phosphate levels in Escherichia coli. Mol. Microbiol. 2016, 100, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitra, A.; Herren, C.D.; Patel, I.R.; Coleman, A.; Mukhopadhyay, S. Integration of AI-2 Based Cell-Cell Signaling with Metabolic Cues in Escherichia coli. PLoS ONE 2016, 11, e0157532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoseki, T.; Kutsukake, K.; Ohnishi, K.; Iino, T. Functional analysis of the flagellar genes in the fliD operon of Salmonella typhimurium. Microbiology 1995, 141, 1715–1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hess, J.F.; Oosawa, K.; Kaplan, N.; Simon, M.I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 1988, 53, 79–87. [Google Scholar] [CrossRef]
- Feng, L.; Liu, B.; Liu, Y.; Ratiner, Y.A.; Hu, B.; Li, D.; Zong, X.; Xiong, W.; Wang, L. A genomic islet mediates flagellar phase variation in Escherichia coli strains carrying the flagellin-specifying locus flk. J. Bacteriol. 2008, 190, 4470–4477. [Google Scholar] [CrossRef] [Green Version]
- Bazurto, J.V.; Downs, D.M. Metabolic network structure and function in bacteria goes beyond conserved enzyme components. Microb. Cell 2016, 3, 260–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kettles, R.A.; Tschowri, N.; Lyons, K.J.; Sharma, P.; Hengge, R.; Webber, M.A.; Grainger, D.C. The Escherichia coli MarA protein regulates the ycgZ-ymgABC operon to inhibit biofilm formation. Mol. Microbiol. 2019, 112, 1609–1625. [Google Scholar] [CrossRef]
- Holden, E.R.; Yasir, M.; Turner, A.K.; Wain, J.; Charles, I.G.; Webber, M.A. Massively parallel transposon mutagenesis identifies temporally essential genes for biofilm formation in Escherichia coli. Microb. Genom. 2021, 7, 000673. [Google Scholar] [CrossRef]
- Gardner, A.M.; Gessner, C.R.; Gardner, P.R. Regulation of the nitric oxide reduction operon (norRVW) in Escherichia coli. Role of NorR and sigma54 in the nitric oxide stress response. J. Biol. Chem. 2003, 278, 10081–10086. [Google Scholar] [CrossRef] [Green Version]
- Quinones-Pena, M.A.; Tavizon, G.; Puente, J.L.; Martinez-Anaya, C.; Hernandez-Chinas, U.; Eslava, C.A. Effects of static magnetic fields on the enteropathogenic Escherichia coli. Bioelectromagnetics 2017, 38, 570–578. [Google Scholar] [CrossRef] [PubMed]
- Brkovic, S.; Postic, S.; Ilic, D. Influence of the magnetic field on microorganisms in the oral cavity. J. Appl. Oral. Sci. 2015, 23, 179–186. [Google Scholar] [CrossRef] [Green Version]
- Pellicer, M.T.; Fernandez, C.; Badia, J.; Aguilar, J.; Lin, E.C.; Baldom, L. Cross-induction of glc and ace operons of Escherichia coli attributable to pathway intersection. Characterization of the glc promoter. J. Biol. Chem. 1999, 274, 1745–1752. [Google Scholar] [CrossRef] [Green Version]
- Dolan, S.K.; Welch, M. The Glyoxylate Shunt, 60 Years On. Annu. Rev. Microbiol. 2018, 72, 309–330. [Google Scholar] [CrossRef] [PubMed]
- Pellicer, M.-T.; Badia, J.; Aguilar, J.; Baldomà, L. glc locus of Escherichia coli: Characterization of genes encoding the subunits of glycolate oxidase and the glc regulator protein. J. Bacteriol. 1996, 178, 2051–2059. [Google Scholar] [CrossRef] [Green Version]
- Kiupakis, A.K.; Reitzer, L. ArgR-independent induction and ArgR-dependent superinduction of the astCADBE operon in Escherichia coli. J. Bacteriol. 2002, 184, 2940–2950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 2016, 40, 133–159. [Google Scholar] [CrossRef] [Green Version]
- Pech-Canul, A.; Nogales, J.; Miranda-Molina, A.; Alvarez, L.; Geiger, O.; Soto, M.J.; Lopez-Lara, I.M. FadD is required for utilization of endogenous fatty acids released from membrane lipids. J. Bacteriol. 2011, 193, 6295–6304. [Google Scholar] [CrossRef] [Green Version]
- Lingens, F. The biosynthesis of aromatic amino acids and its regulation. Angew. Chem. Int. Ed. Engl. 1968, 7, 350–360. [Google Scholar] [CrossRef]
- Yanofsky, C. The different roles of tryptophan transfer RNA in regulating trp operon expression in E. coli versus B. subtilis. Trends Genet. 2004, 20, 367–374. [Google Scholar] [CrossRef]
- Shimazaki, J.; Furukawa, S.; Ogihara, H.; Morinaga, Y. L-Tryptophan prevents Escherichia coli biofilm formation and triggers biofilm degradation. Biochem. Biophys. Res. Commun. 2012, 419, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Maeda, T.; Hong, S.H.; Wood, T.K. Reconfiguring the quorum-sensing regulator SdiA of Escherichia coli to control biofilm formation via indole and N-acylhomoserine lactones. Appl. Environ. Microbiol. 2009, 75, 1703–1716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, G.; Sharma, S.; Sharma, P.; Chandola, D.; Dang, S.; Gupta, S.; Gabrani, R. Escherichia coli biofilm: Development and therapeutic strategies. J. Appl. Microbiol. 2016, 121, 309–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Galloway, W.R.; Hodgkinson, J.T.; Bowden, S.D.; Welch, M.; Spring, D.R. Quorum sensing in Gram-negative bacteria: Small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 2011, 111, 28–67. [Google Scholar] [CrossRef]
- Camilli, A.; Bassler, B.L. Bacterial small-molecule signaling pathways. Science 2006, 311, 1113–1116. [Google Scholar] [CrossRef] [Green Version]
- Schauder, S.; Shokat, K.; Surette, M.G.; Bassler, B.L. The LuxS family of bacterial autoinducers: Biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 2001, 41, 463–476. [Google Scholar] [CrossRef]
- Taga, M.E.; Semmelhack, J.L.; Bassler, B.L. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 2001, 42, 777–793. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Li, J.; March, J.C.; Valdes, J.J.; Bentley, W.E. luxS-dependent gene regulation in Escherichia coli K-12 revealed by genomic expression profiling. J. Bacteriol. 2005, 187, 8350–8360. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Song, H.; Zheng, H.; Ren, Y.; Li, S.; Liu, X.; Yu, W.; Ma, X. Culture of low density E. coli cells in alginate-chitosan microcapsules facilitates stress resistance by up-regulating luxS/AI-2 system. Carbohydr. Polym. 2016, 141, 160–165. [Google Scholar] [CrossRef]
- Xue, T.; Zhao, L.; Sun, H.; Zhou, X.; Sun, B. LsrR-binding site recognition and regulatory characteristics in Escherichia coli AI-2 quorum sensing. Cell Res. 2009, 19, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- Xavier, K.B.; Bassler, B.L. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 2005, 187, 238–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, S.; Jaswal, K.; Shiver, A.L.; Balecha, H.; Patra, T.; Chaba, R. A genome-wide screen in Escherichia coli reveals that ubiquinone is a key antioxidant for metabolism of long-chain fatty acids. J. Biol. Chem. 2017, 292, 20086–20099. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Xie, R.; Xu, X.; Liao, X.; Guo, J.; Fang, Y.; Fang, Z.; Huang, J. Static Magnetic Field Inhibits Growth of Escherichia coli Colonies via Restriction of Carbon Source Utilization. Cells 2022, 11, 827. https://doi.org/10.3390/cells11050827
Li H, Xie R, Xu X, Liao X, Guo J, Fang Y, Fang Z, Huang J. Static Magnetic Field Inhibits Growth of Escherichia coli Colonies via Restriction of Carbon Source Utilization. Cells. 2022; 11(5):827. https://doi.org/10.3390/cells11050827
Chicago/Turabian StyleLi, Haodong, Runnan Xie, Xiang Xu, Xingru Liao, Jiaxin Guo, Yanwen Fang, Zhicai Fang, and Jirong Huang. 2022. "Static Magnetic Field Inhibits Growth of Escherichia coli Colonies via Restriction of Carbon Source Utilization" Cells 11, no. 5: 827. https://doi.org/10.3390/cells11050827






