Machine Learning-Driven Multiobjective Optimization: An Opportunity of Microfluidic Platforms Applied in Cancer Research
Abstract
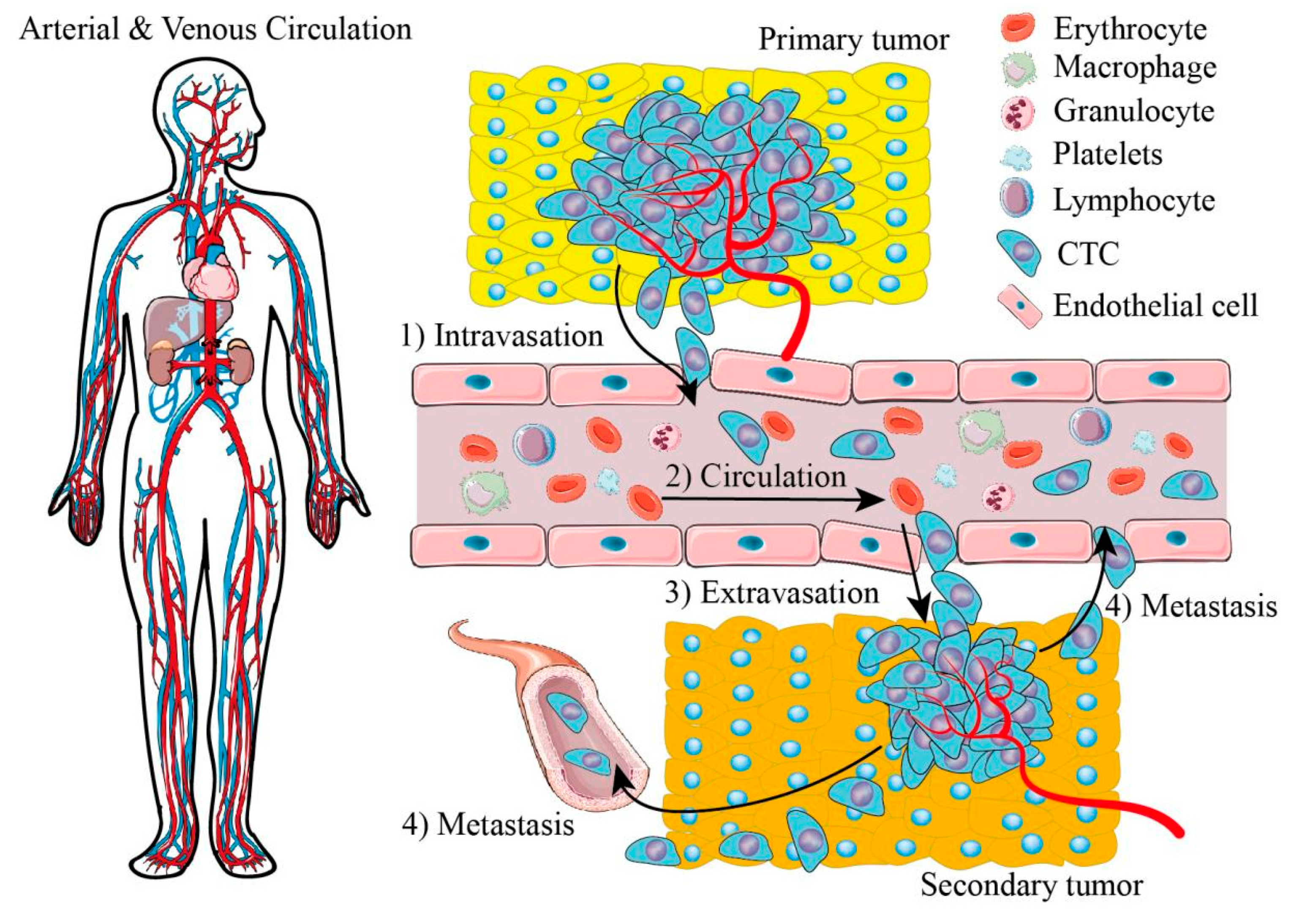
:1. Introduction
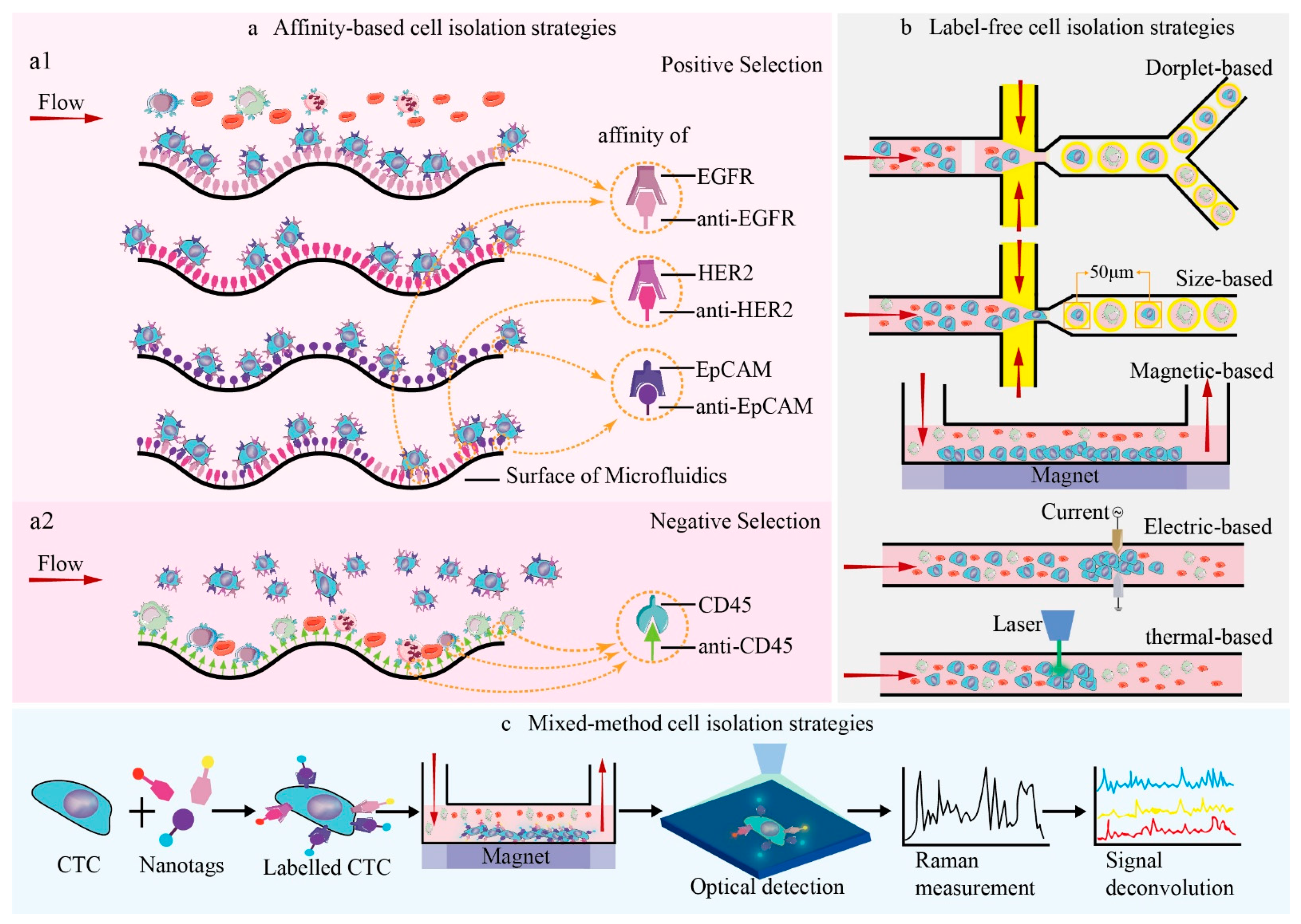
2. Systematic Description
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CTCs | Circulating tumor cells |
| ML | Machine learning |
| PDMS | Polydimethylsiloxane |
| TME | Tumor microenvironment |
| CNN | Convolutional neural network |
| SVM | Support vector machines |
| LR | Linear regression, logistic regression |
| VAE | Variational autoencoder |
| SERS | Surface-enhanced Raman Scattering |
References
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Cho, Y.; Cheng, X.; Yang, S.; Liu, Y.; Liu, Y. Integration of Hierarchical Micro-/Nanostructures in a Microfluidic Chip for Efficient and Selective Isolation of Rare Tumor Cells. Micromachines 2019, 10, 698. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.; Lin, X.; Hui, Y.; Wang, J.; Zhang, Q.; Kong, F. Circulating Tumor Cell Identification Based on Deep Learning. Front. Oncol. 2022, 12, 843879. [Google Scholar] [CrossRef]
- Banaei, N.; Moshfegh, J.; Mohseni-Kabir, A.; Houghton, J.M.; Sun, Y.; Kim, B. Machine Learning Algorithms Enhance the Specificity of Cancer Biomarker Detection Using SERS-Based Immunoassays in Microfluidic Chips. RSC Adv. 2019, 9, 1859–1868. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, S.; Kang, W.; Jiang, S.; Li, K.; Ray, S.; Luther, E.; Ivanov, A.R.; Fu, Y.; Konry, T. Machine Learning-Aided Quantification of Antibody-Based Cancer Immunotherapy by Natural Killer Cells in Microfluidic Droplets. Lab Chip 2020, 20, 2317–2327. [Google Scholar] [CrossRef]
- Byrne, M.B.; Leslie, M.T.; Gaskins, H.R.; Kenis, P.J.A. Methods to Study the Tumor Microenvironment under Controlled Oxygen Conditions. Trends Biotechnol. 2014, 32, 556–563. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Cole, T.; Zhang, Y.; Kim, J.; Tang, S.-Y. Exploiting Machine Learning for Bestowing Intelligence to Microfluidics. Biosens. Bioelectron. 2021, 194, 113666. [Google Scholar] [CrossRef]
- Farshchi, F.; Hasanzadeh, M. Microfluidic Biosensing of Circulating Tumor Cells (CTCs): Recent Progress and Challenges in Efficient Diagnosis of Cancer. Biomed. Pharmacother. 2021, 134, 111153. [Google Scholar] [CrossRef]
- Xu, H.; Liu, X.; Le, W. Recent Advances in Microfluidic Models for Cancer Metastasis Research. TrAC Trends Anal. Chem. 2018, 105, 1–6. [Google Scholar] [CrossRef]
- Chen, P.; Chen, D.; Li, S.; Ou, X.; Liu, B.-F. Microfluidics towards Single Cell Resolution Protein Analysis. TrAC Trends Anal. Chem. 2019, 117, 2–12. [Google Scholar] [CrossRef]
- Whitesides, G.M. The Origins and the Future of Microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, X.; Zare, R.N. Optimizing Chemical Reactions with Deep Reinforcement Learning. ACS Cent. Sci. 2017, 3, 1337–1344. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Wu, Y.; Fu, J.; Gao, Q.; Qiu, J. Developments of 3D Printing Microfluidics and Applications in Chemistry and Biology: A Review. Electroanalysis 2016, 28, 1658–1678. [Google Scholar] [CrossRef]
- Riordon, J.; Sovilj, D.; Sanner, S.; Sinton, D.; Young, E.W.K. Deep Learning with Microfluidics for Biotechnology. Trends Biotechnol. 2019, 37, 310–324. [Google Scholar] [CrossRef]
- Lin, Z.; Luo, G.; Du, W.; Kong, T.; Liu, C.; Liu, Z. Recent Advances in Microfluidic Platforms Applied in Cancer Metastasis: Circulating Tumor Cells’ (CTCs) Isolation and Tumor-On-A-Chip. Small 2020, 16, 1903899. [Google Scholar] [CrossRef]
- Wilson, R.E.; O’Connor, R.; Gallops, C.E.; Kwizera, E.A.; Noroozi, B.; Morshed, B.I.; Wang, Y.; Huang, X. Immunomagnetic Capture and Multiplexed Surface Marker Detection of Circulating Tumor Cells with Magnetic Multicolor Surface-Enhanced Raman Scattering Nanotags. ACS Appl. Mater. Interfaces 2020, 12, 47220–47232. [Google Scholar] [CrossRef]
- Zhang, A.C.; Gu, Y.; Han, Y.; Mei, Z.; Chiu, Y.-J.; Geng, L.; Cho, S.H.; Lo, Y.-H. Computational Cell Analysis for Label-Free Detection of Cell Properties in a Microfluidic Laminar Flow. Analyst 2016, 141, 4142–4150. [Google Scholar] [CrossRef] [Green Version]
- Zeune, L.L.; Boink, Y.E.; van Dalum, G.; Nanou, A.; de Wit, S.; Andree, K.C.; Swennenhuis, J.F.; van Gils, S.A.; Terstappen, L.W.M.M.; Brune, C. Deep Learning of Circulating Tumour Cells. Nat. Mach. Intell. 2020, 2, 124–133. [Google Scholar] [CrossRef]
- White, A.M.; Zhang, Y.; Shamul, J.G.; Xu, J.; Kwizera, E.A.; Jiang, B.; He, X. Deep Learning-Enabled Label-Free On-Chip Detection and Selective Extraction of Cell Aggregate-Laden Hydrogel Microcapsules. Small 2021, 17, 2102868. [Google Scholar] [CrossRef]
- Bhagat, A.A.S.; Bow, H.; Hou, H.W.; Tan, S.J.; Han, J.; Lim, C.T. Microfluidics for Cell Separation. Med. Biol. Eng. Comput. 2010, 48, 999–1014. [Google Scholar] [CrossRef]
- Cima, I.; Wen Yee, C.; Iliescu, F.S.; Min Phyo, W.; Hon Lim, K.; Iliescu, C.; Han Tan, M. Label-Free Isolation of Circulating Tumor Cells in Microfluidic Devices: Current Research and Perspectives. Biomicrofluidics 2013, 7, 011810. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Sethu, P. Microfluidic Adaptation of Density-Gradient Centrifugation for Isolation of Particles and Cells. Bioengineering 2017, 4, 67. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.; Jia, C.-P.; Sun, W.-J.; Wang, W.-T.; Zhang, H.-L.; Cong, H.; Jing, F.-X.; Mao, H.-J.; Jin, Q.-H.; Zhang, Z.; et al. Highly Sensitive Enumeration of Circulating Tumor Cells in Lung Cancer Patients Using a Size-Based Filtration Microfluidic Chip. Biosens. Bioelectron. 2014, 51, 213–218. [Google Scholar] [CrossRef]
- Sun, N.; Li, X.; Wang, Z.; Li, Y.; Pei, R. High-Purity Capture of CTCs Based on Micro-Beads Enhanced Isolation by Size of Epithelial Tumor Cells (ISET) Method. Biosens. Bioelectron. 2018, 102, 157–163. [Google Scholar] [CrossRef]
- Narayanamurthy, V.; Nagarajan, S.; Firus Khan, A.Y.; Samsuri, F.; Sridhar, T.M. Microfluidic Hydrodynamic Trapping for Single Cell Analysis: Mechanisms, Methods and Applications. Anal. Methods 2017, 9, 3751–3772. [Google Scholar] [CrossRef]
- Alshareef, M.; Metrakos, N.; Juarez Perez, E.; Azer, F.; Yang, F.; Yang, X.; Wang, G. Separation of Tumor Cells with Dielectrophoresis-Based Microfluidic Chip. Biomicrofluidics 2013, 7, 011803. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.Y.; Ahmad Kayani, A.B.; Md Ali, M.A.; Kok, C.K.; Majlis, B.Y.; Hoe, S.L.L.; Marzuki, M.; Khoo, A.S.-B.; Ostrikov, K.; Rahman, M.A.; et al. Dielectrophoresis-Based Microfluidic Platforms for Cancer Diagnostics. Biomicrofluidics 2018, 12, 011503. [Google Scholar] [CrossRef]
- Meng, Y.; Asghari, M.; Aslan, M.K.; Yilmaz, A.; Mateescu, B.; Stavrakis, S.; de Mello, A.J. Microfluidics for Extracellular Vesicle Separation and Mimetic Synthesis: Recent Advances and Future Perspectives. Chem. Eng. J. 2021, 404, 126110. [Google Scholar] [CrossRef]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The Present and Future Role of Microfluidics in Biomedical Research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef]
- Stroock, A.D.; Dertinger, S.K.W.; Ajdari, A.; Mezić, I.; Stone, H.A.; Whitesides, G.M. Chaotic Mixer for Microchannels. Science 2002, 295, 647–651. [Google Scholar] [CrossRef] [Green Version]
- Chu, A.; Nguyen, D.; Talathi, S.S.; Wilson, A.C.; Ye, C.; Smith, W.L.; Kaplan, A.D.; Duoss, E.B.; Stolaroff, J.K.; Giera, B. Automated Detection and Sorting of Microencapsulation via Machine Learning. Lab Chip 2019, 19, 1808–1817. [Google Scholar] [CrossRef] [Green Version]
- Shchanikov, S.; Zuev, A.; Bordanov, I.; Danilin, S.; Lukoyanov, V.; Korolev, D.; Belov, A.; Pigareva, Y.; Gladkov, A.; Pimashkin, A.; et al. Designing a Bidirectional, Adaptive Neural Interface Incorporating Machine Learning Capabilities and Memristor-Enhanced Hardware. Chaos Solitons Fractals 2021, 142, 110504. [Google Scholar] [CrossRef]
- Lashkaripour, A.; Rodriguez, C.; Mehdipour, N.; Mardian, R.; McIntyre, D.; Ortiz, L.; Campbell, J.; Densmore, D. Machine Learning Enables Design Automation of Microfluidic Flow-Focusing Droplet Generation. Nat. Commun. 2021, 12, 25. [Google Scholar] [CrossRef]
- Ko, J.; Bhagwat, N.; Yee, S.S.; Ortiz, N.; Sahmoud, A.; Black, T.; Aiello, N.M.; McKenzie, L.; O’Hara, M.; Redlinger, C.; et al. Combining Machine Learning and Nanofluidic Technology To Diagnose Pancreatic Cancer Using Exosomes. ACS Nano 2017, 11, 11182–11193. [Google Scholar] [CrossRef]
- Joshi, K.; Javani, A.; Park, J.; Velasco, V.; Xu, B.; Razorenova, O.; Esfandyarpour, R. A Machine Learning-Assisted Nanoparticle-Printed Biochip for Real-Time Single Cancer Cell Analysis. Adv. Biosyst. 2020, 4, 2000160. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Meng, J.; Ding, G.; Shi, Z.; Wang, R.; Zhang, X. Detection of Non-small Cell Lung Cancer Cells Based on Microfluidic Polarization Microscopic Image Analysis. Electrophoresis 2019, 40, 1202–1211. [Google Scholar] [CrossRef]
- Ahuja, K.; Rather, G.M.; Lin, Z.; Sui, J.; Xie, P.; Le, T.; Bertino, J.R.; Javanmard, M. Toward Point-of-Care Assessment of Patient Response: A Portable Tool for Rapidly Assessing Cancer Drug Efficacy Using Multifrequency Impedance Cytometry and Supervised Machine Learning. Microsyst. Nanoeng. 2019, 5, 34. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Chen, L.; Wang, Y.; Zhang, T.; Chen, Y.-C.; Yoon, E. Label-Free Estimation of Therapeutic Efficacy on 3D Cancer Spheres Using Convolutional Neural Network Image Analysis. Anal. Chem. 2019, 91, 14093–14100. [Google Scholar] [CrossRef]
- Manak, M.S.; Varsanik, J.S.; Hogan, B.J.; Whitfield, M.J.; Su, W.R.; Joshi, N.; Steinke, N.; Min, A.; Berger, D.; Saphirstein, R.J.; et al. Live-Cell Phenotypic-Biomarker Microfluidic Assay for the Risk Stratification of Cancer Patients via Machine Learning. Nat. Biomed. Eng. 2018, 2, 761–772. [Google Scholar] [CrossRef]
- Bachratý, H.; Bachratá, K.; Chovanec, M.; Jančigová, I.; Smiešková, M.; Kovalčíková, K. Applications of Machine Learning for Simulations of Red Blood Cells in Microfluidic Devices. BMC Bioinform. 2020, 21, 90. [Google Scholar] [CrossRef] [Green Version]
- Wan, Z.Y.; Sapsis, T.P. Machine Learning the Kinematics of Spherical Particles in Fluid Flows. J. Fluid Mech. 2018, 857, R2. [Google Scholar] [CrossRef] [Green Version]
- Rizzuto, V.; Mencattini, A.; Álvarez-González, B.; Di Giuseppe, D.; Martinelli, E.; Beneitez-Pastor, D.; Mañú-Pereira, M.D.M.; Lopez-Martinez, M.J.; Samitier, J. Combining Microfluidics with Machine Learning Algorithms for RBC Classification in Rare Hereditary Hemolytic Anemia. Sci. Rep. 2021, 11, 13553. [Google Scholar] [CrossRef]
- Hervé, L.; Kraemer, D.C.A.; Cioni, O.; Mandula, O.; Menneteau, M.; Morales, S.; Allier, C. Alternation of Inverse Problem Approach and Deep Learning for Lens-Free Microscopy Image Reconstruction. Sci. Rep. 2020, 10, 20207. [Google Scholar] [CrossRef]
- Jayan, H.; Pu, H.; Sun, D.-W. Recent Developments in Raman Spectral Analysis of Microbial Single Cells: Techniques and Applications. Crit. Rev. Food Sci. Nutr. 2021, 61, 1–15. [Google Scholar] [CrossRef]
- Xia, L.; Li, G. Recent Progress of Microfluidics in Surface-enhanced Raman Spectroscopic Analysis. J. Sep. Sci. 2021, 44, 1752–1768. [Google Scholar] [CrossRef]
- Kant, K.; Abalde-Cela, S. Surface-Enhanced Raman Scattering Spectroscopy and Microfluidics: Towards Ultrasensitive Label-Free Sensing. Biosensors 2018, 8, 62. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.; Zhang, A.C.; Han, Y.; Li, J.; Chen, C.; Lo, Y. Machine Learning Based Real-Time Image-Guided Cell Sorting and Classification. Cytom. A 2019, 95, 499–509. [Google Scholar] [CrossRef]
- Honrado, C.; Bisegna, P.; Swami, N.S.; Caselli, F. Single-Cell Microfluidic Impedance Cytometry: From Raw Signals to Cell Phenotypes Using Data Analytics. Lab Chip 2021, 21, 22–54. [Google Scholar] [CrossRef]
- Honrado, C.; McGrath, J.S.; Reale, R.; Bisegna, P.; Swami, N.S.; Caselli, F. A Neural Network Approach for Real-Time Particle/Cell Characterization in Microfluidic Impedance Cytometry. Anal. Bioanal. Chem. 2020, 412, 3835–3845. [Google Scholar] [CrossRef]
- Kaushik, M.; Chandra Joshi, R.; Kushwah, A.S.; Gupta, M.K.; Banerjee, M.; Burget, R.; Dutta, M.K. Cytokine Gene Variants and Socio-Demographic Characteristics as Predictors of Cervical Cancer: A Machine Learning Approach. Comput. Biol. Med. 2021, 134, 104559. [Google Scholar] [CrossRef]
- Yang, Z.; LaRiviere, M.J.; Ko, J.; Till, J.E.; Christensen, T.; Yee, S.S.; Black, T.A.; Tien, K.; Lin, A.; Shen, H.; et al. A Multianalyte Panel Consisting of Extracellular Vesicle MiRNAs and MRNAs, CfDNA, and CA19-9 Shows Utility for Diagnosis and Staging of Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2020, 26, 3248–3258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Gong, M.; Xiong, Z.; Zhao, Y.; Xing, D. Immune-Related Prognostic Genes Signatures in the Tumor Microenvironment of Sarcoma. Math. Biosci. Eng. 2021, 18, 2243–2257. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Waldeisen, J.R.; Huang, T.J. “Microfluidic Drifting”—Implementing Three-Dimensional Hydrodynamic Focusing with a Single-Layer Planar Microfluidic Device. Lab Chip 2007, 7, 1260. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Li, S.; Liu, Y. Machine Learning-Driven Multiobjective Optimization: An Opportunity of Microfluidic Platforms Applied in Cancer Research. Cells 2022, 11, 905. https://doi.org/10.3390/cells11050905
Liu Y, Li S, Liu Y. Machine Learning-Driven Multiobjective Optimization: An Opportunity of Microfluidic Platforms Applied in Cancer Research. Cells. 2022; 11(5):905. https://doi.org/10.3390/cells11050905
Chicago/Turabian StyleLiu, Yi, Sijing Li, and Yaling Liu. 2022. "Machine Learning-Driven Multiobjective Optimization: An Opportunity of Microfluidic Platforms Applied in Cancer Research" Cells 11, no. 5: 905. https://doi.org/10.3390/cells11050905
APA StyleLiu, Y., Li, S., & Liu, Y. (2022). Machine Learning-Driven Multiobjective Optimization: An Opportunity of Microfluidic Platforms Applied in Cancer Research. Cells, 11(5), 905. https://doi.org/10.3390/cells11050905





