Abstract
Sweet cherry, an economically important horticultural crop, has strong antioxidant activity. The fruits contain compounds potentially beneficial to human health—particularly anthocyanins, which are synthesized in cytosol and predominantly accumulated in vacuoles. Although anthocyanin levels differ among dark-red, blush, and yellow sweet cherry cultivars, the regulatory mechanism of anthocyanin transport and accumulation is not well understood in this species. In this study, we identified 53 glutathione S-transferase genes (PavGSTs) from sweet cherry and found that PavGST1 expression was well correlated with anthocyanin accumulation in cultivars with different fruit skin colors. TRV-mediated virus-induced silencing of PavGST1 decreased anthocyanin accumulation in sweet cherry fruits and downregulated the expressions of anthocyanin biosynthetic and regulatory genes. In addition, transient overexpression of PavGST1 promoted anthocyanin accumulation. Furthermore, yeast one-hybrid and dual-luciferase assays revealed that PavMYB10.1 and PavMYB75 directly bind to different MYB binding sites of the PavGST1 promoter (MBS-1 and MBS-3) to activate PavGST1 transcription. According to our results, PavGST1 plays a central role in sweet cherry fruit anthocyanin accumulation. Our findings provide novel insights into the coordinative regulatory mechanisms of PavGST1 and PavMYBs in anthocyanin accumulation in sweet cherry.
1. Introduction
Anthocyanins are important natural water-soluble flavonoid pigments widely distributed in leaves, fruits, seeds, and flowers that are responsible for red, purple, black, and blue coloration in these plant organs. Anthocyanins carry out diverse biological and physiological functions that aid pollination and seed dispersal, confer biotic and abiotic stress resistance, and prolong fruit life span in plants [1,2,3]. Given their potent antioxidant properties, such as immunomodulatory, antioxidant, cardio-protective, antithrombotic, and anti-cancer activities [4,5,6,7], many anthocyanins are also human health-promoting food ingredients.
Anthocyanins are biosynthesized by a series of enzyme-encoding structural genes in the flavonoid pathway that uses phenylalanine as the precursor. These genes include early biosynthetic genes encoding phenylalanine ammonia lyase (PAL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), and flavonoid 3’-hydroxylase (F3’H), as well as late biosynthetic genes for dihydroflavonol 4-reductase (DFR), anthocyanin synthase (ANS), and UDP-glucose:flavonoid 3-glucosyltransferase (UFGT), all having specific contributions to anthocyanin biosynthesis [3,8,9,10]. In addition, synthetic anthocyanins in cytosol are transported to vacuoles for storage [11,12,13]. The involvement of three distinct anthocyanin transporters has been proposed for this process: glutathione S-transferase (GST) transporters, ATP-binding cassette (ABC) transporters, and multidrug and toxic compound extrusion (MATE) family transporters [14,15,16,17,18,19].
In this study, we focused on the GST family. Encoded by a large gene family in plants, GSTs play vital roles in various physiological and developmental processes, including resistance to biotic and abiotic stresses, detoxification of xenobiotics and toxic lipid peroxides, glucosinolate biosynthesis and metabolism, and signaling via catalysis of the conjunction of glutathione (GSH) to various electrophilic compounds [20,21,22]. Numerous GSTs have been reported in plants thus far, including 42 in maize [23], 55 in Arabidopsis [21], 79 in rice [22], 61 in orange [24], 90 in tomato [25], 54 in peach [26], 23 in apple [27], and 97 in kiwifruit [28]. In addition, recent studies have functionally demonstrated that some plant GSTs are essential for anthocyanin and proanthocyanidin vacuolar accumulation. The function of these GSTs in vacuolar anthocyanin transport was first ascertained in the maize bronze-2 (bz2, GST-like protein) mutant [29], which was deduced by the visible loss of pigmentation. GSTs with similar functions have been subsequently identified in various plant species; examples include transparent testa 19 (AtTT19) in Arabidopsis [14], anthocyanin 9 (PhAN9) in petunia [30], flavonoid 3 (fl3) in carnation [31], and CMGSTF12 in Brassicaceae [32]. Anthocyanin-related GSTs have recently been identified as essential for fruit or floral pigmentation in fruit crops, including VviGST1 and VviGST4 in grape [33,34], LcGST4 in litchi [35], FvRAP (encoding a GST transporter for anthocyanin) in strawberry [36], AcGST1 in kiwifruit [28], MdGSTF6 in apple [27], and PpGST1 in peach [37]. In Rosaceae, however, the molecular mechanisms underlying the influence of the GST gene family on anthocyanin transport and accumulation are still unclear.
Sweet cherry (Prunus avium L. (Rosaceae)) is an economically important horticultural crop widely cultivated in temperate regions worldwide [38]. Sweet cherry exhibits a variety of fruit skin colors, including yellow, orange, blush, and dark red. These color differences are due to variation in the content of accumulated anthocyanins, which have strong antioxidant activity and potential benefits for human health [38,39]. Previous studies have demonstrated that the PavMYB10.1-PavbHLH-PavWD40 (MBW) protein complex regulates anthocyanin biosynthesis in sweet cherry and that the R2R3-MYB transcription factor PavMYB10.1 plays a key role in determining different fruit skin coloration patterns [39]. Nevertheless, the molecular mechanisms underlying anthocyanin vacuolar sequestration and accumulation in sweet cherry fruit skin remain unknown, and anthocyanin-related GSTs, which play a vital role in anthocyanin vacuolar accumulation, have not yet been clearly identified in sweet cherry.
In our current study, 53 GST genes were identified from the genome sequence of sweet cherry [40]. Among these genes, PavGST1 was very strongly expressed and found to be highly correlated with anthocyanin accumulation in sweet cherry cultivars with different fruit skin colors. The role of PavGST1 in the regulation of vacuolar accumulation of anthocyanins in sweet cherry was subsequently characterized using overexpression and silencing approaches. In addition, the silencing or overexpression of PavGST1 was revealed to alter fruit skin coloration in sweet cherry, thereby indicating the important function of PavGST1 in anthocyanin accumulation. Further experiments demonstrated that PavMYB10.1 and PavMYB75 can directly bind to the PavGST1 promoter to activate the expression of PavGST1. These findings revealing that PavGST1 encodes the pivotal anthocyanin transporter in sweet cherry provide new insights into the mechanisms underlying anthocyanin accumulation in this important fruit crop species.
2. Materials and Methods
2.1. Plant Materials
Sweet cherry cultivars Chunlu, Hongshouqiu, and 13–33 were grown in the resource garden of the National Fruit Tree Germplasm Repository, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences (Zhengzhou, China). The mature fruits of the three cultivars were harvested at 55 days after full bloom. Nicotiana tabacum used in this study was maintained in our laboratory and was grown at 22–25 °C in a greenhouse with a 16 h light/8 h dark cycle and 60–75% relative humidity.
2.2. Anthocyanin, Proanthocyanidin, Flavonol, and Total Flavonoid Extractions and Measurements
Anthocyanins were extracted from sweet cherry fruits and quantified according to the method of Shen et al., 2017 [41]. The peels were extracted with 1% pre-cooled hydrochloric acid-ethanol solution on ice, followed by centrifugation at 10,000× g for 30 min at 4 °C. Total anthocyanin content, calculated as cyanidin-3-glucoside, was measured by a pH differential method. Proanthocyanidins were extracted from sweet cherry fruits and measured using a previously reported phloroglucinolysis protocol [42,43]. The powder sample (0.5 g) from sweet cherry fruit peel was extracted with 10 mL of anhydrous ethanol. After centrifugation, the precipitate was dissolved in 3 mL vanillin (4% w/v) and 1.5 mL concentrated hydrochloric acid. Then, the absorbance of the reaction mixture was measured at 500 nm by an ultraviolet spectrophotometer. Extraction and quantification of total flavonoids was implemented as previously described [44]. Flavonol content was estimated by high performance liquid chromatography as described previously by Jakobek et al., 2007 [45]. The absorbance was measured at 510 nm for flavonoids and at 425 nm for flavonols, respectively. Six biological replicates and at least three technical replicates were used for each measurement. Fruit color was measured with a Hunter Lab Mini Scan XE Plus colorimeter at four equatorial positions evenly distributed around the fruit.
2.3. Quantitative Real-Time RT-PCR (qRT-PCR) Analysis
Total RNA was extracted and purified from sweet cherry fruits using an EASYspin RNA Plant RNA rapid extraction kit (Yuanpinghao Bio, Tianjin, China) according to the manufacturer’s protocol. The samples triturated with liquid nitrogen were dissolved in the lysis buffer, and the supernatant was collected by centrifugation. After the supernatant passed through the adsorption column, the adsorption column was eluted with RNase-free water. First-strand cDNA was synthesized from purified RNA samples using a FastQuant RT (With gDNase) kit (Tiangen Bio, Beijing, China) following the manufacturer’s instructions. qRT-PCR amplifications were carried out using cDNA templates with TransStart Top Green qPCR SuperMix (TransGen Biotech, Beijing, Chain) containing SYBR green fluorescent intercalating dye on an ABI7500 PCR thermocycler (Applied Biosystems, Foster City, CA, USA). The sweet cherry actin gene (Pav_sc0002247.1_g030.1.mk) was used as an internal control. Relative gene expression was calculated using the 2−∆∆Ct method. The gene-specific primers used for qRT-PCR analysis are detailed in Table S2. Six biological replicates and three technical replicates were used for the qRT-PCR analyses.
2.4. Plasmid Construction and VIGS in Sweet Cherry Fruits
A 426-bp PavGST1 fragment was amplified and cloned into a pTRV2 vector, and the generated recombinant plasmid was transformed into Agrobacterium tumefaciens strain GV3101. VIGS in sweet cherry fruit was carried out as previously described [46,47]. Agrobacterium tumefaciens GV3101 strains expressing pTRV1 and pTRV2 or pTRV2-PavGST1 vectors were mixed in a 1:1 ratio and infiltrated using needleless syringes into the basal pedicel of cherry fruits 25 days after full bloom until the whole fruit was permeated. At 15 dpi, 10 random sweet cherry fruits per strain were collected and photographed. The TRV-mediated gene silencing assays, which involved 50 transformed fruits per strain from the same tree each time, were performed with six biological replicates using six 10-year-old trees.
2.5. Vector Construction and Transient Overexpression in Sweet Cherry Fruits
Transient overexpression assays were performed in sweet cherry fruits using the Agrobacterium tumefaciens method. Briefly, full-length PavGST1 coding sequences were amplified from sweet cherry fruit cDNA and cloned into a pCAMBIA3301 vector. The recombinant plasmids (pCAMBIA3301-PavGST1) and the negative control vector (pCAMBIA3301) were introduced into Agrobacterium tumefaciens GV3101 and then infiltrated into sweet cherry fruits. Agrobacterium strain GV3101 was cultured overnight at 28 °C to an OD600 of 0.8–1.0, resuspended in Agrobacterium infiltration buffer (10 mM MgCl2, 10 mM MES (pH 5.6), and 100 µM acetosyringone) to a final OD600 of 0.8, and then infiltrated into sweet cherry fruits at 35 days after full bloom. The fruits were collected and evaluated at 2 and 3 days after infiltration. Transient overexpression assays were carried out with three independent biological replicates, with at least 20 sweet cherry fruits used per replicate.
2.6. Yeast One-Hybrid (Y1H) Assay
A Y1H assay was performed using the Matchmaker Gold Yeast One-Hybrid system (Clontech, Palo Alto, CA, USA) according to the manufacturer’s instructions. The promoter of PavGST1 was cloned into a pAbAi vector to generate a pAbAi-PavGST1 recombinant plasmid, which was then linearized and inserted into the Y1H Gold yeast strain. The coding sequences of PavMYB10.1 and PavMYB75 were amplified and inserted into a pGADT7 vector to respectively generate pGADT7-PavMYB10.1 and pGADT7-PavMYB75 recombinant plasmids. The pGADT7-PavMYB10.1 and pGADT7-PavMYB75 recombinant plasmids were separately transformed into strain Y1HGold containing pAbAi-PavGST1. The resulting yeasts were incubated on SD/-Leu medium supplemented with aureobasidin A (AbA). Each DNA–protein interaction assay consisted of three biological replicates.
2.7. Dual-Luciferase Reporter Assay
Dual-luciferase reporter assays were performed in N. benthamiana leaves as previously described [48]. To form the reporter construct, the promotors of the PavGST1 gene, pPavGST1, pPavGST1m1, pPavGST1m2, and pPavGST1m3, were amplified and independently inserted into pGreenII 0800-LUC vectors. The full-length ORFs of PavMYB10.1 and PavMYB75 were individually cloned into pGreenII 62-SK vectors to generate effector plasmids. Agrobacterium tumefaciens GV3101 containing the constructed effector and reporter plasmids were co-infiltrated into N. benthamiana leaves. The effector plasmid with an empty vector pGreenII 62-SK was used as a control. Two days later, a Dual-Luciferase Reporter Assay kit (Promega, Madison, WI, USA) was used to measure the activity of LUC and REN with a Luminoskan Ascent microplate luminometer (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The dual-luciferase reporter assay was carried out with at least four biological replicates.
2.8. GUS Activity Analysis
The full-length coding sequences of PavMYB10.1 and PavMYB75 were separately cloned into pCAMBIA1302 to generate the effector plasmids. The promoter fragment of PavGST1 was inserted into pBI121 in place of the 35S promoter coding region to generate the reporter plasmid. The recombinant effector plasmid and reporter plasmid were then used to co-transform tobacco leaves via Agrobacterium tumefaciens strain GV3101. Two days after infiltration, GUS activity was measured as previously described [49]. The assay included at least three biological replicates.
2.9. Statistical Analysis
All data are presented as means ± standard deviation of at least three independent experiments. Statistically significant differences between means were determined using a t-test or one-way analysis of variance (ANOVA) at the 5% significance level in SPSS version 17.0 (SPSS Inc., Chicago, IL, USA). Graphs were constructed with Origin 9.1 (Microcal Software Inc., Northampton, MA, USA).
3. Results
3.1. Anthocyanin Accumulation in Sweet Cherry Fruit Skin during Fruit Development and Ripening
To investigate the basis of fruit skin coloration in sweet cherry, we evaluated the phenotypes and flavonoid composition of three cultivars with different fruit skin colors: dark red Chunlu, blush Hongshouqiu, and yellow 13–33. The developmental process of Chunlu, Hongshouqiu, and 13–33 sweet cherry fruits is shown in Figure 1A. During early fruit growth and development (1–4 weeks after full bloom (WAFB)), fruit skin color did not significantly differ among Chunlu, Hongshouqiu, and 13–33. After the yellow-white stage of sweet cherry fruit growth and development (5 WAFB), significant differences in fruit skin color were observed among the three cultivars (Figure 1A). The anthocyanin content of Chunlu increased rapidly with fruit development (from 5 to 9 weeks) and was higher than in Hongshouqiu and 13–33. The anthocyanin content of Hongshouqiu was low and increased slowly from 5 to 9 weeks, whereas no anthocyanins were detected in 13–33 (Figure 1B). Similarly, the proanthocyanidin and especially total flavonoid contents of Chunlu fruits were significantly higher than those of Hongshouqiu and 13–33 during later fruit development and ripening (7–9 WAFB). No significant difference in flavonol contents was detected among Chunlu, Hongshouqiu, and 13–33 (Figure 1C). These results suggest that differences in fruit skin color in sweet cherry are due to variations in anthocyanin accumulation.
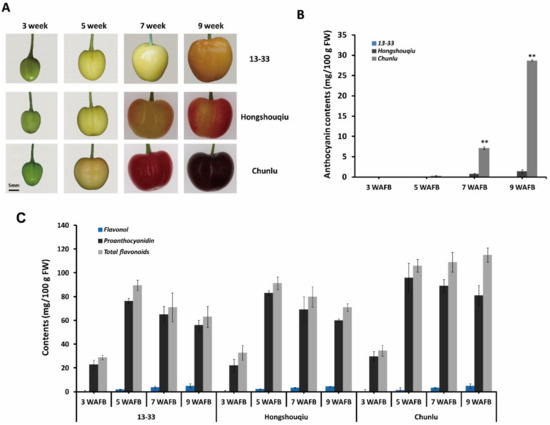
Figure 1.
Developing fruits (A) and anthocyanin (B), proanthocyanidin (C), flavonol (C), and total flavonoid (C) contents measured 3, 5, 7, and 9 weeks after full bloom (WAFB) in sweet cherry varieties Chunlu, Hongshouqiu, and 13–33. Scale bar indicates 5 mm. Values are means ± SD from six independent biological replicates. ** Significant differences as calculated using the Student’s t-test at p < 0.01.
3.2. Strong Correlation between PavGST1 Transcript Levels and Anthocyanin Accumulation in Sweet Cherry
To investigate the correlation between GSTs and anthocyanin accumulation, we examined transcript levels of 53 sweet cherry GST genes at different stages in the cultivars Chunlu, Hongshouqiu, and 13–33 by quantitative real-time RT-PCR (qRT-PCR). Among the 53 PavGSTs, Pav_sc0001124.1_g450.1.mk (named PavGST1) was more highly expressed in Chunlu fruits than those of Hongshouqiu and 13–33, and fruit transcript levels of PavGST1 in Hongshouqiu were significantly higher than in 13–33 (Figure 2). Moreover, the expression level of PavGST1 rapidly and gradually increased during later fruit development and ripening (5–9 WAFB) in Chunlu and Hongshouqiu fruits, respectively, consistent with the anthocyanin contents of their fruit skins. In contrast, PavGST1 expression was not detected during fruit growth, development, or ripening in sweet cherry cultivar 13–33 (Figure 2). These results indicate that PavGST1 probably participates in anthocyanin accumulation in sweet cherry.
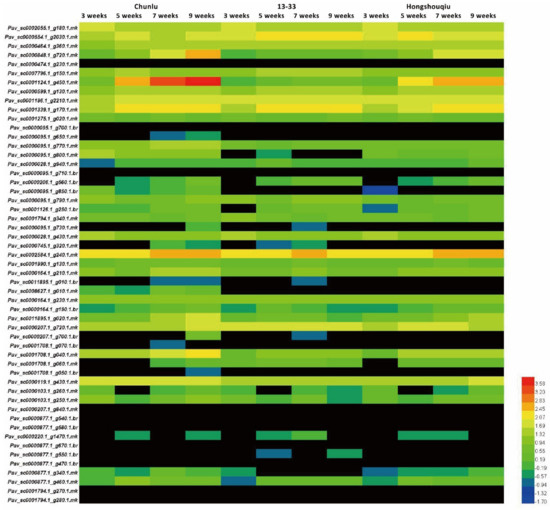
Figure 2.
Expression patterns of 53 PavGST genes in developing fruits of sweet cherry varieties Chunlu, Hongshouqiu, and 13–33.
3.3. Significantly Decreased Anthocyanin Accumulation in Sweet Cherry Fruits following Silencing of PavGST1 by VIGS
To determine whether PavGST1 is essential for anthocyanin accumulation in sweet cherry fruits during fruit growth and development, we performed virus-induced gene silencing (VIGS) to knock down the expression of the PavGST1 gene in sweet cherry fruits. According to qRT-PCR, the expression of PavGST1 was dramatically reduced in PavGST1-silenced fruits compared with control fruits (TRV) at 15 days post-inoculation (dpi), thus indicating that PavGST1 was efficiently silenced (Figure 3A). Compared with control fruits, PavGST1-silenced fruits had visible fruit skin color defects at 15 and 25 dpi (Figure 3B). The epidermis of PavGST1-silenced fruits was green-yellow or pink around the injection site at 15 and 25 dpi, whereas that of control fruits was dark or blackish red (Figure 3B). This result indicates that transient knockdown of PavGST1 dramatically reduced sweet cherry fruit coloration. At the same time, anthocyanin contents around the injection site of PavGST1-silenced fruits were significantly decreased compared with those of control fruits at 15 and 25 dpi (Figure 3C). These results suggest that PavGST1 plays an important role in anthocyanin accumulation in sweet cherry.
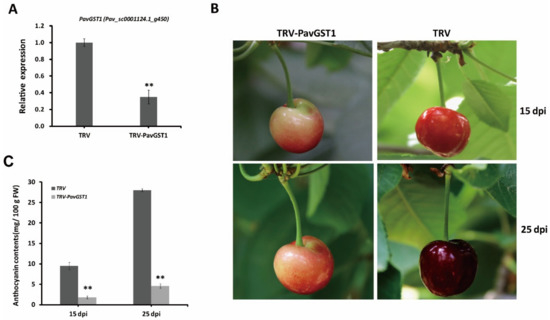
Figure 3.
Effect of PavGST1 silencing on anthocyanin accumulation in sweet cherry fruits. (A) PavGST1 transcript levels in PavGST1-silenced and control fruits relative to Pavactin at 15 days post-inoculation (dpi), as determined by qRT-PCR. (B) Phenotypes of PavGST1-silenced and control fruits at 15 and 25 dpi in Chunlu sweet cherry varieties. (C) Changes in anthocyanin content in PavGST1-silenced and control fruits at 15 and 25 dpi. Values are means ± SD from six independent replicates. Statistical significance was determined according to one-way analysis of variance (ANOVA, **, p ˂ 0.01).
3.4. Downregulation of Anthocyanin Biosynthetic Structural Genes in PavGST1-Silenced Sweet Cherry Fruits
To further understand the role of PavGST1 in anthocyanin accumulation in sweet cherry, we used qRT-PCR to measure transcript levels of key structural genes in the anthocyanin biosynthetic pathway, including PavPAL, PavC4H, Pav4CL, PavCHS, PavCHI, PavF3H, PavFLS, PavDFR, PavANS, PavLAR, PavANR, and PavUFGT, in PavGST1-silenced fruits and control fruits at 15 and 25 dpi. As shown in Figure 4, the expressions of early structural genes in the anthocyanin biosynthesis pathway, including PavPAL, PavC4H, Pav4CL, PavCHS, PavCHI, and PavF3H, were significantly downregulated in PavGST1-silenced fruits compared with control fruits at 15 and 25 dpi (Figure 4). In contrast, transcription of the flavonol-biosynthesis key structural gene PavFLS was upregulated by the silencing of PavGST1, particularly at 25 dpi (Figure 4). Compared with the controls, transcript levels of late biosynthetic genes that specifically contribute to anthocyanin biosynthesis, namely, PavDFR, PavANS, and PavUFGT, were significantly downregulated in PavGST1-silenced sweet cherry fruits. In contrast, expression levels of the late PA biosynthetic genes PavLAR and PavANR were slightly higher in PavGST1-silenced sweet cherry fruits than in control fruits at 15 and 25 dpi (Figure 4).
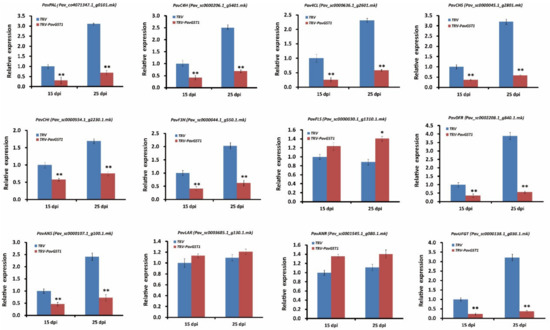
Figure 4.
Transcript levels of anthocyanin pathway genes PavPAL, PavC4H, Pav4CL, PavCHS, PavCHI, PavF3H, PavFLS, PavDFR, PavANS, PavLAR, PavANR, and PavUFGT in PavGST1-silenced sweet cherry fruits and control fruits at 15 and 25 dpi. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate-CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase, LAR, leucoanthocyanidin reductase; ANR, anthocyanidin reductase; UFGT, UDP-glucose:flavonoid-3-O-glucosyltransferase. Values are means ± SD from six independent replicates. Significant differences among means were determined by one-way analysis of variance (ANOVA, *, p < 0.05, **, p < 0.01).
3.5. Promotion of Anthocyanin Biosynthesis in Sweet Cherry Fruits by Transient Overexpression of PavGST1
To further validate that PavGST1 is essential for sweet cherry fruit coloration, we transiently overexpressed this gene in sweet cherry fruits. The overexpression recombinant plasmid p35S::PavGST1 and the pCAMBIA3301 empty vector (as a control) were separately introduced into sweet cherry fruits via Agrobacterium-mediated transformation. Two days after inoculation, a clear red color was observed around the injection site of skins of PavGST1 transiently overexpressed fruits, whereas no obvious red color was detected around the injection sites of control fruits (Figure 5A). Compared with the empty vector control, intense red pigmentation was observed around the injection site of skins of PavGST1-overexpressing sweet cherry fruits 3 days after infiltration (Figure 5A). The expression of PavGST1 was markedly upregulated in sweet cherry fruits transiently overexpressing PavGST1 compared with control fruits (Figure 5B). Additionally, anthocyanin contents as well as the abundance of PavGST1 protein were significantly higher in sweet cherry fruits transiently overexpressing PavGST1 compared with empty vector control fruits at 48 and 72 h after inoculation (Figure 5C). We also measured transcript levels of anthocyanin biosynthetic structural genes by qRT-PCR. Transcript levels of the structural genes PavPAL, PavC4H, Pav4CL, PavCHS, PavCHI, PavF3H, PavDFR, PavANS, and PavUFGT were dramatically higher in PavGST1-overexpressing fruits than in control fruits, whereas expression levels of proanthocyanidin or flavonol biosynthetic structural genes PavLAR, PavANR, and PavFLS were almost the same between fruits transiently overexpressing PavGST1 and control fruits (Figure 5D). These results further confirm that PavGST1 promotes anthocyanin accumulation in sweet cherry.
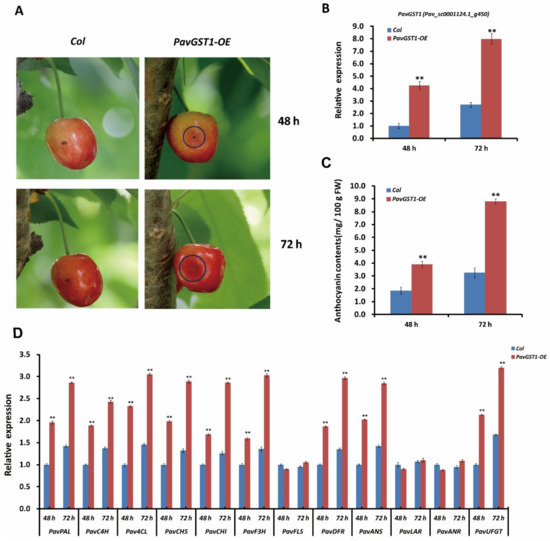
Figure 5.
Enhanced anthocyanin accumulation in sweet cherry fruits resulting from transient expression of PavGST1. (A) Phenotypes of PavGST1 transiently overexpressing fruits and control fruits recorded 48 and 72 h after inoculation in Chunlu sweet cherry varieties. (B) Results of qRT-PCR analysis of PavGST1 expression levels in PavGST1 transiently overexpressing fruits and control fruits. The Pavactin gene was used as an internal control. (C) Anthocyanin contents of PavGST1 transiently overexpressing fruits compared with the control. (D) Transcript levels of anthocyanin pathway genes in PavGST1 transiently overexpressing fruits and control fruits at 48 and 72 h after inoculation. Values represent means ± SD from six independent replicates. Statistical significance was determined using Student’s t-test (**, p ˂ 0.01).
3.6. Regulation of PavMYB10.1 and PavMYB75 Expression Levels by PavGST1 and Their Direct Binding to the PavGST1 Promoter
Multiple studies have demonstrated that four R2R3-MYB TF family members, namely, AtMYB75/PAP1, AtMYB90/PAP2, AtMYB113, and AtMYB114, regulate anthocyanin biosynthesis in Arabidopsis. The MYB-bHLH-WD40 (MBW) protein complex has been confirmed as central to the regulation of anthocyanin accumulation in many plant species. To evaluate the interaction between PavGST1 and PavMYBs, we analyzed expression levels of four Arabidopsis homologous anthocyanin-related R2R3-MYB TF family members—PavMYB75/PAP1 (Pav_sc0000464.1_g100), PavMYB10.1 (Pav_sc0000464.1_g130), PavMYB113 (Pav_sc0000464.1_g250), and PavMYB114 (Pav_sc0000464.1_g210), as well as PavbHLH3 (Pav_sc0000146.1_g300) and PavWD40 (Pav_sc0000138.1_g560)—in PavGST1-silenced sweet cherry fruits and control fruits or in PavGST1-overexpressing fruits and control fruits. No PavMYB113 or PavMYB114 gene expression was detected in sweet cherry fruits (Figure S1). Interestingly, PavMYB10.1 expression was more highly upregulated in PavGST1-overexpressing fruits than control fruits; in contrast, transcript levels of PavMYB10.1 in PavGST1-silenced sweet cherry fruits were significantly lower than in control fruits (Figure 6A). Moreover, PavMYB75 exhibited expression levels similar to those of PavMYB10.1 in PavGST1-silenced sweet cherry fruits and PavGST1-overexpressing fruits, whereas the expressions of PavbHLH3 and PavWD40 did not significantly differ between PavGST1-silenced sweet cherry fruits or PavGST1-overexpressing fruits and control fruits (Figure 6A).
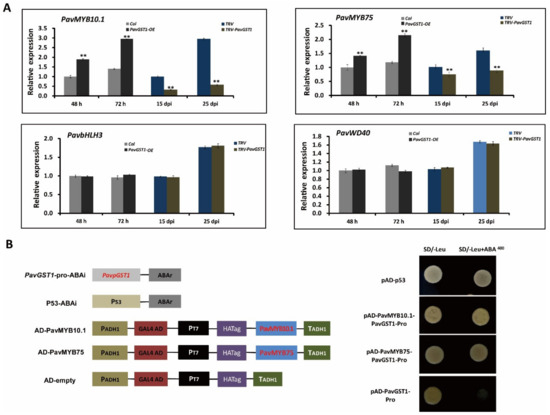
Figure 6.
Data showing that PavMYB10.1 and PavMYB75 expression levels are regulated by PavGST1 and their direct binding to the PavGST1 promoter. (A) Changes in transcript levels of PavMYB75, PavMYB10.1, PavbHLH3, and PavWD40 genes in PavGST1-overexpressing and PavGST1-silenced sweet cherry fruits. (B) Results of yeast one-hybrid (Y1H) assays showing PavMYB10.1 and PavMYB75 individually bound to the PavGST1 promoter. Values represent means ± SD from six independent replicates. Statistical significance was determined according to one-way analysis of variance (ANOVA, **, p < 0.01).
To further explore the correlation between PavMYB10.1 or PavMYB75 and PavGST1, the 2500 bp upstream region of the PavGST1 promoter was cloned and then analyzed using PlantCARE online tools. Predicted and putative cis-regulatory elements of the PavGST1 promoter region, such as the TATA-box, CAAT-box, A-box, and G-box, are listed in Table S1. We also detected several major light-responsive elements (G-Box, GATA-box, GT1-motif, and TCCC-motif); hormone-responsive elements, including methyl jasmonate (MeJA)-responsive elements (CGTCA-motif, TGACG-motif), ABA-responsive elements (ABRE, TGACG-motif), and an auxin-responsive element (TGA-element); and defense and stress-responsive elements (TC-rich repeats), which suggests that the expression of PavGST1 is controlled by light, hormones, and abiotic stresses. In addition, three MYB binding site (MBS) elements (CAACCA) were located at −1464 bp, −1781 bp, and −2154 bp upstream of the transcriptional start site (Table S1).
Next, yeast one-hybrid assays were performed to assess whether PavGST1 is directly regulated by PavMYB10.1 and PavMYB75 in sweet cherry. No growth of transformant yeast cells co-expressing the PavGST1 promoter and the AD-Empty vector was observed on SD/-Leu medium supplemented with AbA. In contrast, the resistance reporter gene AbA driven by the PavGST1 promoter and the PavMYB10.1- or PavMYB75-AD recombinant plasmid were co-expressed in yeast cells, and the yeast transformant cells were able to grow on SD/-Leu medium supplemented with AbA (Figure 6B). These results indicate that PavMYB10.1 and PavMYB75 directly bind to the promoter regions of PavGST1 in vitro.
3.7. Direct Activation of PavGST1 Expression by Both PavMYB10.1 and PavMYB75
To validate whether both PavMYB10.1 and PavMYB75 affect the transcriptional activity of PavGST1, we performed GUS activity and dual-luciferase (Luc) transient expression assays. For the GUS assay, the reporter plasmid PavGST1-Pro::GUS and the effector plasmids p35S::PavMYB10.1, p35S::PavMYB75, or p35S::PavMYB10.1-PavbHLH3 were constructed and transformed into tobacco leaves (Figure 7A). GUS activities were significantly higher in tobacco leaves co-transformed with PavGST1-Pro-GUS/CaMV35S-PavMYB10.1, PavGST1-Pro-GUS/CaMV35S-PavMYB75, or PavGST1-Pro-GUS/CaMV35S-PavMYB10.1-PavbHLH3 than in leaves co-transformed with PaPG1Pro-GUS/CaMV35S-Empty; this was especially the case for leaves co-transformed with PavGST1-Pro-GUS/CaMV35S-PavMYB10.1-PavbHLH3 (Figure 7B). In addition, PavMYB10.1 co-transformed into tobacco leaves exhibited higher transactivation effects on the PavGST1 promoter (Figure 7B) than did co-transformed PavGST1-Pro-GUS/CaMV35S-PavMYB75. We subsequently carried out a dual-luciferase (Luc) transient expression assay in tobacco leaves with a dual-reporter construct system containing PavGST1 promoter-driven LUC and CaMV35S promoter-driven Renilla luciferase (REN) and used an effector plasmid expressing PavMYB10.1, PavMYB75, or PavMYB10.1+PavbHLH3 driven by the CaMV35S promoter to determine the activation effect of PavMYB10.1 and PavMYB75 on PavGST1 expression (Figure 7C). Compared with the control, the LUC/REN ratio was significantly increased when the PavGST1 promoter-LUC reporter was co-transfected into tobacco leaves along with PavMYB10.1, PavMYB75, or PavMYB10.1+PavbHLH3 (Figure 7D); this was especially true for PavMYB10.1+PavbHLH3, which resulted in the highest LUC/REN ratio. Taken together, these results confirm that PavMYB10.1 and PavMYB75 can positively activate the transcription of PavGST1.
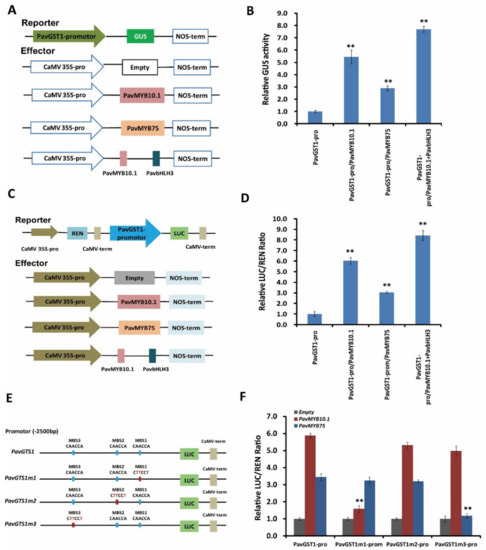
Figure 7.
Direct activation of the PavGST1 promoter by PavMYB10.1 and PavMYB75. (A) Effector and reporter vector construction diagrams for GUS activity assays. (B) Results of GUS activity assays showing that PavMYB10.1, PavMYB75, and PavMYB10+PavbHLH3 all significantly activated the promoter of PavGST1. (C) Schematic representation of effector and reporter vectors used for dual-luciferase assays. (D) Effects of PavMYB10.1, PavMYB75, and PavMYB10+PavbHLH3 on PavGST1 promoter activity according to dual-luciferase assays. (E) Schematic diagram of PavGST1 promoter motif mutations. (F) Effects of PavMYB10.1 and PavMYB75 on the activities of original and mutated promoters of PavGST1 in dual-luciferase reporter assays. Each experiment included three replicates. Values represent means ± SD from six independent replicates. Significant differences were determined using Student’s t-test (**, p < 0.01).
To explore whether the differential effects of PavMYB10.1 and PavMYB75 on the transcriptional activity of PavGST1 are due to distinct motifs in the PavGST1 promoter, we divided the full-length PavGST1 promoter into three fragments (PavGST1m1–PavGST1m3) relative to the three MBSs (Figure 7E) and carried out dual-luciferase assays to measure the PavGST1 promoter activity of each fragment. As shown in Figure 7F, changing the MBS1 element in the PavGST1 promoter from CAACCA to CTTCCT (PavGST1m1) resulted in significantly reduced transactivation activity by PavMYB10.1, but not by PavMYB75, relative to the non-mutated PavGST1 promoter (PavGST1). Similarly, mutation of the MBS3 element from CAACCA to CTTCCT in the PavGST1 promoter (PavGST1m3) significantly reduced (by 63%) the transactivation activity of PavMYB75 compared with the original PavGST1 promoter (Figure 7F), whereas the transactivation activity of PavMYB10.1 did not significantly differ between MBS3-mutated and non-mutated PavGST1 promoters (Figure 7F). Furthermore, PavMYB10.1 and PavMYB75 exerted no obvious transactivation activities when the MBS2 element of the PavGST1 promoter was changed from CAACCA to CTTCCT (PavGST1m2) (Figure 7F). These results confirm that PavMYB10.1 and PavMYB75 proteins specifically bind to MBS1 and MBS3, respectively, in the PavGST1 promoter.
4. Discussion
Fruit color is an important agronomic trait that largely determines sweet cherry fruit quality, and a red fruit skin color, which is mainly due to anthocyanin accumulation, is an important target in sweet cherry breeding [39]. Accumulated anthocyanins not only contribute to sweet cherry fruit coloration, but also protect against certain diseases and are beneficial to human health. Elucidation of the biosynthetic processes involved in anthocyanin accumulation would therefore be a major contribution to the improvement of sweet cherry fruit quality.
4.1. The Essential Role of PavGST1 Encoding GST in Anthocyanin Transport and Accumulation in Sweet Cherry
As is well known, anthocyanins are biosynthesized in the cytoplasm and accumulated in vacuoles. Previous research has revealed that GST-mediated transport plays a pivotal role in anthocyanin accumulation in many plant species, including maize [29], petunia [30], perilla [49], Arabidopsis [14,50], grape [33], litchi [35], apple [27], kiwifruit [28], and peach [37]. In the present study, we identified 53 GST genes and analyzed their expressions in three sweet cherry cultivars with different skin colors, namely, Chunlu, Hongshouqiu, and 13–33. We found a GST gene (PavGST1, Pav_sc0001124.1_g450.1.mk) whose expression was consistent with anthocyanin accumulation in fruit skins of the different sweet cherry cultivars, thus indicating that PavGST1 may play an important role in anthocyanin transport in sweet cherry. When we transiently overexpressed PavGST1 and knocked down PavGST1 expression in sweet cherry fruits, anthocyanin accumulations were significantly changed. Our results demonstrate that PavGST1 encoding GST is essential for anthocyanin transport and accumulation. The functional role of GSTs in anthocyanin transport and accumulation has been ascertained by studying their loss-of-function mutants, such as bz2 (bronze-2) from maize [29], an9 (anthocyanin 9) from petunia [30], fl3 (flavonoid 3) from carnation [31], tt19 (transparent testa 19) from Arabidopsis [14], and rap (reduced anthocyanin in petioles) from strawberry [36].
At the same time, several GSTs in fruit tree species have been thoroughly studied through genetic approaches. These investigations in peach, apple, kiwifruit, and litchi have demonstrated that a GST member can be simultaneously responsible for anthocyanin sequestration and accumulation [27,28,35,37]. Heterologous expression of PpGST1 [37] (Prunus persica), VviGST4 [34] (Vitis vinifera), LcGST4 [35] (Litchi chinensis), FaRAP [36] (Fragaria ananassa), MdGSTF6 [27] (Malus domestica), and AcGST1 [28] (Actinidia chinensis) in an Arabidopsis tt19 mutant has been found to functionally complement the anthocyanin-less phenotype in vegetative tissues, thus indicating that these anthocyanin-related GST genes possess similar, conserved functions in the regulation of anthocyanin transport and accumulation. Our findings similarly indicate that transient overexpression of PavGST1 promotes anthocyanin accumulation in sweet cherry fruits.
Our study has also revealed that the major flavonoids in sweet cherry fruits are proanthocyanidins, anthocyanins, and flavonols and that (especially) flavonol and anthocyanin contents differ significantly among sweet cherry cultivars with different fruit skin colors. Among the 53 analyzed GST genes, PavGST1 was the only one whose expression was highly correlated with different sweet cherry fruit skin colors. We hypothesize that PavGST1 is related to both anthocyanin and proanthocyanidin accumulation. In Arabidopsis, TT19 participates in the transport and accumulation of both anthocyanins and proanthocyanidins, which has been inferred from the loss of TT19 function in an Arabidopsis mutant phenotype [51]. VviGST4 and AcGST1 are involved in both anthocyanin and proanthocyanidin accumulation as well, as these two genes functionally complement anthocyanin-less and PA-deficient phenotypes in vegetative tissues and seed coats, respectively, of the Arabidopsis tt19 mutant [28,34].
4.2. A Putative Model for PavGST1 Expression during Sweet Cherry Fruit Skin Coloration
Several previous studies have revealed that MYB1/A/10 (an MYB transcription factor) can directly bind to anthocyanin transport-related GST gene promoters and activate their transcription. In Arabidopsis, overexpression of the PAP1 gene (a R2R3-MYB transcription factor) activates most genes related to anthocyanin biosynthesis and transportation, including AtTT19, AtDFR, and AtUFGT [52]. In apple, MdMYB1 directly regulates and activates MdGSTF6 expression [27]. In peach, PpMYB10.1 specifically recognizes the MBS1 element of the PpGST1 promoter to directly activate the expression of this gene to regulate anthocyanin accumulation [37]. In strawberry, the expression of FvRAP is dramatically induced by the overexpression of FvMYB10 and significantly reduced in the fruit of myb10 varieties [36]. Similar results have also been observed in tea, lilies, litchi, and kiwifruit [35,36,53,54]. According to our study, both PavMYB10.1 and PavMYB75 can directly activate the promoter activity of PavGST1. In addition, we found that PavMYB10.1 and PavMYB75 proteins specifically bind to different MBS elements in the PavGST1 promoter sequence, which suggests that expression of PavGST1 is regulated via different R2R3 MYB transcription factors. Furthermore, transient overexpression of PavGST1 in sweet cherry accelerates PavMYB10.1 and PavMYB75 transcription. We thus speculate that the coordinated roles of PavGST1 in anthocyanin transport and PavMYB10.1 and PavMYB75 in anthocyanin biosynthesis ultimately control sweet cherry fruit skin coloration [55,56]. In strawberry, overexpression of FvMYB10 in the myb10−rap− double-mutant blocks red color development [36], which suggests that the function of FvMYB10 in fruit anthocyanin accumulation depends on FvGST/RAP.
On the basis of the above-mentioned results, we propose a functional model to explain the role of PavGST1 in anthocyanin accumulation in sweet cherry fruits (Figure 8). In our model, two R2R3-MYB transcription factors, PavMYB10.1 and PavMYB75, directly regulate PavGST1 expression by specifically binding to different MBSs of the PavGST1 promoter region and act as positive regulators of PavGST1 to promote anthocyanin accumulation. PavGST1 can accelerate the expression of PavMYB10.1 and PavMYB75, which might enhance anthocyanin accumulation. We conclude that PavMYB10.1 not only controls anthocyanin synthesis but also regulates anthocyanin transport in sweet cherry. Additional research is needed to determine whether the PavMYB75 gene regulates anthocyanin accumulation in sweet cherry.
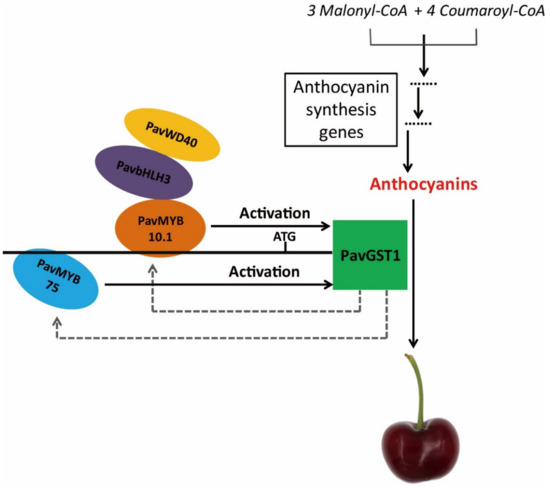
Figure 8.
Proposed model of the role of PavMYB10.1 and PavMYB75 transcription factors in the positive regulation of anthocyanin accumulation by PavGST1 in sweet cherry. In this model, PavMYB10.1 and PavMYB75 bind to different MBS recognition sites of the PavGST1 promoter to activate PavGST1 transcription.
In conclusion, the transient silencing and overexpression analyses of PavGST1 in this study have revealed that PavGST1, a GST gene, plays an indispensable role in anthocyanin transport and accumulation in sweet cherry fruits. More specifically, PavMYB10.1 and PavMYB75 directly bind to the promoter of PavGST1 to enhance its transcriptional activity. Our study has provided novel insights into the molecular mechanisms by which PavMYB10.1 and PavMYB75 regulate the activity of PavGST1 during anthocyanin transport and accumulation in sweet cherry.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells11071170/s1, Figure S1: Expression profiles of anthocyanin-related R2R3-MYB TF family members during sweet cherry fruit growth and development; Table S1: Cis-acting elements found in the upstream region of PavGST1 gene: Table S2: Primers used in this study.
Author Contributions
X.Q. and M.L. conceived the project. X.Q. designed the experiments. X.Q., C.L., Y.D., L.C., and L.S. performed the experiments. X.Q. analyzed the results and wrote the manuscript. M.L. provided scientific suggestions and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant from the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (grant no. CAAS-ASTIP-2021-ZFRI, the Natural Science Foundation of Henan Province of China (grant nos. 202300410533 and 212300410419), and National Natural Science Foundation of China (3210180675).
Conflicts of Interest
The authors declare no conflict of interest.
References
- He, J.; Giusti, M.M. Anthocyanins: Natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Butelli, E.; De Stefano, R.; Schoonbeek, H.J.; Magusin, A.; Pagliarani, C.; Wellner, N.; Hill, L.; Orzaez, D.; Granell, A.; et al. Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr. Biol. 2013, 23, 1094–1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagchi, D.; Bagchi, M.; Stohs, S.J.; Das, D.K.; Ray, S.D.; Kuszynski, C.A.; Joshi, S.S.; Pruess, H.G. Free radicals and grape seed proanthocyanidin extract: Importance in human health and disease prevention. Toxicology 2000, 148, 187–197. [Google Scholar] [CrossRef]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Clifford, T.; Howatson, G.; West, D.J.; Stevenson, E.J. The potential benefits of red beetroot supplementation in health and disease. Nutrients 2015, 7, 2801–2822. [Google Scholar] [CrossRef]
- Petroni, K.; Pilu, R.; Tonelli, C. Anthocyanins in corn: A wealth of genes for human health. Planta 2014, 240, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Holton, T.A.; Cornish, E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef]
- Xie, D.Y.; Sharma, S.B.; Paiva, N.L.; Ferreira, D.; Dixon, R.A. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 2003, 299, 396–399. [Google Scholar] [CrossRef] [Green Version]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef]
- Grotewold, E.; Davies, K. Trafficking and sequestration of anthocyanins. Nat. Prod. Commun. 2008, 3, 1251–1258. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Dixon, R.A. The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci. 2010, 15, 72–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez, C.; Conejero, G.; Torregrosa, L.; Cheynier, V.; Terrier, N.; Ageorges, A. In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant J. 2011, 67, 960–970. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, H.; Huang, J.R. Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant. 2012, 5, 387–400. [Google Scholar] [CrossRef]
- Muñoz, C.; Hoffmann, T.; Escobar, N.M.; Ludemann, F.; Botella, M.A.; Valpuesta, V.; Schwab, W. The strawberry fruit Fra a allergen functions in flavonoid biosynthesis. Mol. Plant. 2010, 3, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Francisco, R.M.; Regalado, A.; Ageorges, A.; Burla, B.J.; Bassin, B.; Eisenach, C.; Zarrouk, O.; Vialet, S.; Marlin, T.; Chaves, M.M.; et al. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell 2013, 25, 1840–1854. [Google Scholar] [CrossRef] [Green Version]
- Marinova, K.; Pourcel, L.; Weder, B.; Schwarz, M.; Barron, D.; Routaboul, J.M.; Debeaujon, I.; Klein, M. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 2007, 19, 2023–2038. [Google Scholar] [CrossRef] [Green Version]
- Gomez, C.; Terrier, N.; Torregrosa, L.; Vialet, S.; Fournier-Level, A.; Verries, C.; Souquet, J.; Mazauric, J.; Klein, M.; Cheynier, V.; et al. Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol. 2009, 150, 402–415. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Huhman, D.; Shadle, G.; He, X.Z.; Sumner, L.W.; Tang, Y.; Dixon, R.A. MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell 2011, 23, 1536–1555. [Google Scholar] [CrossRef] [Green Version]
- Marrs, K.A. The functions and regulation of glutathione S-transferases in plants. Annu. Rev. Plant Biol. 1996, 47, 127–158. [Google Scholar] [CrossRef]
- Sappl, P.G.; Carroll, A.J.; Clifton, R.; Lister, R.; Whelan, J.; Harvey Millar, A.; Singh, K.B. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J. 2009, 58, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Ghanashyam, C.; Bhattacharjee, A. Comprehensive expression analysis suggests overlapping and specific roles of rice glutathione S-transferase genes during development and stress responses. BMC Genom. 2010, 11, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGonigle, B.; Keeler, S.J.; Lau, S.M.; Koeppe, M.K.; O’Keefe, D.P.A. genomics approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize. Plant Physiol. 2000, 124, 1105–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Licciardello, C.; D’Agostino, N.; Traini, A.; Recupero, G.R.; Frusciante, L.; Chiusano, M.L. Characterization of the glutathione S-transferase gene family through ESTs and expression analyses within common and pigmented cultivars of Citrus sinensis (L.) Osbeck. BMC Plant Biol. 2014, 14, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Islam, S.; Rahman, I.A.; Islam, T.; Ghosh, A. Genome-wide identification and expression analysis of glutathione S-transferase gene family in tomato: Gaining an insight to their physiological and stress-specific roles. PLoS ONE 2017, 12, e0187504. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Dong, W.Q.; Wang, K.; Zhang, B.; Allan, A.C.; Wang, K.L.; Chen, K.S.; Xu, C.J. Differential sensitivity of fruit pigmentation to ultraviolet light between two peach cultivars. Front. Plant Sci. 2017, 8, 1552. [Google Scholar] [CrossRef]
- Jiang, S.; Chen, M.; He, N.; Chen, X.L.; Wang, N.; Sun, Q.G.; Zhang, T.L.; Xu, H.F.; Fang, H.C.; Wang, Y.C.; et al. MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Hortic. Res. 2019, 6, 40. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Qi, Y.; Zhang, A.; Wu, H.; Liu, Z.; Ren, X. Molecular cloning and functional characterization of AcGST1, an anthocyanin-related glutathione S-transferase gene in kiwifruit (Actinidia chinensis). Plant Mol. Biol. 2019, 100, 451–465. [Google Scholar] [CrossRef]
- Marrs, K.A.; Alfenito, M.R.; Lloyd, A.M.; Walbot, V. A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 1995, 375, 397–400. [Google Scholar] [CrossRef]
- Mueller, L.A.; Goodman, C.D.; Silady, R.A.; Walbot, V. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein1. Plant Physiol. 2000, 123, 1561–1570. [Google Scholar] [CrossRef] [Green Version]
- Larsen, E.S.; Alfenito, M.R.; Briggs, W.R.; Walbot, V. A carnation anthocyanin mutant is complemented by the glutathione S-transferases encoded by maize Bz2 and petunia An9. Plant Cell Rep. 2003, 21, 900–904. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, Y.; Zhang, M.; Cai, F.; Qin, J.; Wang, Q.; Liu, C.; Wang, G.; Xu, L.; Yang, L.; et al. Molecular cloning and functional characterization of a glutathione S-transferase involved in both anthocyanin and proanthocyanidin accumulation in Camelina sativa (Brassicaceae). Genet. Mol. Res. 2012, 11, 4711–4719. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.; Curtin, C.; Bézier, A.; Franco, C.; Zhang, W. Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J. Exp. Bot. 2008, 59, 3621–3634. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Díaz, R.; Madrid-Espinoza, J.; Salinas-Cornejo, J.; González-Villanueva, E.; Ruiz-Lara, S. Differential roles for VviGST1, VviGST3, and VviGST4 in proanthocyanidin and anthocyanin transport in Vitis vinifera. Front. Plant Sci. 2016, 7, 1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Zhao, J.; Lai, B.; Qin, Y.; Wang, H.; Hu, G. LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn. Plant Cell Rep. 2016, 35, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Dai, C.; Li, Y.; Feng, J.; Liu, Z.; Kang, C. Reduced Anthocyanins in Petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. J. Exp. Bot. 2018, 69, 2595–2608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Dong, W.; Zhu, Y.; Allan, A.C.; Wang, K.L.; Xu, C. PpGST1, an anthocyanin-related glutathione S-transferase gene, is essential for fruit coloration in peach. Plant Biotechnol. J. 2020, 18, 1284–1295. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Liu, X.; Zhong, F.; Tian, R.; Zhang, K.; Zhang, X.; Li, T. Comparative study of phenolic compounds and antioxidant activity in different species of cherries. J. Food Sci. 2011, 76, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Wang, H.; Li, M.; Wang, J.; Yang, Y.; Zhang, X.; Yan, G.H.; Zhang, H.; Liu, J.S.; Zhang, K. The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef] [Green Version]
- Shirasawa, K.; Isuzugawa, K.; Ikenaga, M.; Saito, Y.; Yamamoto, T.; Hirakawa, H.; Isobe, S. The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res. 2017, 24, 499–508. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Guo, X.; Zhao, D.; Zhang, Q.; Jiang, Y.; Wang, Y.; Peng, X.; Wei, Y.; Zhai, Z.; Zhao, W.; et al. Cloning and expression profiling of the PacSnRK2 and PacPP2C gene families during fruit development, ABA treatment, and dehydration stress in sweet cherry. Plant Physiol. Biochem. 2017, 119, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.A.; Jones, G.P. Analysis of proanthocyanidin cleavage products following acid-catalysis in the presence of excess phloroglucinol. J. Agric. Food Chem. 2001, 49, 1740–1746. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, K.; Maoz, I.; Lewinsohn, E.; Lerno, L.; Ebeler, S.E.; Lichter, A. Girdling of table grapes at fruit set can divert the phenylpropanoid pathway towards accumulation of proanthocyanidins and change the volatile composition. Plant Sci. 2020, 296, 110495. [Google Scholar] [CrossRef] [PubMed]
- Awad, M.A.; de Jager, A.; van Westing, L.M. Flavonoid and chlorogenic acid levels in apple fruit: Characterisation of variation. Sci. Hortic. 2000, 83, 249–263. [Google Scholar] [CrossRef]
- Jakobek, L.; Seruga, M.; Novak, I.; Medvidovic-Kosanovic, M. Flavonols, phenolic acids and antioxidant activity of some red fruits. Dtsch. Lebensm. Rundschau. 2007, 103, 369–377. [Google Scholar]
- Fu, D.Q.; Zhu, B.Z.; Zhu, H.L.; Jiang, W.B.; Luo, Y.B. Virus-induced gene silencing in tomato fruit. Plant J. 2005, 43, 299–308. [Google Scholar] [CrossRef]
- Qi, X.; Liu, C.; Song, L.; Li, Y.; Li, M. PaCYP78A9, a cytochrome P450, regulates fruit size in sweet cherry (Prunus avium L.). Front. Plant Sci. 2017, 8, 2076. [Google Scholar] [CrossRef] [Green Version]
- Qi, X.; Liu, C.; Song, L.; Li, M. PaMADS7, a MADS-box transcription factor, regulates sweet cherry fruit ripening and softening. Plant Sci. 2020, 301, 110634. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Yamazaki, M.; Shibata, M.; Nishiyama, Y.; Springob, K.; Kitayama, M.; Shimada, N.; Aoki, T.; Ayabe, S.; Saito, K. Differential gene expression profiles of red and green forms of Perilla frutescens leading to comprehensive identification of anthocyanin biosynthetic genes. FEBS J. 2008, 275, 3494–3502. [Google Scholar] [CrossRef]
- Kitamura, S.; Shikazono, N.; Tanaka, A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 2004, 37, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Wangwattana, B.; Koyama, Y.; Nishiyama, Y.; Kitayama, M.; Yamazaki, M.; Saito, K. Characterization of PAP1-upregulated glutathione S-transferase genes in Arabidopsis thaliana. Plant Biotechnol. 2008, 25, 191–196. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Xu, L.; Xu, H.; Yang, P.; He, G.; Tang, Y.; Qi, X.; Song, M.; Ming, J. LhGST is an anthocyanin-related glutathione S-transferase gene in Asiatic hybrid lilies (Lilium spp.). Plant Cell Rep. 2021, 40, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Wang, L.; Zhang, Y.; Ruan, L.; Li, H.; Wu, L.; Xu, L.; Zhang, C.; Zhou, X.G.; Cheng, H.; et al. A coupled role for CsMYB75 and CsGSTF1 in anthocyanin hyperaccumulation in purple tea. Plant J. 2019, 97, 825–840. [Google Scholar] [CrossRef] [PubMed]
- Teng, S.; Keurentjes, J.; Bentsink, L.; Koornneef, M.; Smeekens, S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005, 139, 1840–1852. [Google Scholar] [CrossRef] [Green Version]
- Zheng, T.; Tan, W.; Yang, H.; Zhang, L.E.; Li, T.; Liu, B.; Zhang, D.; Lin, H. Regulation of anthocyanin accumulation via MYB75/HAT1/TPL-mediated transcriptional repression. PLoS Genet. 2019, 15, e1007993. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).