SIN-Like Pathway Kinases Regulate the End of Mitosis in the Methylotrophic Yeast Ogataea polymorpha
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Plasmids
2.2. Yeast Growth Conditions and General Methods
2.3. Microscopy
2.4. RNA Analysis
2.5. Yeast Cell Extracts and Immunoblotting
3. Results
3.1. OpHCD1 and OpHCD2 Encode Kinases Similar to S. pombe SIN Kinases
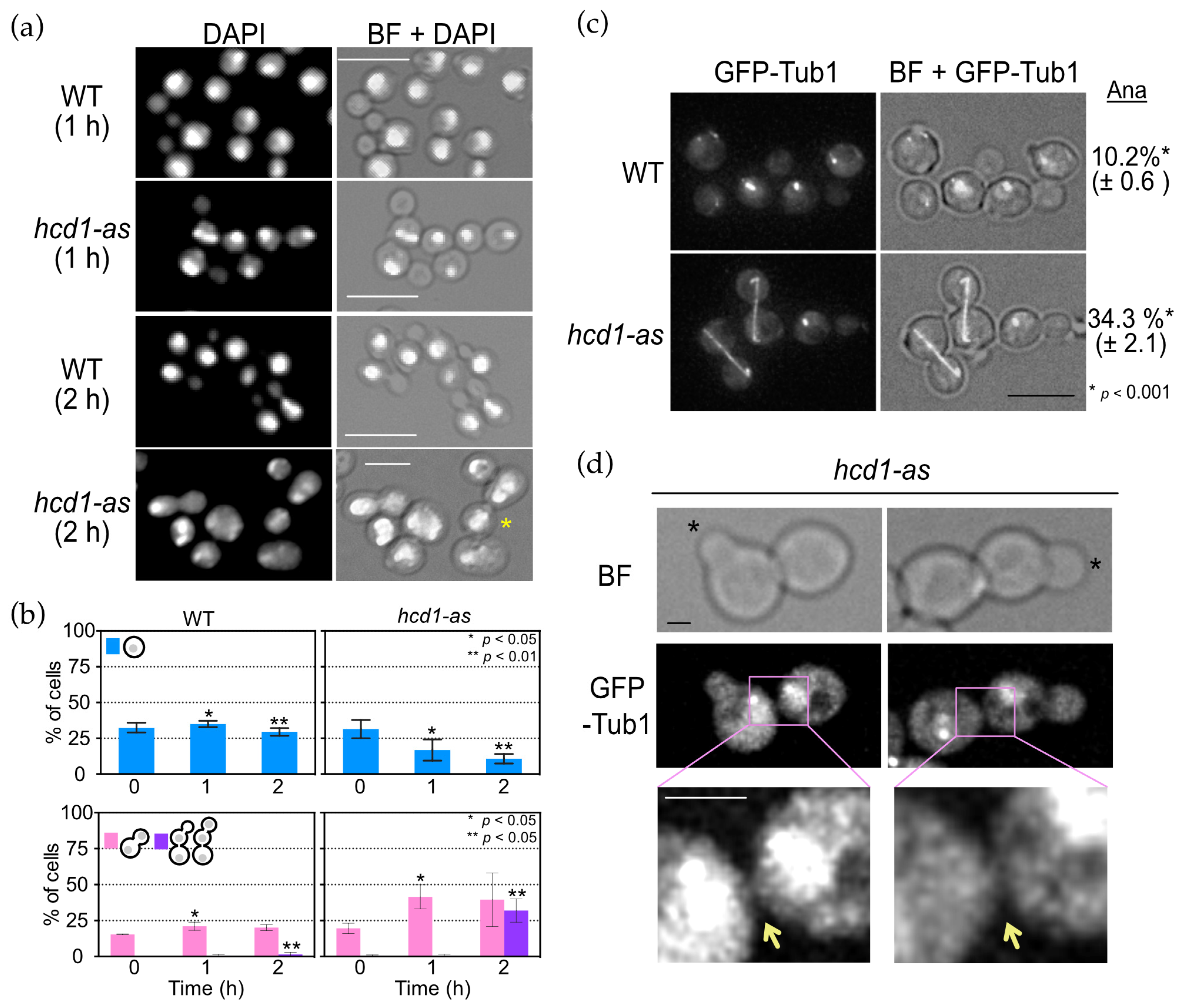
3.2. OpHcd1 Plays Roles in Both Mitosis and Cytokinesis
3.3. Optimisation of iAID System for Analysis of Hcd2 Function
3.4. OpHcd2 Functions in Mitotic Exit and Cytokinesis
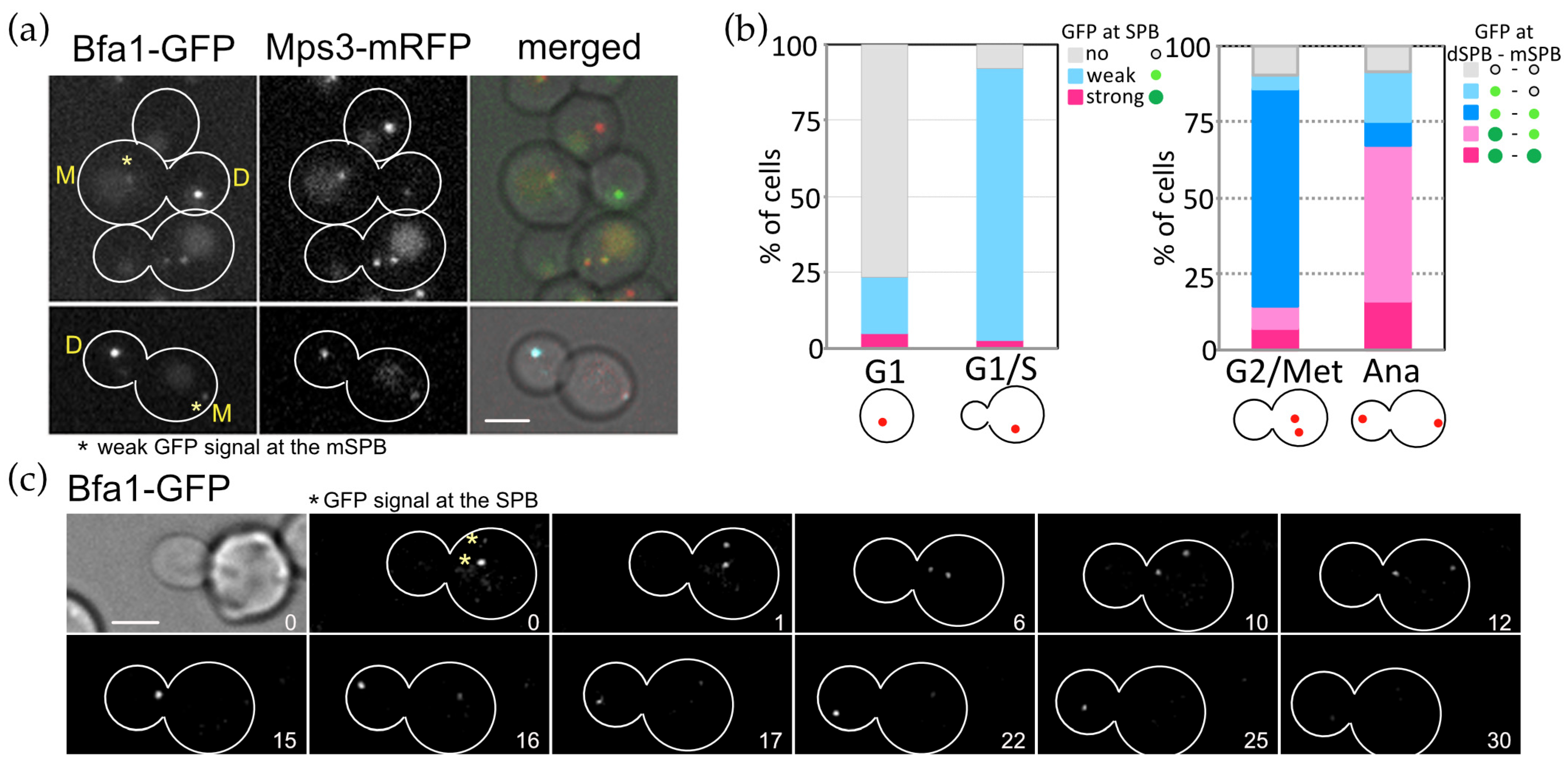
3.5. OpHcd1 and OpHcd2 Preferentially Associate with the SPB in the Mother Cell Body in Late Anaphase
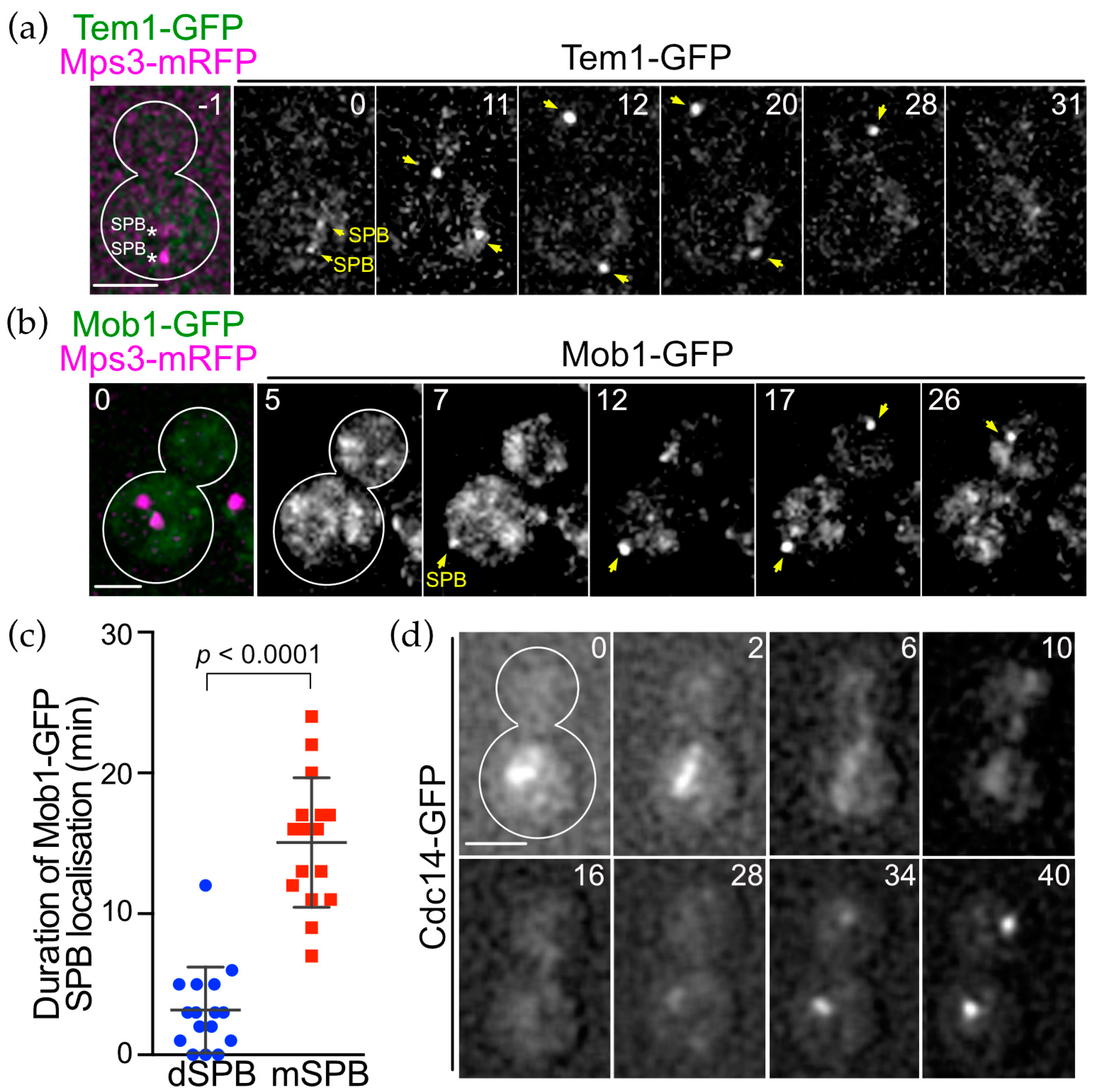
3.6. SPB Association of Conserved MEN Core Components Are Regulated Spatially and Temporally during the Cell Cycle of O. polymorpha
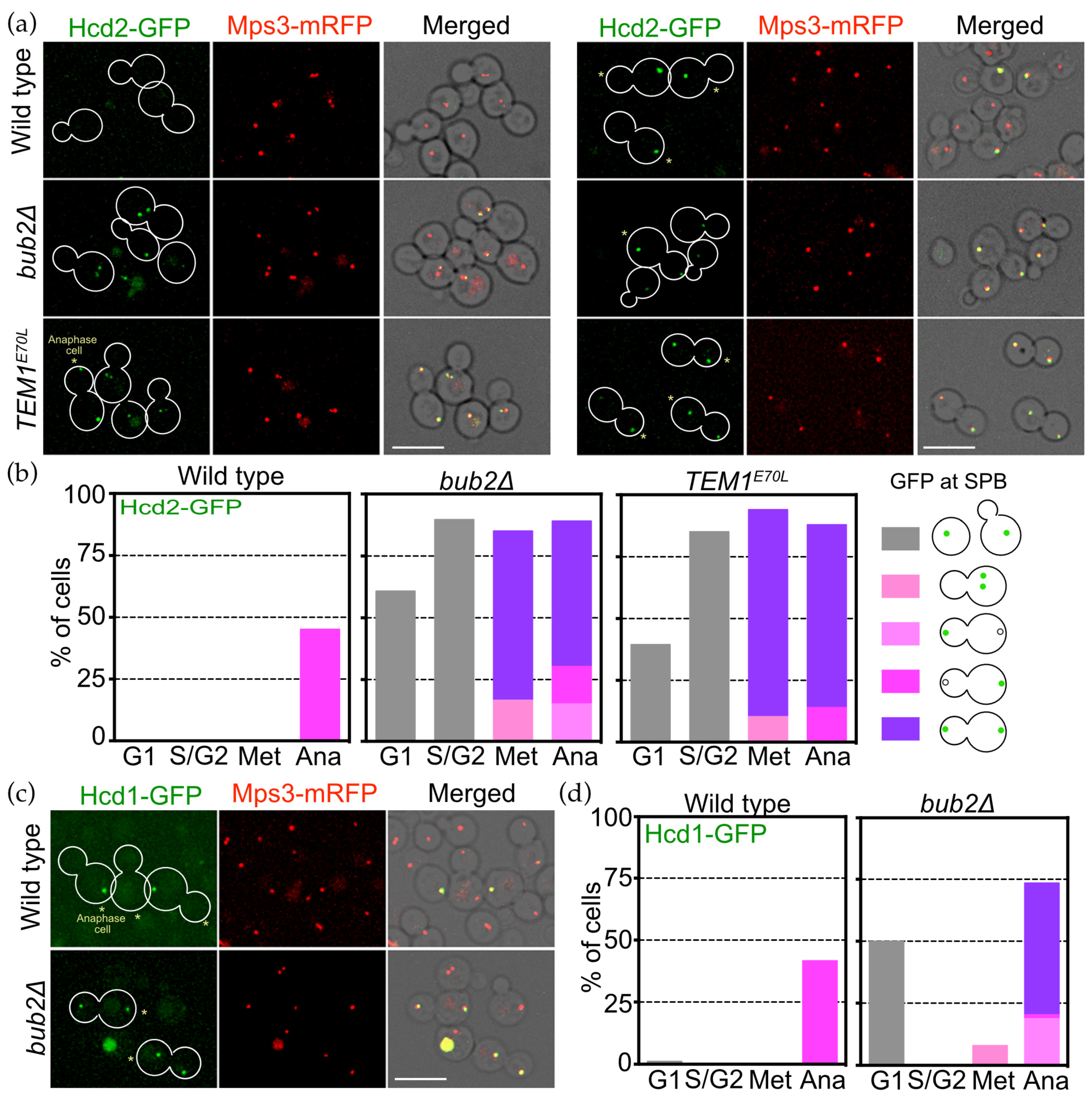
3.7. Bub2 Inhibits SPB Association of Hcd1 and Hcd2 in Daughter Cells
3.8. Polo-Like Mitotic Kinase Cdc5 Restricts the SPB Association of Hcd1 to Anaphase
4. Discussion
4.1. OpHcd1 and OpHcd2 Are Required for Mitotic Exit and Cytokinesis
4.2. ME-Signalling Pathway Senses Tem1 Activity and Cell Cycle Phase
4.3. Asymmetric SPB Localisation of the ME-Signalling Pathway in O. polymorpha
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baro, B.; Queralt, E.; Monje-Casas, F. Regulation of Mitotic Exit in Saccharomyces cerevisiae. Methods Mol. Biol. 2017, 1505, 3–17. [Google Scholar] [PubMed]
- Meitinger, F.; Palani, S.; Pereira, G. The power of MEN in cytokinesis. Cell Cycle 2012, 11, 219–228. [Google Scholar] [CrossRef]
- Jaspersen, S.L.; Charles, J.F.; Tinker-Kulberg, R.L.; Morgan, D.O. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell 1998, 9, 2803–2817. [Google Scholar] [CrossRef] [PubMed]
- Luca, F.C.; Winey, M. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell 1998, 9, 29–46. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Komarnitsky, S.I.; Chiang, Y.C.; Luca, F.C.; Chen, J.; Toyn, J.H.; Winey, M.; Johnston, L.H.; Denis, C.L. DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol. Cell. Biol. 1998, 18, 2100–2107. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.; Haridas, S.; Wolfe, K.H.; Lopes, M.R.; Hittinger, C.T.; Goeker, M.; Salamov, A.A.; Wisecaver, J.H.; Long, T.M.; Calvey, C.H.; et al. Comparative genomics of biotechnologically important yeasts. Proc. Natl. Acad. Sci. USA 2016, 113, 9882–9887. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.-X.; Zhou, X.; Kominek, J.; Kurtzman, C.P.; Hittinger, C.T.; Rokas, A. Reconstructing the Backbone of the Saccharomycotina Yeast Phylogeny Using Genome-Scale Data. G3 2016, 6, 3927–3939. [Google Scholar] [CrossRef] [PubMed]
- Guertin, D.A.; Chang, L.; Irshad, F.; Gould, K.L.; McCollum, D. The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J. 2000, 19, 1803–1815. [Google Scholar] [CrossRef]
- Milne, S.W.; Cheetham, J.; Lloyd, D.; Shaw, S.; Moore, K.; Paszkiewicz, K.H.; Aves, S.J.; Bates, S. Role of Candida albicans Tem1 in mitotic exit and cytokinesis. Fungal Genet. Biol. 2014, 69, 84–95. [Google Scholar] [CrossRef][Green Version]
- Bates, S. Candida albicans Cdc15 is essential for mitotic exit and cytokinesis. Sci. Rep. 2018, 8, 8899. [Google Scholar] [CrossRef]
- Kim, J.-M.; Zeng, C.J.T.; Nayak, T.; Shao, R.; Huang, A.-C.; Oakley, B.R.; Liu, B. Timely septation requires SNAD-dependent spindle pole body localization of the septation initiation network components in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 2009, 20, 2874–2884. [Google Scholar] [CrossRef] [PubMed]
- Bardin, A.J.; Visintin, R.; Amon, A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 2000, 102, 21–31. [Google Scholar] [CrossRef]
- Pereira, G.; Höfken, T.; Grindlay, J.; Manson, C.; Schiebel, E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell 2000, 6, 1–10. [Google Scholar] [CrossRef]
- Rock, J.M.; Amon, A. Cdc15 integrates Tem1 GTPase-mediated spatial signals with Polo kinase-mediated temporal cues to activate mitotic exit. Genes Dev. 2011, 25, 1943–1954. [Google Scholar] [CrossRef] [PubMed]
- Visintin, R.; Amon, A. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol. Biol. Cell 2001, 12, 2961–2974. [Google Scholar] [CrossRef]
- Stoepel, J.; Ottey, M.A.; Kurischko, C.; Hieter, P.; Luca, F.C. The mitotic exit network Mob1p-Dbf2p kinase complex localizes to the nucleus and regulates passenger protein localization. Mol. Biol. Cell 2005, 16, 5465–5479. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mohl, D.A.; Huddleston, M.J.; Collingwood, T.S.; Annan, R.S.; Deshaies, R.J. Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J. Cell Biol. 2009, 184, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Höfken, T.; Schiebel, E. A role for cell polarity proteins in mitotic exit. EMBO J. 2002, 21, 4851–4862. [Google Scholar] [CrossRef]
- Pereira, G.; Schiebel, E. Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol. Cell 2005, 19, 209–221. [Google Scholar] [CrossRef]
- D’Aquino, K.E.; Monje-Casas, F.; Paulson, J.; Reiser, V.; Charles, G.M.; Lai, L.; Shokat, K.M.; Amon, A. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol. Cell 2005, 19, 223–234. [Google Scholar] [CrossRef]
- Burke, D.J. Interpreting spatial information and regulating mitosis in response to spindle orientation. Genes Dev. 2009, 23, 1613–1618. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.Y.; Amon, A. Spindle position is coordinated with cell-cycle progression through establishment of mitotic exit-activating and -inhibitory zones. Mol. Cell 2010, 39, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Falk, J.E.; Tsuchiya, D.; Verdaasdonk, J.; Lacefield, S.; Bloom, K.; Amon, A. Spatial signals link exit from mitosis to spindle position. Elife 2016, 5, 159. [Google Scholar] [CrossRef] [PubMed]
- Caydasi, A.K.; Pereira, G. SPOC alert—When chromosomes get the wrong direction. Exp. Cell Res. 2012, 318, 1421–1427. [Google Scholar] [CrossRef]
- Maekawa, H.; Kaneko, Y. Inversion of the chromosomal region between two mating type loci switches the mating type in Hansenula polymorpha. PLoS Genet. 2014, 10, e1004796. [Google Scholar] [CrossRef]
- Maekawa, H.; Neuner, A.; Rüthnick, D.; Schiebel, E.; Pereira, G.; Kaneko, Y. Polo-like kinase Cdc5 regulates Spc72 recruitment to spindle pole body in the methylotrophic yeast Ogataea polymorpha. Elife 2017, 6, e24340. [Google Scholar] [CrossRef]
- Lu, S.F.; Tolstorukov, I.I.; Anamnart, S.; Kaneko, Y.; Harashima, S. Cloning, sequencing, and functional analysis of H-OLE1 gene encoding delta9-fatty acid desaturase in Hansenula polymorpha. Appl. Microbiol. Biotechnol. 2000, 54, 499–509. [Google Scholar] [CrossRef]
- Janke, C.; Magiera, M.M.; Rathfelder, N.; Taxis, C.; Reber, S.; Maekawa, H.; Moreno-Borchart, A.; Doenges, G.; Schwob, E.; Schiebel, E.; et al. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast 2004, 21, 947–962. [Google Scholar] [CrossRef]
- Saraya, R.; Krikken, A.M.; Kiel, J.A.; Baerends, R.J.S.; Veenhuis, M.; Klei, I.J. Novel genetic tools for Hansenula polymorpha. FEMS Yeast Res. 2012, 12, 271–278. [Google Scholar] [CrossRef]
- Faber, K.N.; Haima, P.; Harder, W.; Veenhuis, M.; AB, G. Highly-efficient electrotransformation of the yeast Hansenula polymorpha. Curr. Genet. 1994, 25, 305–310. [Google Scholar] [CrossRef]
- Bellí, G.; Garí, E.; Aldea, M.; Herrero, E. Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast 1998, 14, 1127–1138. [Google Scholar] [CrossRef]
- Sherman, F. Getting started with yeast. Meth. Enzymol. 1991, 194, 3–21. [Google Scholar]
- Chen, H.; Clyborne, W.K.; Sedat, J.W.; Agard, D.A. PRIISM: An integrated system for display and analysis of 3-D microscope images. In Proceedings of the SPIE/IS&T 1992 Symposium on Electronic Imaging: Science and Technology, San Jose, CA, USA, 9–14 February 1992; Volume 1660, pp. 784–790. [Google Scholar]
- Knop, M.; Siegers, K.; Pereira, G.; Zachariae, W.; Winsor, B.; Nasmyth, K.; Schiebel, E. Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast 1999, 15, 963–972. [Google Scholar] [CrossRef]
- Bishop, A.C.; Ubersax, J.A.; Petsch, D.T.; Matheos, D.P.; Gray, N.S.; Blethrow, J.; Shimizu, E.; Tsien, J.Z.; Schultz, P.G.; Rose, M.D.; et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 2000, 407, 395–401. [Google Scholar] [CrossRef]
- Yesbolatova, A.; Saito, Y.; Kitamoto, N.; Makino-Itou, H.; Ajima, R.; Nakano, R.; Nakaoka, H.; Fukui, K.; Gamo, K.; Tominari, Y.; et al. The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nat. Commun. 2020, 11, 5701–5713. [Google Scholar] [CrossRef]
- Fesquet, D.; Fitzpatrick, P.J.; Johnson, A.L.; Kramer, K.M.; Toyn, J.H.; Johnston, L.H. A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. EMBO J. 1999, 18, 2424–2434. [Google Scholar] [CrossRef]
- Pereira, G.; Manson, C.; Grindlay, J.; Schiebel, E. Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J. Cell Biol. 2002, 157, 367–379. [Google Scholar] [CrossRef]
- Scarfone, I.; Venturetti, M.; Hotz, M.; Lengefeld, J.; Barral, Y.; Piatti, S. Asymmetry of the Budding Yeast Tem1 GTPase at Spindle Poles Is Required for Spindle Positioning But Not for Mitotic Exit. PLoS Genet. 2015, 11, e1004938. [Google Scholar] [CrossRef]
- Bardin, A.J.; Amon, A. Men and sin: What’s the difference? Nat. Rev. Mol. Cell Biol. 2001, 2, 815–826. [Google Scholar] [CrossRef]
- González-Novo, A.; Labrador, L.; Pablo-Hernando, M.E.; Correa-Bordes, J.; Sánchez, M.; Jiménez, J.; de Aldana, C.R.V. Dbf2 is essential for cytokinesis and correct mitotic spindle formation in Candida albicans. Mol. Microbiol. 2009, 72, 1364–1378. [Google Scholar] [CrossRef]
- Finley, K.R.; Bouchonville, K.J.; Quick, A.; Berman, J. Dynein-dependent nuclear dynamics affect morphogenesis in Candida albicans by means of the Bub2p spindle checkpoint. J. Cell Sci. 2008, 121, 724. [Google Scholar] [CrossRef]
- Fraschini, R.; D’Ambrosio, C.; Venturetti, M.; Lucchini, G.; Piatti, S. Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J. Cell Biol. 2006, 172, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, H.; Priest, C.; Lechner, J.; Pereira, G.; Schiebel, E. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J. Cell Biol. 2007, 179, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Caydasi, A.K.; Pereira, G. Spindle Alignment Regulates the Dynamic Association of Checkpoint Proteins with Yeast Spindle Pole Bodies. Dev. Cell 2009, 16, 146–156. [Google Scholar] [CrossRef]
- Caydasi, A.K.; Lohel, M.; Grünert, G.; Dittrich, P.; Pereira, G.; Ibrahim, B. A dynamical model of the spindle position checkpoint. Mol. Syst. Biol. 2012, 8, 582. [Google Scholar] [CrossRef]
- Valerio-Santiago, M.; Monje-Casas, F. Tem1 localization to the spindle pole bodies is essential for mitotic exit and impairs spindle checkpoint function. J. Cell Biol. 2011, 192, 599–614. [Google Scholar] [CrossRef]
- Falk, J.E.; Campbell, I.W.; Joyce, K.; Whalen, J.; Seshan, A.; Amon, A. LTE1 promotes exit from mitosis by multiple mechanisms. Mol. Biol. Cell 2016, 27, 3991–4001. [Google Scholar] [CrossRef]
- Bertazzi, D.T.; Kurtulmus, B.; Pereira, G. The cortical protein Lte1 promotes mitotic exit by inhibiting the spindle position checkpoint kinase Kin4. J. Cell Biol. 2011, 193, 1033–1048. [Google Scholar] [CrossRef]
- Lengefeld, J.; Hotz, M.; Rollins, M.; Baetz, K.; Barral, Y. Budding yeast Wee1 distinguishes spindle pole bodies to guide their pattern of age-dependent segregation. Nat. Publ. Group 2017, 19, 941–951. [Google Scholar] [CrossRef]
- Hotz, M.; Barral, Y. The Mitotic Exit Network: New turns on old pathways. Trends Cell Biol. 2014, 24, 145–152. [Google Scholar] [CrossRef]
- Campbell, I.W.; Zhou, X.; Amon, A. Spindle pole bodies function as signal amplifiers in the Mitotic Exit Network. Mol. Biol. Cell 2020, 31, 906–916. [Google Scholar] [CrossRef] [PubMed]
- Sohrmann, M.; Schmidt, S.; Hagan, I.; Simanis, V. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev. 1998, 12, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maekawa, H.; Jiangyan, S.; Takegawa, K.; Pereira, G. SIN-Like Pathway Kinases Regulate the End of Mitosis in the Methylotrophic Yeast Ogataea polymorpha. Cells 2022, 11, 1519. https://doi.org/10.3390/cells11091519
Maekawa H, Jiangyan S, Takegawa K, Pereira G. SIN-Like Pathway Kinases Regulate the End of Mitosis in the Methylotrophic Yeast Ogataea polymorpha. Cells. 2022; 11(9):1519. https://doi.org/10.3390/cells11091519
Chicago/Turabian StyleMaekawa, Hiromi, Shen Jiangyan, Kaoru Takegawa, and Gislene Pereira. 2022. "SIN-Like Pathway Kinases Regulate the End of Mitosis in the Methylotrophic Yeast Ogataea polymorpha" Cells 11, no. 9: 1519. https://doi.org/10.3390/cells11091519
APA StyleMaekawa, H., Jiangyan, S., Takegawa, K., & Pereira, G. (2022). SIN-Like Pathway Kinases Regulate the End of Mitosis in the Methylotrophic Yeast Ogataea polymorpha. Cells, 11(9), 1519. https://doi.org/10.3390/cells11091519





